Development and Application of Novel SSR Markers to Assess the Genetic Diversity and Population Structure of Phacelia secunda Along an Altitudinal Gradient in the Central Chile Andes
Abstract
1. Introduction
2. Results
2.1. Microsatellite Screening
2.2. Microsatellite Loci Characterization
2.3. Transferability to Phacelia brachyantha
2.4. Within-Population Genetic Diversity Along the Altitudinal Gradient
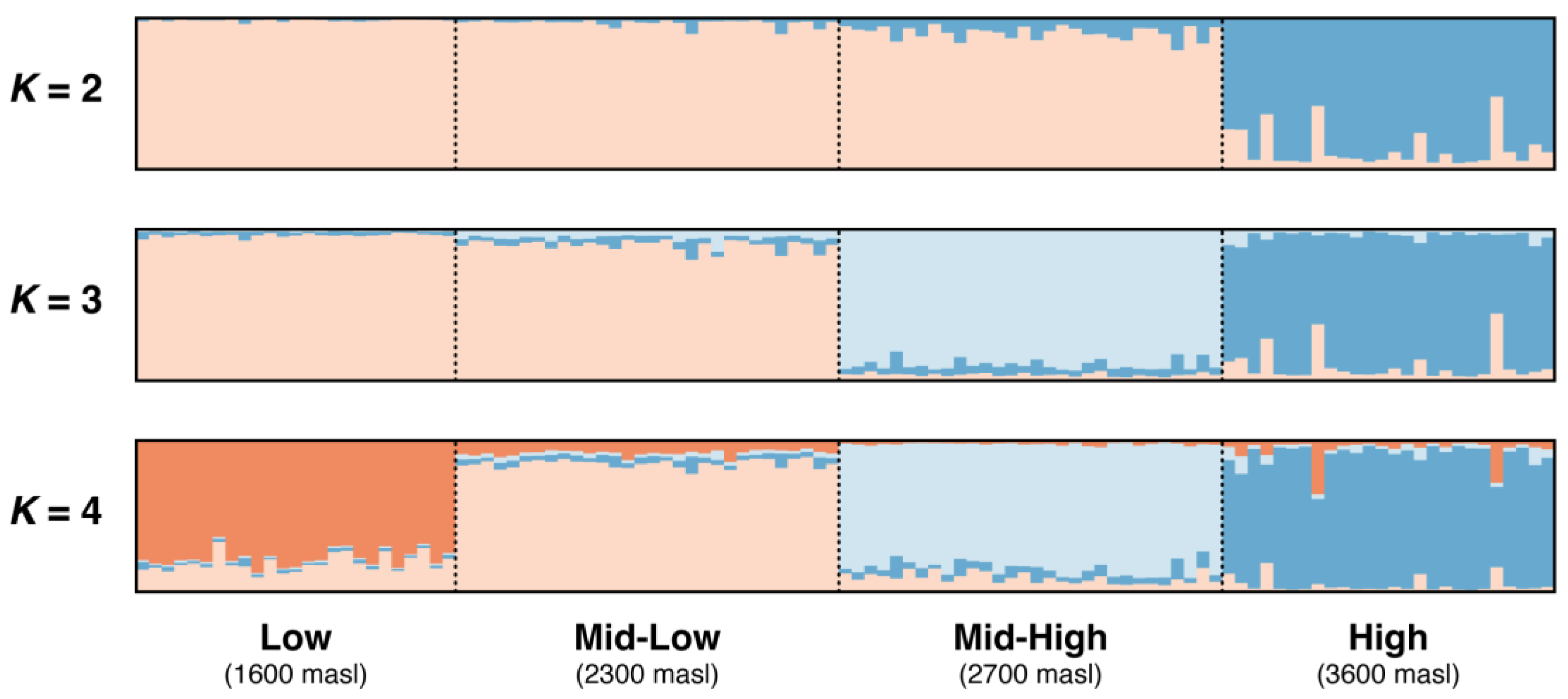
2.5. Genetic Structure and Gene Flow Along the Altitudinal Gradient
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction and Genome Sequencing
4.3. PCR Analyses and Genotyping
4.4. Microsatellite Loci Characterization
4.5. Transferability Assessment
4.6. Within-Population Genetic Diversity Along the Altitudinal Gradient
4.7. Genetic Structure and Gene Flow Along the Altitudinal Gradient
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HTS | High-throughput sequencing |
| SSRs | Simple sequence repeats |
| SNPs | Single nucleotide polymorphisms |
| HWE | Hardy–Weinberg equilibrium |
| LD | Linkage disequilibrium |
References
- Cavieres, L.A. Variación morfológica de Phacelia secunda J.F. Gmel. (Hydrophyllaceae) a lo largo de un gradiente altitudinal en Chile central. Gayana Bot. 2000, 57, 89–96. [Google Scholar] [CrossRef]
- Molina-Montenegro, M.A.; Cavieres, L.A. Variación altitudinal de los atributos morfo-fisiológicos en dos especies de plantas alto-andinas y sus implicancias contra la fotoinhibición. Gayana Bot. 2010, 67, 1–11. [Google Scholar] [CrossRef]
- Hernández-Fuentes, C.; Galmés, J.; Bravo, L.A.; Cavieres, L.A. Elevation provenance affects photosynthesis and its acclimation to temperature in the high-Andes alpine herb Phacelia secunda. Plant Biol. 2023, 25, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Hahn, T.; Kettle, C.J.; Ghazoul, J.; Frei, E.R.; Matter, P.; Pluess, A.R. Patterns of genetic variation across altitude in three plant species of semi-dry grasslands. PLoS ONE 2012, 7, e41608. [Google Scholar] [CrossRef]
- Byars, S.G.; Parsons, Y.; Hoffmann, A.A. Effect of altitude on the genetic structure of an Alpine grass, Poa hiemata. Ann. Bot. 2009, 103, 885–899. [Google Scholar] [CrossRef]
- Reisch, C.; Rosbakh, S. Patterns of genetic variation in European plant species depend on altitude. Divers. Distrib. 2021, 27, 157–163. [Google Scholar] [CrossRef]
- Arroyo, M.T.K.; Armesto, J.J.; Primack, R.B. Community studies in pollination ecology in the high temperate Andes of central Chile II. Effect of temperature on visitation rates and pollination possibilities. Plant Syst. Evol. 1985, 149, 187–203. [Google Scholar] [CrossRef]
- Kudo, G. Outcrossing syndrome in alpine plants: Implications for flowering phenology and pollination success. Ecol. Res. 2022, 37, 288–300. [Google Scholar] [CrossRef]
- Wirth, L.R.; Graf, R.; Gugerli, F.; Landergott, U.; Holderegger, R. Lower selfing rate at higher altitudes in the alpine plant Eritrichium nanum (Boraginaceae). Am. J. Bot. 2010, 97, 899–901. [Google Scholar]
- Arroyo, M.T.K.; Armesto, J.J.; Villagrán, C. Plant phenological patterns in the high Andean Cordillera of Central Chile. J. Ecol. 1981, 69, 205–233. [Google Scholar] [CrossRef]
- Kudo, G.; Ida, T.Y. Early onset of spring increases the phenological mismatch between plants and pollinators. Ecology 2013, 94, 2311–2320. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.S.; Athrey, G.; Leberg, P.L. Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Ecol. Evol. 2021, 11, 15800–15814. [Google Scholar] [CrossRef]
- Jones, A.G.; Small, C.M.; Paczolt, K.A.; Ratterman, N.L. A practical guide to methods of parentage analysis. Mol. Ecol. Resour. 2010, 10, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Love, J.M.; Ferris, K.G. Local adaptation to an altitudinal gradient: The interplay between mean phenotypic trait variation and phenotypic plasticity in Mimulus laciniatus. Perspect. Plant Ecol. Evol. Syst. 2024, 63, 125795. [Google Scholar] [CrossRef]
- Snell-Rood, E.C.; Ehlman, S.M. Ecology and evolution of plasticity. In Phenotypic Plasticity and Evolution; CRC Press: Boca Raton, FL, USA, 2021; pp. 139–160. [Google Scholar]
- Forester, B.R.; Jones, M.R.; Joost, S.; Landguth, E.L.; Lasky, J.R. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 2016, 25, 104–120. [Google Scholar] [CrossRef]
- Escobar-Sandoval, M.; Pâques, L.; Fonti, P.; Martinez-Meier, A.; Rozenberg, P. Phenotypic plasticity of European larch radial growth and wood density along a 1,000 m elevational gradient. Plant-Environ. Interact. 2021, 2, 45–60. [Google Scholar] [CrossRef]
- Ohsawa, T.; Ide, Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 2008, 17, 152–163. [Google Scholar] [CrossRef]
- Vega-Vela, N.E.; Chacón, M.I. Genetic structure along an altitudinal gradient in Lippia origanoides, a promising aromatic plant species restricted to semiarid areas in northern South America. Ecol. Evol. 2012, 2, 2669–2681. [Google Scholar] [CrossRef]
- Daco, L.; Colling, G.; Matthies, D. Clinal variation in quantitative traits but not in evolutionary potential along elevational and latitudinal gradients in the widespread Anthyllis vulneraria. Am. J. Bot. 2024, 111, e16360. [Google Scholar] [CrossRef] [PubMed]
- Dar, T.U.H.; Mangral, Z.A.; Islam, S.U.; Tariq, L.; Dar, R.; Majeed, A.; Goel, S. Genetic variation and population structure of Rhododendron anthopogon along an altitudinal gradient: A case study from Himalaya. Plant. Mol. Biol. Rep. 2004, 42, 650–664. [Google Scholar] [CrossRef]
- Shi, M.-M.; Michalski, S.G.; Chen, X.-Y.; Durka, W. Isolation by elevation: Genetic Structure at neutral and putatively non-neutral loci in a dominant tree of Subtropical Forests, Castanopsis eyrei. PLoS ONE 2011, 6, e21302. [Google Scholar] [CrossRef]
- Tyagi, A.; Singh, S.; Mishra, P.; Singh, A.; Tripathi, A.M.; Jena, S.N.; Roy, S. Genetic diversity and population structure of Arabidopsis thaliana along an altitudinal gradient. AoB Plants 2015, 15, plv145. [Google Scholar] [CrossRef]
- Korbecka, G.; Wolff, K. Characterization of nine microsatellite loci in Cynoglossum officinale (Boraginaceae). Mol. Ecol. Notes 2004, 4, 229–230. [Google Scholar] [CrossRef]
- Ahmad, M.; Lazic, D.; Hansel-Hohl, K.; Lexter, C.; Sehr, E.M. Development of novel microsatellite markers for Alkanna tinctoria by comparative transcriptomics. Appl. Plant Sci. 2019, 7, e11296. [Google Scholar] [CrossRef]
- Herrera, C.; Bazaga, P. Adding a third dimension to the edge of a species’ range: Altitude and genetic structuring in mountainous landscapes. Heredity 2008, 100, 275–285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arroyo, M.T.K.; Tamburrino, Í.; Pliscoff, P.; Robles, V.; Colldecarrera, M.; Guerrero, P.C. Flowering phenology adjustment and flower longevity in a South American alpine species. Plants 2021, 10, 461. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Gonzalo-Turpin, H.; Hazard, L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. J. Ecol. 2009, 97, 742–751. [Google Scholar] [CrossRef]
- Sambatti, J.B.M.; Rice, K.J. Local adaptation, patterns of selection, and gene flow in the Californian serpentine sunflower (Helianthus exilis). Evolution 2006, 60, 696–710. [Google Scholar] [CrossRef] [PubMed]
- de Villemereuil, P.; Mouterde, M.; Gaggiotti, O.E.; Till-Bottraud, I. Patterns of phenotypic plasticity and local adaptation in the wide elevation range of the alpine plant Arabis alpina. J. Ecol. 2018, 106, 1952–1971. [Google Scholar] [CrossRef]
- Sun, Y.-Q.; Zhao, W.; Xu, C.-Q.; Xu, Y.; El-Kassaby, Y.A.; De La Torre, A.R.; Mao, J.-F. Genetic variation related to high elevation adaptation revealed by common garden experiments in Pinus yunnanensis. Front. Genet. 2020, 10, 1405. [Google Scholar] [CrossRef]
- de Villemereuil, P.; Gaggiotti, O.E.; Goudet, J. Common garden experiments to study local adaptation need to account for population structure. J. Ecol. 2022, 110, 1005–1009. [Google Scholar] [CrossRef]
- Singh, A.; Verma, A.K.; Kumar, S.; Bag, S.K.; Roy, S. Genome-wide DNA methylation and their transgenerational pattern differ in Arabidopsis thaliana populations originated along the elevation of West Himalaya. BMC Plant Biol. 2024, 24, 936. [Google Scholar] [CrossRef]
- Dudash, M.R.; Berg, J.A.; Zimmer, E.A. Progeny array analysis to estimate outcrossing rates, inbreeding coefficients, and inbreeding depression among native, naturalized, and invasive populations of Mimulus guttatus (Phrymaceae). Front. Plant Sci. 2024, 15, 1411868. [Google Scholar] [CrossRef] [PubMed]
- Meglécz, E.; Pech, N.; Gilles, A.; Dubut, V.; Hingamp, P.; Trilles, A.; Grenier, R.; Martin, J.F. QDD version 3.1: A User friendly computer program for microsatellite selection and primer design revisited: Experimental validation of variables determining genotyping success rate. Mol. Ecol. Resour. 2014, 14, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 15, 421. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucl. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetic software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- van Oosterhout, C.; Hutchinson, W.F.; Willis, D.P.M.; Shipley, P.F. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Brookfield, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol. 1996, 5, 4534–4555. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. 2012. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, M.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Meirmans, P.G. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 2006, 60, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Chesser, R. Estimation of fixation indexes and gene diversities. Ann. Hum. Genet. 1983, 47, 253–259. [Google Scholar] [CrossRef]

| Population | Elevation (m) | A | AE | HO | HE | F | |
|---|---|---|---|---|---|---|---|
| Low | 1600 | Mean | 4.985 a | 2.810 a | 0.394 a | 0.524 a | 0.237 a |
| SE | 0.529 | 0.388 | 0.045 | 0.055 | 0.067 | ||
| Mid-Low | 2300 | Mean | 4.947 a | 2.587 a | 0.504 b | 0.526 a | 0.025 b |
| SE | 0.543 | 0.294 | 0.049 | 0.050 | 0.052 | ||
| Mid-High | 2800 | Mean | 4.842 a | 2.847 ab | 0.472 a | 0.523 a | 0.062 b |
| SE | 0.563 | 0.407 | 0.051 | 0.054 | 0.065 | ||
| High | 3500 | Mean | 4.263 b | 2.053 b | 0.387 a | 0.434 b | 0.117 ab |
| SE | 0.396 | 0.186 | 0.055 | 0.051 | 0.064 | ||
| Total | All | Mean | 4.737 | 2.574 | 0.439 | 0.502 | 0.110 |
| SE | 0.253 | 0.166 | 0.026 | 0.026 | 0.032 |
| Source of Variation | d.f. | Sum of Squares | Mean Square | Estimated Variance | Percentage of Variation |
|---|---|---|---|---|---|
| (A) K = 2—Group 1: Low + Mid-Low + Mid-High; Group 2: High | |||||
| Among populations | 1 | 34.880 | 34.880 | 0.375 | 7% |
| Within populations | 220 | 1095.692 | 4.980 | 4.980 | 93% |
| Total | 221 | 1130.572 | 5.356 | 100% | |
| FST = 0.070, p = 0.001; FST max = 0.490; F′ST = 0.143, p = 0.001 | |||||
| (B) K = 3—Group 1: Low + Mid-Low; Group 2: Mid-High; Group 3: High | |||||
| Among populations | 2 | 56.986 | 28.493 | 0.339 | 6% |
| Within populations | 219 | 1073.586 | 4.902 | 4.902 | 94% |
| Total | 221 | 1130.572 | 5.241 | 100% | |
| FST = 0.065, p = 0.001; FST max = 0.487; F′ST = 0.133, p = 0.001 | |||||
| (C) K = 4—Each sampling site is considered a population | |||||
| Among populations | 3 | 69.062 | 23.021 | 0.328 | 6% |
| Within populations | 218 | 1061.511 | 4.869 | 4.869 | 94% |
| Total | 221 | 1130.572 | 5.197 | 100% | |
| FST = 0.063, p = 0.001; FST max = 0.488; F′ST = 0.129, p = 0.001 | |||||
| (A) K = 2 | ||||
| Population | Low + Mid-Low + Mid-High (1600 + 2300 + 2800 m) | High (3600 m) | ||
| Low + Mid-Low + Mid-High (1600 + 2300 + 2800 m) | – | 4.767 | ||
| High (3600 m) | 0.143 | – | ||
| (B) K = 3 | ||||
| Population | Low + Mid-Low (1600 + 2300 m) | Mid-High (2800 m) | High (3600 m) | |
| Low + Mid-Low (1600 + 2300 m) | – | 5.552 | 2.538 | |
| Mid-High (2800 m) | 0.089 | – | 2.539 | |
| High (3600 m) | 0.165 | 0.162 | – | |
| (C) K = 4 | ||||
| Population | Low (1600 m) | Mid-Low (2300 m) | Mid-High (2800 m) | High (3600 m) |
| Low (1600 m) | – | 9.621 | 4.026 | 2.050 |
| Mid-Low (2300 m) | 0.053 | – | 5.586 | 2.409 |
| Mid-High (2800 m) | 0.117 | 0.088 | – | 2.539 |
| High (3600 m) | 0.192 | 0.196 | 0.162 | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Díaz, C.; Ortíz-Sepúlveda, A.; Valladares, M.A.; Farias-Cantillana, D.; Molina-Montenegro, M.A.; Ballesteros, G.I. Development and Application of Novel SSR Markers to Assess the Genetic Diversity and Population Structure of Phacelia secunda Along an Altitudinal Gradient in the Central Chile Andes. Plants 2025, 14, 1135. https://doi.org/10.3390/plants14071135
Torres-Díaz C, Ortíz-Sepúlveda A, Valladares MA, Farias-Cantillana D, Molina-Montenegro MA, Ballesteros GI. Development and Application of Novel SSR Markers to Assess the Genetic Diversity and Population Structure of Phacelia secunda Along an Altitudinal Gradient in the Central Chile Andes. Plants. 2025; 14(7):1135. https://doi.org/10.3390/plants14071135
Chicago/Turabian StyleTorres-Díaz, Cristian, Ana Ortíz-Sepúlveda, Moisés A. Valladares, Darío Farias-Cantillana, Marco A. Molina-Montenegro, and Gabriel I. Ballesteros. 2025. "Development and Application of Novel SSR Markers to Assess the Genetic Diversity and Population Structure of Phacelia secunda Along an Altitudinal Gradient in the Central Chile Andes" Plants 14, no. 7: 1135. https://doi.org/10.3390/plants14071135
APA StyleTorres-Díaz, C., Ortíz-Sepúlveda, A., Valladares, M. A., Farias-Cantillana, D., Molina-Montenegro, M. A., & Ballesteros, G. I. (2025). Development and Application of Novel SSR Markers to Assess the Genetic Diversity and Population Structure of Phacelia secunda Along an Altitudinal Gradient in the Central Chile Andes. Plants, 14(7), 1135. https://doi.org/10.3390/plants14071135






