Transcriptome Analysis Reveals the Role of Plant Hormone Signal Transduction Pathways in the Drought Stress Response of Hemerocallis middendorffii
Abstract
1. Introduction
2. Results
2.1. Transcriptome Sequencing Data and Assembly Statistics
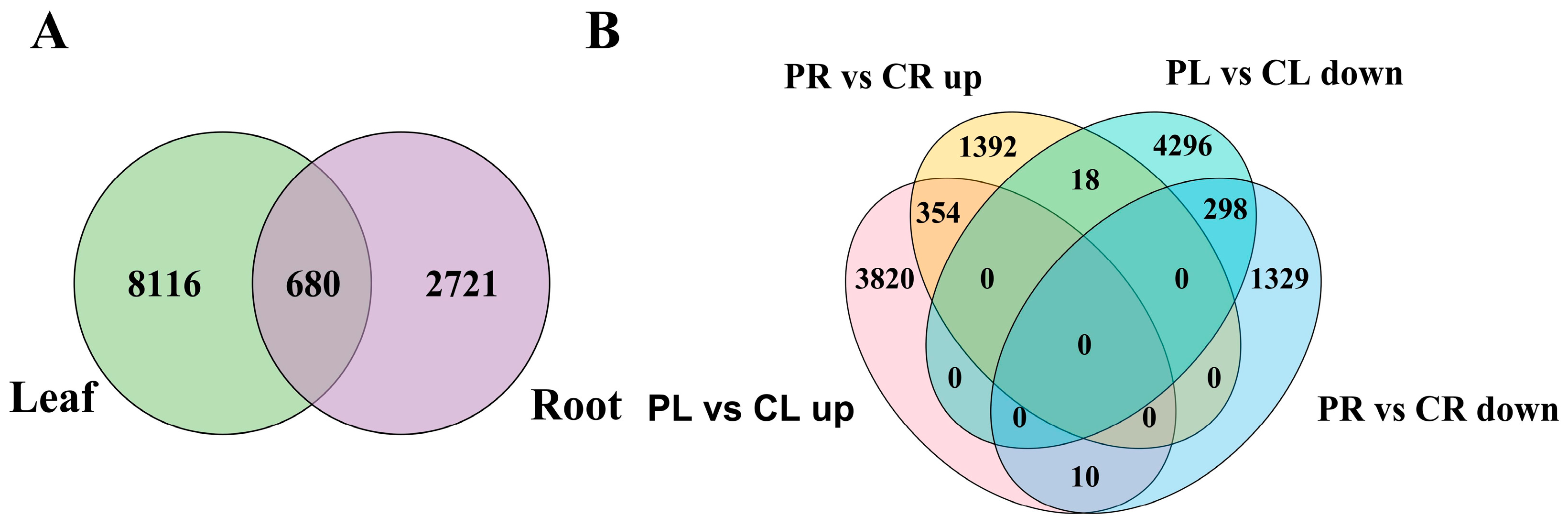
2.2. Analysis of DEGs
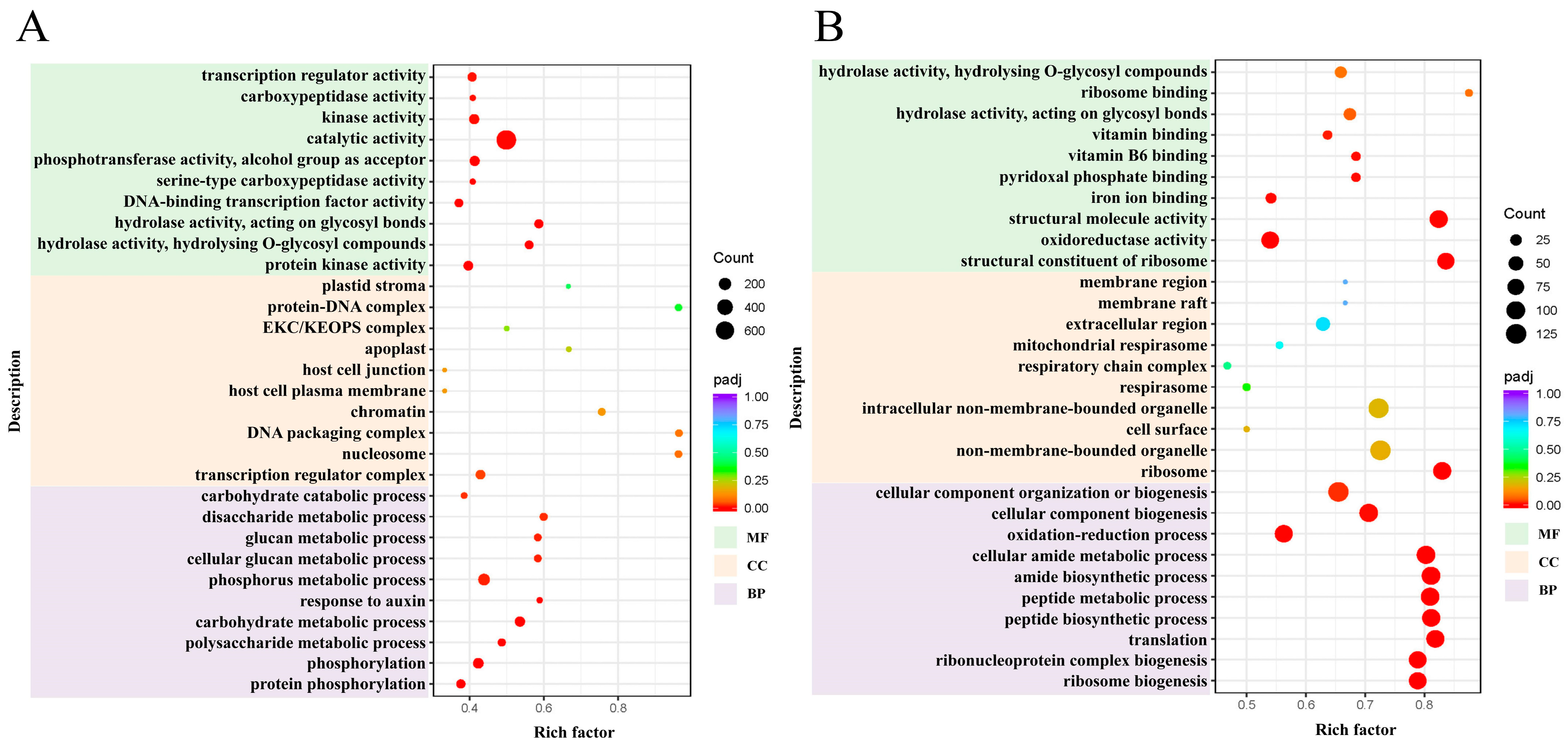
2.3. Gene Ontology (GO) Enrichment Analysis of DEGs
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of DEGs
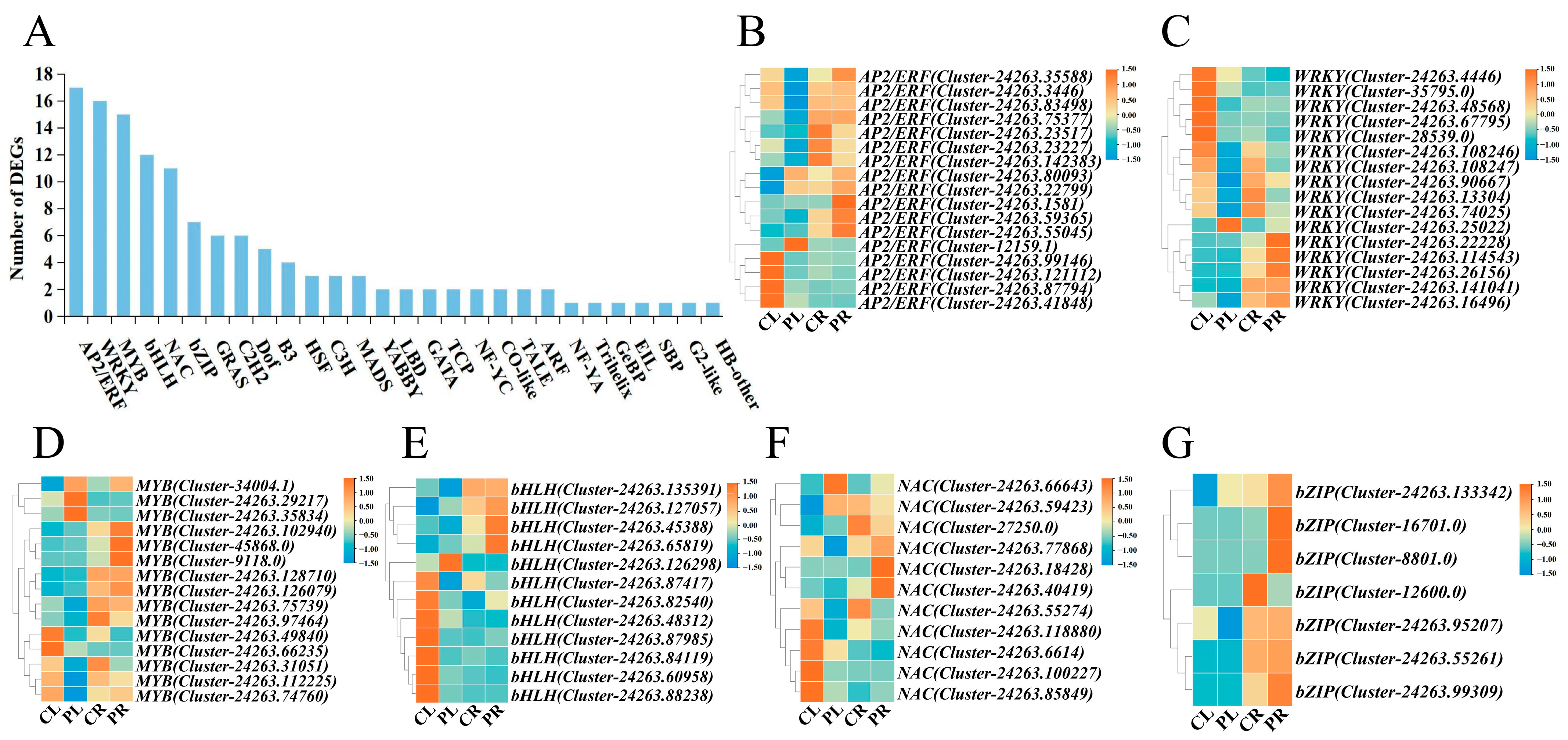
2.5. Identification and Analyzation of Differentially Expressed TFs
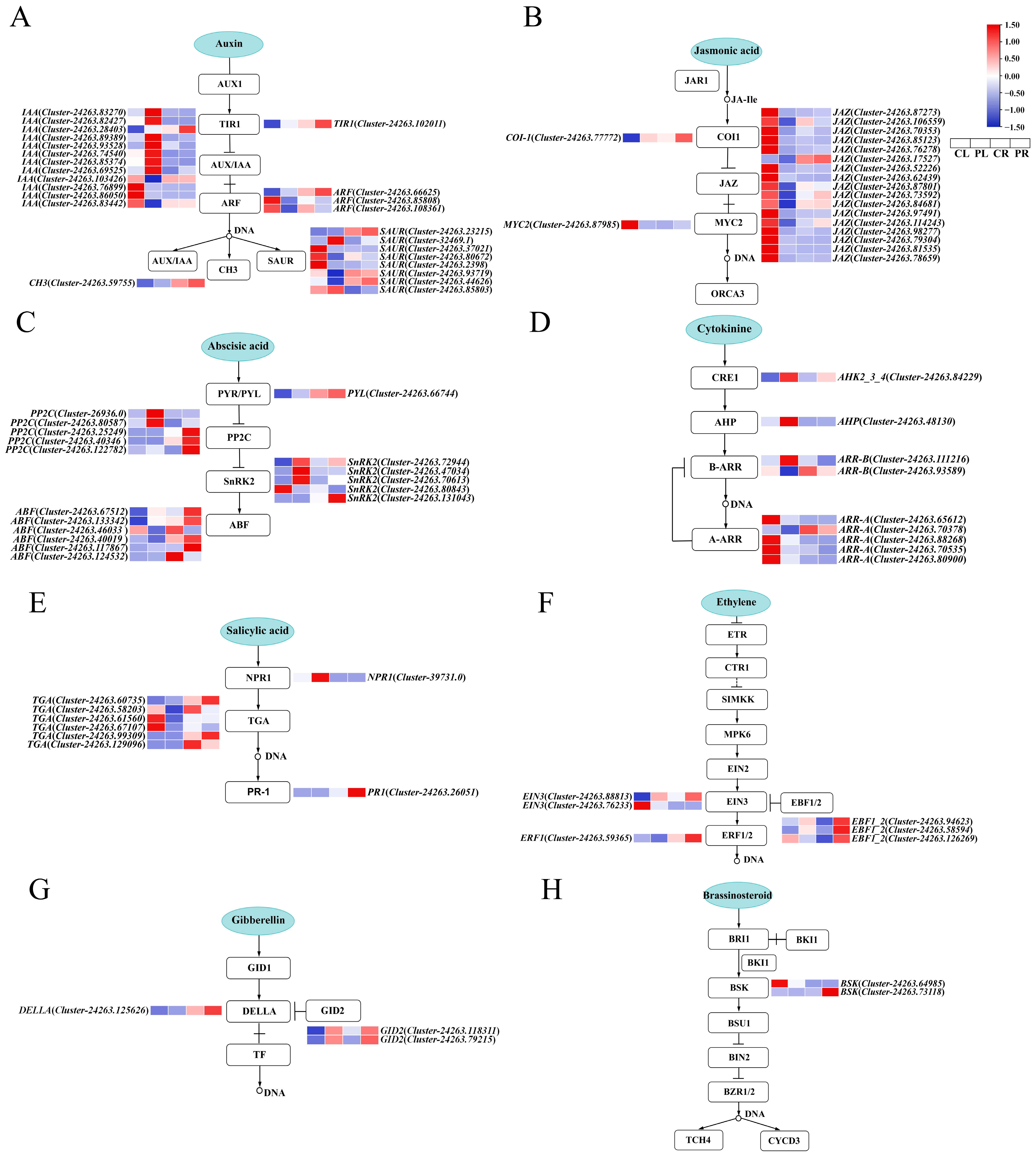
2.6. Analysis of the Expression Pattern of Relevant DEGs on the Plant Hormone Signal Transduction Pathway
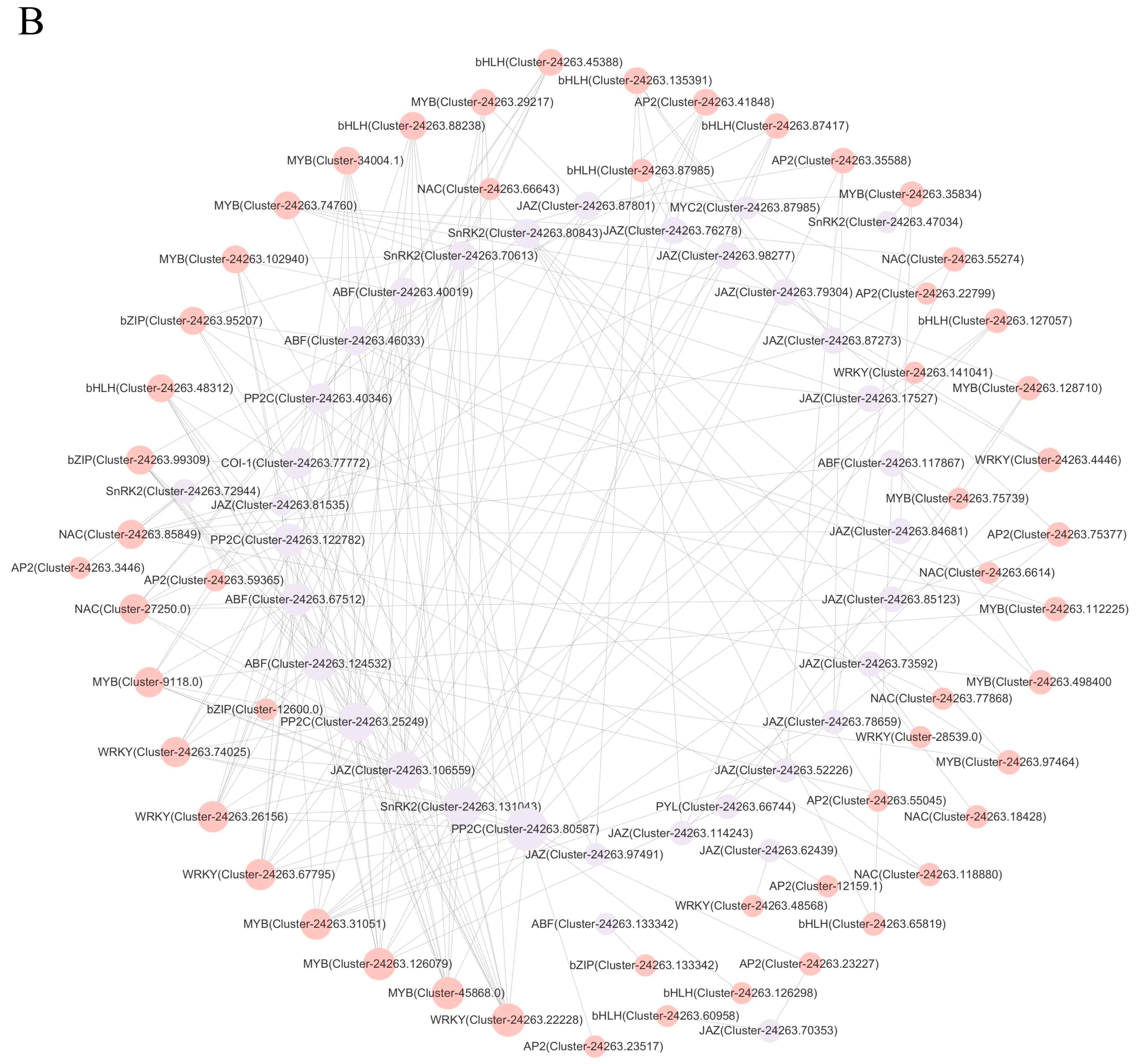
2.7. Correlational Network Analysis of Functional Genes Associated with ABA and JA Signal Transduction Pathways in Correlation with TFs
2.8. Validation of RNA-Seq Data by qRT-PCR of DEGs
3. Discussion
4. Materials and Methods
4.1. Material and Drought Stress Treatment
4.2. Total RNA Extraction, cDNA Library Construction, Sequencing and De Novo Assembly from the Hemerocallis middendorffii
4.3. Analysis of Transcriptome Data
4.4. Validation of Gene qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Q.; Lu, X. Studies on the physiological response of Hemerocallis middendorffii to two types of drought stresses. Int. J. Mol. Sci. 2024, 25, 13733. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.G.; Liu, X. Genetic diversity and DNA fingerprinting of Hemerocallis spp. accessions based on EST-SSR markers. Genet. Resour. Crop Evol. 2023, 70, 2033–2046. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Gao, Y.K. Molecular characterization and expression patterns of the HkSVP gene reveal distinct roles in inflorescence structure and floral organ development in Hemerocallis fulva. Int. J. Mol. Sci. 2021, 22, 12010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.M.; Yang, W. Evaluation of aromatic characteristics and potential applications of Hemerocallis L. based on the analytic hierarchy process. Molecules 2024, 29, 2712. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Q.X. Responsive mechanism of Hemerocallis citrina Baroni to complex saline-alkali stress revealed by photosynthetic characteristics and antioxidant regulation. Plant Cell Rep. 2024, 43, 176. [Google Scholar] [CrossRef]
- Zhang, B.X.; Li, Z.B. Impact of Cd and Pb on the photosynthetic and antioxidant systems of Hemerocallis citrina Baroni as revealed by physiological and transcriptomic analyses. Plant Cell Rep. 2024, 43, 226. [Google Scholar] [CrossRef]
- Yang, X.Y.; Lu, M.Q. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Gong, Z.Z.; Xiong, L.M. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Zhang, Y.; Jing, X. Plants’ response to abiotic stress: Mechanisms and strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Shivhare, R.; Lata, C. Assessment of pearl millet genotypes for drought stress tolerance at early and late seedling stages. Acta Physiol. Plant. 2019, 41, 39. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S. Drought tolerance in plants: Physiological and molecular responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.Y.; Yang, W.B. An integrated framework for drought stress in plants. Int. J. Mol. Sci. 2024, 25, 9347. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Li, X.M. Current views of drought research: Experimental methods, adaptation mechanisms and regulatory strategies. Front. Plant Sci. 2024, 15, 1371895. [Google Scholar] [CrossRef]
- Min, X.Y.; Lin, X.S. Coordinated mechanisms of leaves and roots in response to drought stress underlying full-length transcriptome profiling in Vicia sativa L. BMC Plant Biol. 2020, 20, 165. [Google Scholar] [CrossRef]
- Asakura, H.; Yamakawa, T. Transcriptomic and metabolomic analyses provide insights into the upregulation of fatty acid and phospholipid metabolism in tomato fruit under drought stress. J. Agric. Food Chem. 2021, 69, 2894–2905. [Google Scholar] [CrossRef]
- Danquah, A.; De Zelicourt, A. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, F.F. Balancing growth and adaptation to stress: Crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 2020, 43, 2325–2335. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Khan, S.A.; Li, M.Z. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Yang, S.Q.; Xu, K. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef]
- Luo, X.Y.; Li, C. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 2019, 39, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Raherison Elie, S.M.; Giguère, I. Modular organization of the white spruce (Picea glauca) transcriptome reveals functional organization and evolutionary signatures. New Phytol. 2015, 207, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Wang, Y.J. Transcriptomic and physiological analyses identifying Lanzhou lily (Lilium davidii var. unicolor) drought adaptation strategies. Hortic. Plant J. 2023, 9, 145–157. [Google Scholar] [CrossRef]
- Li, W.; Fu, L.F. Physiological characteristic changes and full-length transcriptome of rose (Rosa chinensis) roots and leaves in response to drought stress. Plant Cell Physiol. 2020, 61, 2153–2166. [Google Scholar] [CrossRef]
- Zhang, J.W.; Huang, D.Z. Evaluation of drought resistance and transcriptome analysis for the identification of drought-responsive genes in Iris germanica. Sci. Rep. 2021, 11, 16308. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghang, S.E.; Ullah, F. Transcriptome analysis of drought-resistant and drought-sensitive sorghum (Sorghum bicolor) genotypes in response to PEG-induced drought stress. Int. J. Mol. Sci. 2020, 21, 772. [Google Scholar] [CrossRef]
- Dou, L.R.; He, K.K. The E3 ligase MREL57 modulates microtubule stability and stomatal closure in response to ABA. Nat. Commun. 2021, 12, 2181. [Google Scholar] [CrossRef]
- Huang, B.L.; Li, X. Transcriptomic analysis of Eruca vesicaria subs. sativa lines with contrasting tolerance to polyethylene glycol-simulated drought stress. BMC Plant Biol. 2019, 19, 419. [Google Scholar] [CrossRef]
- Bashir, K.; Matsui, A. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, 658. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, D. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2023, 13, 1030890. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Huang, S.Q. Comparative transcriptome sequencing analysis and functional identification of a NAM-2-like gene in jute (Corchorus capsularis L.). Plant Physiol. Biochem. 2021, 161, 25–35. [Google Scholar] [CrossRef]
- François, P.; Arbes, H. RiboDoc: A docker-based package for ribosome profiling analysis. Comput. Struct. Biotechnol. J. 2021, 19, 2851–2860. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Bhalla, R. Over-expression of OSRIP18 increases drought and salt tolerance in transgenic rice plants. Transgenic Res. 2012, 21, 785–795. [Google Scholar] [CrossRef]
- Lqbal, S.; Wang, X.K. Phytohormones trigger drought tolerance in crop plants: Outlook and future perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef]
- Kim, T.H. Mechanism of ABA signal transduction: Agricultural highlights for improving drought tolerance. J. Plant Biol. 2014, 57, 1–8. [Google Scholar] [CrossRef]
- Belda-Palazόn, B.; Adamo, M. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Wang, L.Z.; Hu, W. Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9. Plant Sci. 2015, 237, 33–45. [Google Scholar] [CrossRef]
- Liang, D.; Ni, Z.Y. Exogenous melatonin promotes biomass accumulation and photosynthesis of kiwifruit seedlings under drought stress. Sci. Hortic. 2019, 246, 34–43. [Google Scholar] [CrossRef]
- Goossens, J.; Fernández-Calvo, P. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Liu, N.; Staswick, P.E. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ. 2016, 39, 2515–2529. [Google Scholar] [CrossRef]
- Lei, H.X.; Zhang, H.F. Physiological and transcriptomic analyses of roots from Panax ginseng C. A. Meyer under drought stress. Ind. Crop. Prod. 2023, 191, 115858. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2019–2042. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plantarum. 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Kong, L.F.; Song, Q. The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA-mediated signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.W. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019, 100, 384–398. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Z. MdMYB44-like positively regulates salt and drought tolerance via the MdPYL8-MdPP2CA module in apple. Plant J. 2023, 118, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Joo, J. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.J.; Zhang, J. RtNAC055 promotes drought tolerance via a stomatal closure pathway linked to methyl jasmonate/hydrogen peroxide signaling in Reaumuria trigyna. Hortic. Res. 2024, 11, uhae001. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, X.K.; Gao, J.H. Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti- inflammatory effects. Mol. Plant. 2022, 15, 1367–1383. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-deltadeltact method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Yu, H.; Lu, S.; Bai, Y.; Meng, Y.; Chen, L.; Wu, L.; Zhou, Y. Transcriptome Analysis Reveals the Role of Plant Hormone Signal Transduction Pathways in the Drought Stress Response of Hemerocallis middendorffii. Plants 2025, 14, 1082. https://doi.org/10.3390/plants14071082
Qian Y, Yu H, Lu S, Bai Y, Meng Y, Chen L, Wu L, Zhou Y. Transcriptome Analysis Reveals the Role of Plant Hormone Signal Transduction Pathways in the Drought Stress Response of Hemerocallis middendorffii. Plants. 2025; 14(7):1082. https://doi.org/10.3390/plants14071082
Chicago/Turabian StyleQian, Ying, Haihang Yu, Siyu Lu, Yun Bai, Yuan Meng, Lifei Chen, Lin Wu, and Yunwei Zhou. 2025. "Transcriptome Analysis Reveals the Role of Plant Hormone Signal Transduction Pathways in the Drought Stress Response of Hemerocallis middendorffii" Plants 14, no. 7: 1082. https://doi.org/10.3390/plants14071082
APA StyleQian, Y., Yu, H., Lu, S., Bai, Y., Meng, Y., Chen, L., Wu, L., & Zhou, Y. (2025). Transcriptome Analysis Reveals the Role of Plant Hormone Signal Transduction Pathways in the Drought Stress Response of Hemerocallis middendorffii. Plants, 14(7), 1082. https://doi.org/10.3390/plants14071082






