Multiple Dataset-Based Insights into the Phylogeny and Phylogeography of the Genus Exbucklandia (Hamamelidaceae): Additional Evidence on the Evolutionary History of Tropical Plants
Abstract
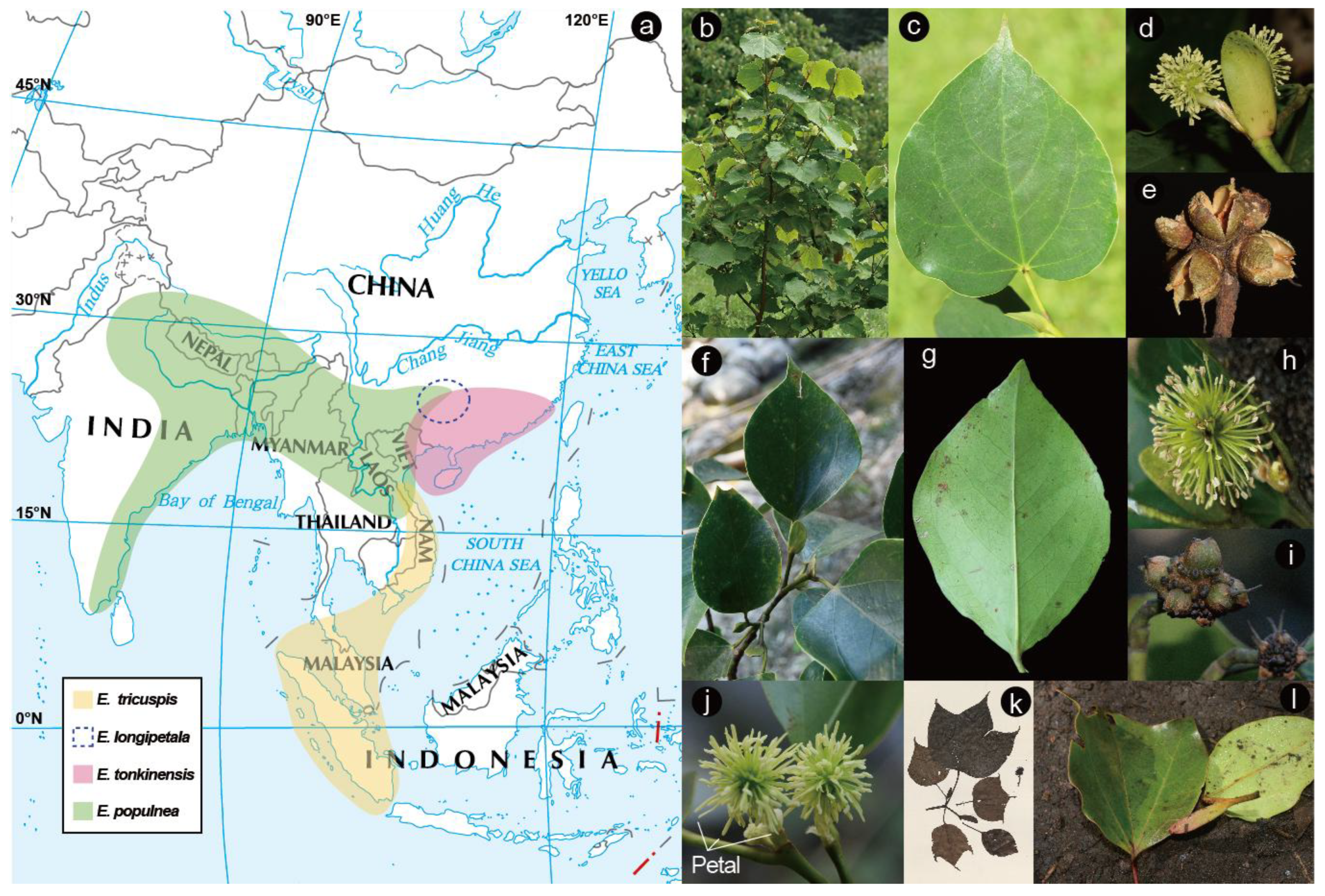
1. Introduction
2. Results
2.1. Haplotype Network of the Combined Chloroplast Regions
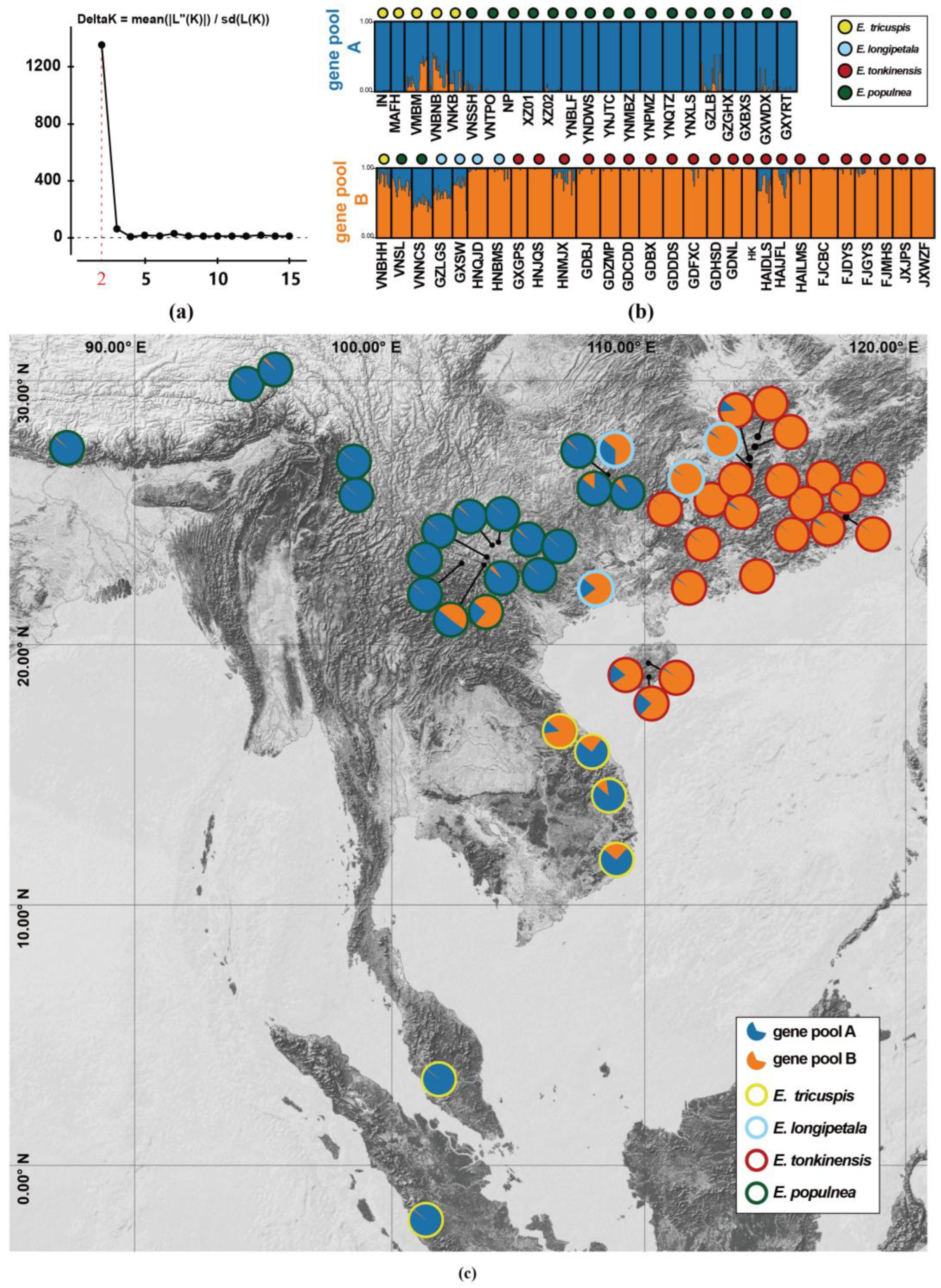
2.2. Population Genetics Based on SSR Loci
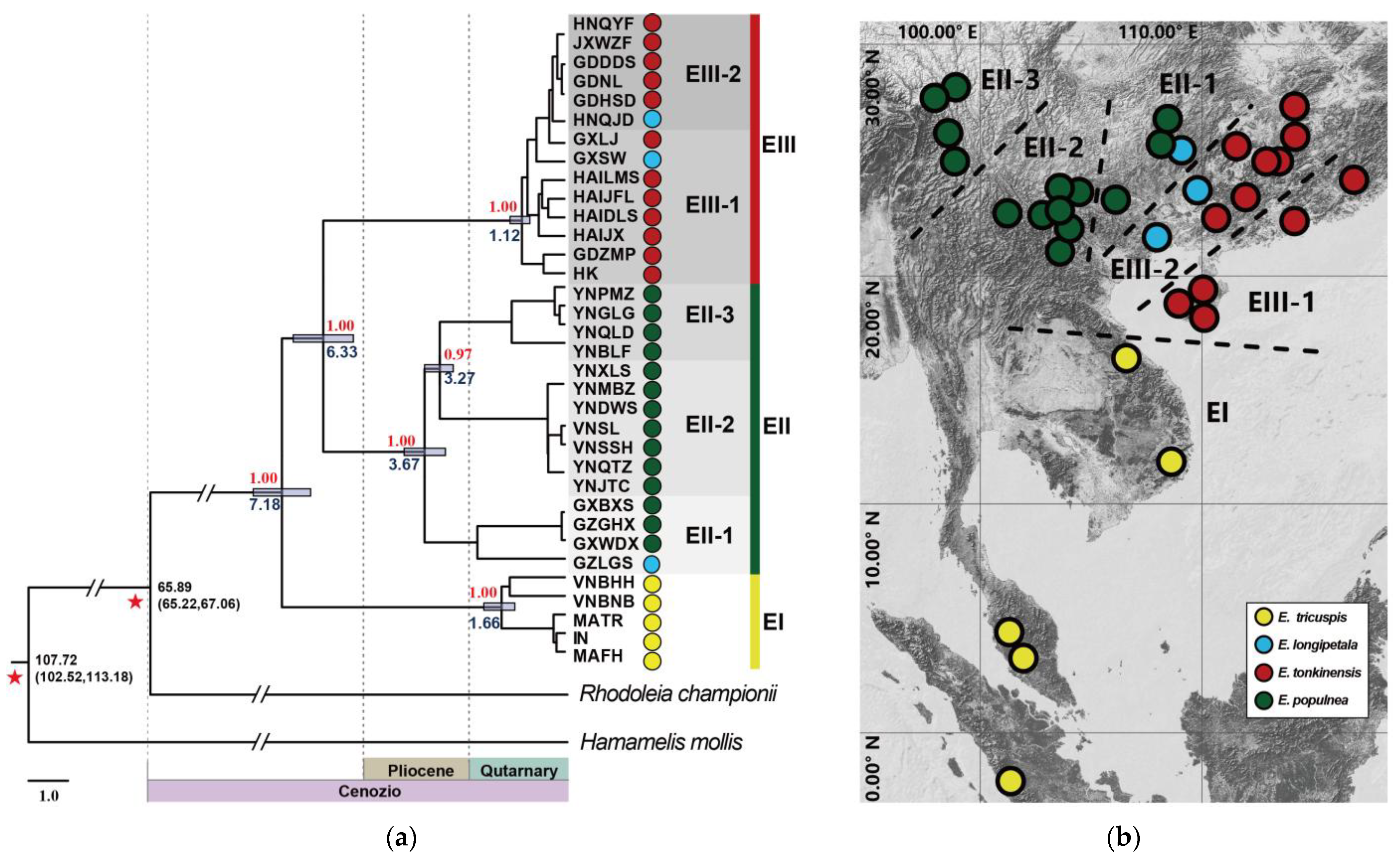
2.3. Phylogenetic Trees and Divergence Time Estimation
3. Discussion
3.1. The Origin and Dispersal History of Exbucklandia by Seed Dispersal
3.2. The Southward Expansion of E. populnea and E. tonkinensis Through Pollen Dispersal
3.3. Taxonomic Implication of Exbucklandia Species
3.4. The Genetic Diversity and Conservation of Exbucklandia Species
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
- To construct a plastid tree with high support, 1 representative individual per population was selected for plastid genome sequencing, so that we could assemble complete chloroplast genome sequences to construct a plastid tree.
- To construct a plastid haplotype network, we amplified four cpDNA fragments in 8 individuals per population, and the results showed that most populations harbored only one haplotype, suggesting that 8 individuals per population were sufficient.
- For obtaining nuclear data, we selected populations containing ≥10 individuals to amplify with SSR primers to ensure the reliability of genetic diversity and population structure analyses.
4.2. Plastid Region Amplification and Haplotype Network Construction
4.3. SSR Amplification, Inspection, and Diversity Analysis
4.4. Shallow Plastid Genome Sequencing and Assembly
4.5. Phylogenetic Tree Construction and Divergence Time Estimation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verstappen, H.T. Quaternary climatic changes and natural environment in SE Asia. GeoJournal 1980, 4, 45–54. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-X.; Fu, C.-X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenetics Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Liu, Z.; Utescher, T.; Jin, J.; Shu, J.; Li, Y.; Liu, Y.-S.C. Revisiting the Paleogene climate pattern of East Asia: A synthetic review. Earth-Sci. Rev. 2014, 139, 213–230. [Google Scholar] [CrossRef]
- Davis, M.B.; Shaw, R.G. Range shifts and adaptive responses to Quaternary climate change. Science 2001, 292, 673–679. [Google Scholar] [CrossRef]
- Carstens, B.C.; Knowles, L.L. Shifting distributions and speciation: Species divergence during rapid climate change. Mol. Ecol. 2007, 16, 619–627. [Google Scholar] [CrossRef]
- Gibbard, P.L.; Head, M.J. The quaternary period. In Geologic Time Scale 2020; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1217–1255. [Google Scholar]
- Avise, J. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Soltis, D.E.; Soltis, P.S.; Albert, V.A.; Oppenheimer, D.G.; DePamphilis, C.W.; Ma, H.; Frohlich, M.W.; Theißen, G. Missing links: The genetic architecture of flower and floral diversification. Trends Plant Sci. 2002, 7, 22–31. [Google Scholar] [CrossRef]
- Futuyma, D.J. Progress on the origin of species. PLoS Biol. 2005, 3, e62. [Google Scholar] [CrossRef]
- Lomolino, M.V.; Sax, D.F.; Riddle, B.R.; Brown, J.H. The island rule and a research agenda for studying ecogeographical patterns. J. Biogeogr. 2006, 33, 1503–1510. [Google Scholar] [CrossRef]
- Friedman, W.E.; Ryerson, K.C. Reconstructing the ancestral female gametophyte of angiosperms: Insights from Amborella and other ancient lineages of flowering plants. Am. J. Bot. 2009, 96, 129–143. [Google Scholar] [CrossRef]
- Hickerson, M.J.; Carstens, B.C.; Cavender-Bares, J.; Crandall, K.A.; Graham, C.H.; Johnson, J.B.; Rissler, L.; Victoriano, P.F.; Yoder, A.D. Phylogeography’s past, present, and future: 10 years after. Mol. Phylogenetics Evol. 2010, 54, 291–301. [Google Scholar]
- Sun, X.; Wang, P. How old is the Asian monsoon system?—Palaeobotanical records from China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 222, 181–222. [Google Scholar]
- Guo, Z.T.; Sun, B.; Zhang, Z.S.; Peng, S.Z.; Xiao, G.Q.; Ge, J.Y.; Hao, Q.Z.; Qiao, Y.S.; Liang, M.Y.; Liu, J.F. A major reorganization of Asian climate by the early Miocene. Clim. Past 2008, 4, 153–174. [Google Scholar] [CrossRef]
- Abe, M.; Hori, M.; Yasunari, T.; Kitoh, A. Effects of the Tibetan Plateau on the onsetof the summer monsoon in South Asia: The role of the air-sea interaction. J. Geophys. Res. Atmos. 2013, 118, 1760–1776. [Google Scholar]
- Chen, Y.S.; Deng, T.; Zhou, Z.; Sun, H. Is the East Asian flora ancient or not? Natl. Sci. Rev. 2018, 5, 920–932. [Google Scholar]
- Lu, L.M.; Mao, L.F.; Yang, T.; Ye, J.F.; Liu, B.; Li, H.L.; Sun, M.; Miller, J.T.; Mathews, S.; Hu, H.H.; et al. Evolutionary history of the angiosperm flora of China. Nature 2018, 554, 234–238. [Google Scholar] [CrossRef]
- Li, S.F.; Valdes, P.J.; Farnsworth, A.; Davies-Barnard, T.; Su, T.; Lunt, D.J.; Spicer, R.A.; Liu, J.; Deng, W.Y.D.; Huang, J. Orographic evolution of northern Tibet shaped vegetation and plant diversity in eastern Asia. Sci. Adv. 2021, 7, eabc7741. [Google Scholar]
- Qian, H.; Ricklefs, R.E. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 2000, 407, 180–182. [Google Scholar] [CrossRef]
- Harrison, S.P.; Yu, G.; Takahara, H.; Prentice, I.C. Diversity of temperate plants in east Asia. Nature 2001, 413, 129–130. [Google Scholar] [CrossRef]
- Manchester, S.R.; Chen, Z.D.; Lu, A.M.; Uemura, K. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the Northern Hemisphere. J. Syst. Evol. 2009, 47, 1–42. [Google Scholar]
- Ye, J.W.; Tian, B.; Li, D.Z. Monsoon intensification in East Asia triggered the evolution of its flora. Front. Plant Sci. 2022, 13, 1046538. [Google Scholar]
- Endress, P.K. Hamamelidaceae. In Flowering Plants·Dicotyledons: Magnoliid, Hamamelid and Caryophyllid Families; Springer: Berlin/Heidelberg, Germany, 1993; pp. 322–331. [Google Scholar]
- Zhang, Z.-Y.; Lu, A.-M. Hamamelidaceae: Geographic distribution, fossil history and origin. J. Syst. Evol. 1995, 33, 313–339. [Google Scholar]
- Bailey, C.D. Plant Systematics: A Phylogenetic Approach. Cladistics-Int. J. Willi Hennig Soc. 2010, 24, 848–850. [Google Scholar]
- Group, A.P.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Brown, R.W. Alterations in some fossil and living floras. J. Wash. Acad. Sci. 1946, 36, 344–355. [Google Scholar]
- Lakhanpal, R.N. The Rujada flora of west central Oregon. In University of California Publications in Geological Sciences; University of California Pres: Berkeley, CA, USA, 1958; Volume 35, pp. 1–65. [Google Scholar]
- Meyer, H. The Oligocene Lyons flora of northwestern Oregon. Ore Bin 1973, 35, 37–51. [Google Scholar]
- Baghai, N.L.; Jorstad, R.B. Paleontology, paleoclimatology and paleoecology of the late Middle Miocene Musselshell Creek flora, Clearwater County Idaho. A preliminary study of a new fossil flora. Palaios 1995, 10, 424–436. [Google Scholar]
- Manchester, S.R. Biogeographical relationships of North American tertiary floras. Ann. Mo. Bot. Gard. 1999, 86, 472–522. [Google Scholar]
- Zhou, Z.K.; Crepet, W.L.; Nixon, K.C. The earliest fossil evidence of the Hamamelidaceae: Late Cretaceous (Turonian) inflorescences and fruits of Altingioideae. Am. J. Bot. 2001, 88, 753–766. [Google Scholar]
- Pigg, K.B.; Wehr, W.C. Tertiary flowers, fruits, and seeds of Washington State and adjacent areas-Part III. Wash. Geol. 2002, 30, 3–16. [Google Scholar]
- Wu, J.; Sun, B.; Liu, Y.-S.C.; Xie, S.; Lin, Z. A new species of Exbucklandia (Hamamelidaceae) from the Pliocene of China and its paleoclimatic significance. Rev. Palaeobot. Palynol. 2009, 155, 32–41. [Google Scholar] [CrossRef]
- Shi, G.L. Fossil Plants from the Oligocene Ningming Formation of Guangxi, and a Preliminary Palaeoclimatic Reconstruction of the Flora. Ph.D. Thesis, Nanjing Institute of Geology and Palaeontology, Nanjing, China, 2010. (Chinese with English Abstract). [Google Scholar]
- Huang, J.; Shi, G.L.; Su, T.; Zhou, Z.K. Miocene Exbucklandia (Hamamelidaceae) from Yunnan, China and its biogeographic and palaeoecologic implications. Rev. Palaeobot. Palynol. 2017, 244, 96–106. [Google Scholar] [CrossRef]
- Vink, W. Hamamelidaceae. In Flora Malesiana-Series 1, Spermatophyta; Noordhoff-Kolff: Djakarta, Indonesia, 1955; Volume 5, pp. 363–379. [Google Scholar]
- Qiu, Y.L.; Chase, M.W.; Hoot, S.B.; Conti, E. Phylogenetics of the Hamamelidae and their allies: Parsimony analyses of nucleotide sequences of the plastid gene rbcL. Int. J. Plant Sci. 1998, 159, 891–905. [Google Scholar] [CrossRef]
- Li, J.; Bogle, A.L.; Klein, A.S. Phylogenetic relationships of the Hamamelidaceae inferred from sequences of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Am. J. Bot. 1999, 86, 1027–1037. [Google Scholar] [CrossRef]
- Maslova, N.P.; Herman, A.B. New finds of fossil hamamelids and data on the phylogenetic relationships between the Platanaceae and Hamamelidaceae. Paleontol. J. 2004, 38, 563–575. [Google Scholar]
- Jia, L.; Wang, S.; Hu, J.; Miao, K.; Huang, Y.; Ji, Y. Plastid phylogenomics and fossil evidence provide new insights into the evolutionary complexity of the ‘woody clade’ in Saxifragales. BMC Plant Biol. 2024, 24, 277. [Google Scholar] [CrossRef]
- Chang, H.T. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1979; Volume 35. [Google Scholar]
- Chang, H.T. A Revision of the Hamamelidaceous Flora of China. Acta Sci. Nat. Univ. Sunyatseni 1973, 1, 58–75. [Google Scholar]
- Chang, H.T. Notulae Plantarum Austro-Sinicarum. Acta Sci. Nat. Univ. Sunyatseni 1959, 4, 19–48. [Google Scholar]
- Garden, M.B. Tropicos.org. Available online: https://tropicos.org (accessed on 31 December 2024).
- WFO. The WFO Plant List. Available online: https://www.worldfloraonline.org/ (accessed on 31 December 2024).
- Turland, N.J.; Wiersema, J.H.; Barrie, F.R.; Greuter, W.; Hawksworth, D.L.; Herendeen, P.S.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code), Proceedings of the Nineteenth International Botanical Congress Shenzhen, China, 23–29 July 2017; Koeltz Botanical Books: Oberreifenberg, Germany, 2018. [Google Scholar]
- Lecompte, O.; Tardieu-Blot, M.L.; Cuong, V.V. Saxifragaceae, Crypteroniaceae, Droseraceae, Hamamelidaceae, Haloragaceae, Rhizophoraceae, Sonneratiaceae, Punicaceae; National Museum of Natural History: Paris, France, 1965; Volume 4. [Google Scholar]
- Huang, C.; Yin, Q.; Khadka, D.; Meng, K.; Fan, Q.; Chen, S.; Liao, W. Identification and development of microsatellite (SSRs) makers of Exbucklandia (HAMAMELIDACEAE) by high-throughput sequencing. Mol. Biol. Rep. 2019, 46, 3381–3386. [Google Scholar] [CrossRef]
- Xiang, X.; Xiang, K.; Ortiz, R.D.C.; Jabbour, F.; Wang, W. Integrating palaeontological and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamelidaceae. J. Biogeogr. 2019, 46, 2622–2631. [Google Scholar] [CrossRef]
- Tarullo, C.; Rose, J.; Sytsma, K.; Drew, B.T. Using a supermatrix approach to explore phylogenetic relationships, divergence times, and historical biogeography of Saxifragales. Turk. J. Bot. 2021, 45, 440–456. [Google Scholar]
- Xu, L.L.; Yu, R.M.; Lin, X.R.; Zhang, B.W.; Li, N.; Lin, K.; Zhang, D.Y.; Bai, W.N. Different rates of pollen and seed gene flow cause branch-length and geographic cytonuclear discordance within Asian butternuts. New Phytol. 2021, 232, 388–403. [Google Scholar] [PubMed]
- Huang, D.I.; Hefer, C.A.; Kolosova, N.; Douglas, C.J.; Cronk, Q.C.B. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 2014, 204, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Ennos, R. Estimating the relative rates of pollen and seed migration among plant populations. Heredity 1994, 72, 250–259. [Google Scholar] [CrossRef]
- Lu, H.Y.; Guo, Z.T. Evolution of the monsoon and dry climate in East Asia during late Cenozoic: A review. Sci. China Earth Sci. 2014, 57, 70–79. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar]
- Zachos, J.C.; Shackleton, N.J.; Revenaugh, J.S.; Pälike, H.; Flower, B.P. Climate response to orbital forcing across the Oligocene-Miocene boundary. Science 2001, 292, 274–278. [Google Scholar]
- Irwin, D.E. Phylogeographic breaks without geographic barriers to gene flow. Evolution 2002, 56, 2383–2394. [Google Scholar]
- Yang, Y.Y.; Qu, X.J.; Zhang, R.; Stull, G.W.; Yi, T.S. Plastid phylogenomic analyses of Fagales reveal signatures of conflict and ancient chloroplast capture. Mol. Phylogenetics Evol. 2021, 163, 107232. [Google Scholar]
- Liu, L.X.; Du, Y.X.; Folk, R.A.; Wang, S.Y.; Li, P. Plastome Evolution in Saxifragaceae and Multiple Plastid Capture Events Involving Heuchera and Tiarella. Front. Plant Sci. 2020, 11, 361. [Google Scholar] [CrossRef]
- Fehrer, J.; Gemeinholzer, B.; Chrtek, J.; Bräutigam, S. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenetics Evol. 2007, 42, 347–361. [Google Scholar]
- Yudai, O.; Noriyuki, F.; Michio, W.; Atsushi, K.; Manabu, I.; Mikio, W.; Noriaki, M.; Makoto, K. Nonuniform concerted evolution and chloroplast capture: Heterogeneity of observed introgression patterns in three molecular data partition phylogenies of Asian Mitella (saxifragaceae). Mol. Biol. Evol. 2005, 22, 285–296. [Google Scholar]
- Acosta, M.C.; Premoli, A.C. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenetics Evol. 2010, 54, 235–242. [Google Scholar]
- Chen, S.; Milne, R.; Zhou, R.; Meng, K.; Yin, Q.; Guo, W.; Ma, Y.; Mao, K.; Xu, K.; Kim, Y.D.; et al. When tropical and subtropical congeners met: Multiple ancient hybridization events within Eriobotrya in the Yunnan-Guizhou Plateau, a tropical-subtropical transition area in China. Mol. Ecol. 2022, 31, 1543–1561. [Google Scholar] [CrossRef]
- Lin, C.; Zhao, W.; Chen, Z.; Fan, Q.; Xu, K. Ilex danxiaensis (Aquifoliaceae), a Distinct New Tree Species Endemic to Danxia Mountain in Guangdong Province, China, Based on Molecular and Morphological Evidence. Forests 2023, 14, 583. [Google Scholar] [CrossRef]
- Westoby, M.; Leishman, M.; Lord, J. Comparative ecology of seed size and dispersal. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1996, 351, 1309–1318. [Google Scholar]
- Moles, A.T.; Ackerly, D.D.; Webb, C.O.; Tweddle, J.C.; Dickie, J.B.; Westoby, M. A brief history of seed size. Science 2005, 307, 576–580. [Google Scholar]
- De La Torre, A.R.; Roberts, D.R.; Aitken, S.N. Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol. Ecol. 2014, 23, 2046–2059. [Google Scholar]
- An, X.Y.; Ping, H.E.; Hong, L.X. The flowering phenology and reproductive features of the endangered plant Disanthus cercidifolius var. longipes H. T. Chang (Hamamelidaceae). Acta Ecol. Sin. 2004, 24, 14–21. [Google Scholar]
- Yu, Y.; Fan, Q.; Shen, R.; Guo, W.; Jin, J.; Cui, D.; Liao, W. Genetic Variability and Population Structure of Disanthus cercidifolius subsp. longipes (Hamamelidaceae) Based on AFLP Analysis. PLoS ONE 2014, 9, e107769. [Google Scholar]
- Li, B.J.; Wang, J.Y.; Liu, Z.J.; Zhuang, X.Y.; Huang, J.X. Genetic diversity and ex situ conservation of Loropetalum subcordatum, an endangered species endemic to China. BMC Genet. 2018, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Yuan, J.; Yin, G.; Yang, J.; Zou, W. Genetic diversity and structure among natural populations of Mytilaria laosensis (Hamamelidaceae) revealed by microsatellite markers. Silvae Genet. 2018, 67, 93–98. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Hu, Y.; Zhuang, X.; Xu, W.; Li, P.; Wang, Z. Mining and characterization of novel EST-SSR markers of Parrotia subaequalis (Hamamelidaceae) from the first Illumina-based transcriptome datasets. PLoS ONE 2019, 14, e0215874. [Google Scholar]
- Yu, N.; Li, R.S.; Dong, M.; Yang, J.C. Genetic diversity and paternity analysis of Mytilaria laosensis hybrids using microsatellite markers provide insight into the breeding system. Ind. Crops Prod. 2023, 191, 115974. [Google Scholar]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot5177. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Bioinform. Methods Protoc. 1999, 132, 71–91. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar]
- Smouse, R.P.P.; Peakall, R. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Hug, L.A.; Roger, A.J. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol. Biol. Evol. 2007, 24, 1889–1897. [Google Scholar] [CrossRef]
- Endress, P.K.; Friis, E.M. Archamamelis, hamamelidalean flowers from the Upper Cretaceous of Sweden. Plant Syst. Evol. 1991, 175, 101–114. [Google Scholar] [CrossRef]
- Mai, D.H. The fossils of Rhodoleia champion (Hamamelidaceae) in Europe. Acta Palaeobot. 2001, 41, 161–176. [Google Scholar]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Ecol. Evol. 2007, 7, 214. [Google Scholar] [CrossRef]

| Species | Pop ID | NT | I | HO | HE | F |
|---|---|---|---|---|---|---|
| E. tricuspis | IN | 33 | 0.334 | 0.211 | 0.218 | 0.034 |
| MAFH | 25 | 0.168 | 0.105 | 0.106 | 0.054 | |
| VMBM | 98 | 1.118 | 0.360 | 0.588 | 0.347 | |
| VNBHH | 40 | 0.379 | 0.244 | 0.238 | 0.044 | |
| VNBNB | 98 | 1.056 | 0.297 | 0.524 | 0.387 | |
| VNKB | 80 | 0.897 | 0.289 | 0.464 | 0.422 | |
| E. populnea | NP | 38 | 0.428 | 0.277 | 0.271 | −0.031 |
| XZ01 | 23 | 0.200 | 0.115 | 0.121 | −0.010 | |
| XZ02 | 34 | 0.393 | 0.222 | 0.241 | 0.051 | |
| YNBLF | 53 | 0.649 | 0.347 | 0.383 | 0.105 | |
| YNDWS | 48 | 0.561 | 0.310 | 0.340 | 0.029 | |
| YNJTC | 57 | 0.616 | 0.360 | 0.367 | 0.017 | |
| YNMBZ | 46 | 0.556 | 0.276 | 0.343 | 0.149 | |
| YNPMZ | 47 | 0.536 | 0.280 | 0.321 | 0.098 | |
| YNQTZ | 45 | 0.594 | 0.314 | 0.386 | 0.201 | |
| YNXLS | 54 | 0.590 | 0.267 | 0.333 | 0.190 | |
| GZLB | 66 | 0.777 | 0.357 | 0.441 | 0.192 | |
| GZGHX | 46 | 0.479 | 0.273 | 0.290 | 0.025 | |
| GXBXS | 49 | 0.592 | 0.290 | 0.362 | 0.157 | |
| GXWDX | 61 | 0.663 | 0.254 | 0.382 | 0.326 | |
| GXYRT | 37 | 0.319 | 0.221 | 0.201 | −0.087 | |
| VNSL | 47 | 0.409 | 0.207 | 0.225 | 0.064 | |
| VNNCS | 41 | 0.380 | 0.226 | 0.220 | −0.017 | |
| VNSSH | 106 | 1.220 | 0.386 | 0.613 | 0.371 | |
| VNTPO | 90 | 0.920 | 0.300 | 0.482 | 0.355 | |
| E. longipetala | GZLGS | 39 | 0.429 | 0.324 | 0.278 | −0.158 |
| GXSW | 50 | 0.687 | 0.375 | 0.436 | 0.147 | |
| HNQJD | 50 | 0.542 | 0.202 | 0.326 | 0.344 | |
| HNBMS | 69 | 0.779 | 0.315 | 0.434 | 0.266 | |
| E. tonkinensis | GXGPS | 60 | 0.684 | 0.321 | 0.393 | 0.265 |
| HNJQS | 62 | 0.675 | 0.392 | 0.387 | 0.063 | |
| HNMJX | 78 | 0.946 | 0.369 | 0.522 | 0.339 | |
| GDBJ | 50 | 0.581 | 0.315 | 0.355 | 0.072 | |
| GDZMP | 84 | 0.940 | 0.338 | 0.500 | 0.344 | |
| GDCDD | 58 | 0.618 | 0.383 | 0.358 | −0.096 | |
| GDBX | 90 | 1.017 | 0.476 | 0.543 | 0.129 | |
| GDDDS | 45 | 0.492 | 0.255 | 0.300 | 0.192 | |
| GDFXC | 67 | 0.740 | 0.264 | 0.413 | 0.324 | |
| GDHSD | 73 | 0.904 | 0.342 | 0.513 | 0.337 | |
| GDNL | 56 | 0.775 | 0.424 | 0.475 | 0.131 | |
| HK | 57 | 0.717 | 0.387 | 0.421 | 0.072 | |
| HAIDLS | 87 | 1.106 | 0.397 | 0.600 | 0.307 | |
| HAIJFL | 79 | 0.971 | 0.419 | 0.526 | 0.233 | |
| HAILMS | 67 | 0.789 | 0.410 | 0.448 | 0.089 | |
| FJCBC | 35 | 0.356 | 0.164 | 0.235 | 0.246 | |
| FJDYS | 47 | 0.427 | 0.193 | 0.250 | 0.229 | |
| FJGYS | 64 | 0.652 | 0.296 | 0.354 | 0.128 | |
| FJMHS | 41 | 0.445 | 0.267 | 0.277 | 0.024 | |
| JXJPS | 46 | 0.586 | 0.341 | 0.367 | 0.060 | |
| JXWZF | 61 | 0.763 | 0.427 | 0.455 | 0.081 | |
| Total | 0.649± | 0.303± | 0.372± | 0.152± | ||
| 0.242 | 0.079 | 0.118 | 0.141 |
| FIS | FIT | FST | NM | |
|---|---|---|---|---|
| E. populnea | 0.126 ± 0.026 | 0.609 ± 0.041 | 0.557 ± 0.04 | 0.248 ± 0.035 |
| E. tonkinensis | 0.18 ± 0.038 | 0.533 ± 0.031 | 0.432 ± 0.023 | 0.367 ± 0.037 |
| E. tricuspis | 0.283 ± 0.07 | 0.643 ± 0.045 | 0.507 ± 0.032 | 0.283 ± 0.036 |
| E. longipetala | 0.116 ± 0.063 | 0.584 ± 0.035 | 0.503 ± 0.042 | 0.321 ± 0.048 |
| Genus Exbucklandia | 0.189 ± 0.033 | 0.621 ± 0.027 | 0.536 ± 0.022 | 0.232 ± 0.02 |
| Pop ID | Country | Research Contents (Number of Individuals) | Pop ID | Country | Research Contents (Number of Individuals) | ||||
|---|---|---|---|---|---|---|---|---|---|
| A * | B ** | C *** | A * | B ** | C *** | ||||
| E. tonkinensis | E. populnea | ||||||||
| FJMHS | China | ★(8) | ●(15) | XZ01 | China | ★(8) | ●(22) | ||
| FJGYS | China | ★(8) | ●(23) | XZ02 | China | ★(8) | ●(18) | ||
| FJCBC | China | ★(8) | ●(27) | YNQTZ | China | ▲(1) | ★(8) | ●(20) | |
| FJDYS | China | ★(8) | ●(17) | YNXLS | China | ▲(1) | ★(8) | ●(18) | |
| GDBJ | China | ★(8) | ●(24) | YNDWS | China | ▲(1) | ★(8) | ●(16) | |
| GDHSD | China | ▲(1) | ★(8) | ●(16) | YNMBZ | China | ▲(1) | ★(8) | ●(19) |
| GDDDS | China | ▲(1) | ★(8) | ●(20) | YNJTC | China | ▲(1) | ★(8) | ●(23) |
| GDWZS | China | ★(8) | YNQLD | China | ▲(1) | ★(8) | |||
| GDNL | China | ▲(1) | ★(8) | ●(19) | YNGLG | China | ▲(1) | ★(8) | |
| GDCDD | China | ★(8) | ●(19) | YNYFS | China | ★(8) | |||
| GDZMP | China | ▲(1) | ★(8) | ●(20) | YNPMZ | China | ▲(1) | ★(8) | ●(23) |
| GDFXC | China | ★(8) | ●(24) | YNHQZ | China | ★(8) | |||
| GDBX | China | ★(8) | ●(24) | YNBLF | China | ▲(1) | ★(8) | ●(21) | |
| GXLJ | China | ▲(1) | NP | Nepal | ★(8) | ●(16) | |||
| GXGPS | China | ●(16) | VNTPO | Vietnam | ★(8) | ●(24) | |||
| HAIJFL | China | ▲(1) | ★(8) | ●(19) | VNNCS | Vietnam | ★(8) | ●(20) | |
| HAIJX | China | ▲(1) | ★(8) | VNSSH | Vietnam | ▲(1) | ★(8) | ●(19) | |
| HAILMS | China | ▲(1) | ★(8) | ●(21) | VNSL | Vietnam | ▲(1) | ★(8) | ●(20) |
| HAIDLS | China | ▲(1) | ★(8) | ●(15) | E. tricuspis | ||||
| HK | China | ▲(1) | ★(8) | ●(15) | IN | Indonesia | ▲(1) | ★(8) | ●(16) |
| HNMJX | China | ★(8) | ●(24) | MATR | Malaysia | ▲(1) | ★(8) | ||
| HNQYF | China | ▲(1) | MAFH | Malaysia | ▲(1) | ★(8) | ●(14) | ||
| HNJQS | China | ★(8) | ●(25) | VNKB | Vietnam | ★(8) | ●(16) | ||
| JXWZF | China | ▲(1) | ★(8) | ●(20) | VNBNB | Vietnam | ▲(1) | ★(8) | ●(21) |
| JXJPS | China | ★(8) | ●(19) | VNBHH | Vietnam | ▲(1) | ★(8) | ●(15) | |
| E. populnea | VMBM | Vietnam | ★(8) | ●(25) | |||||
| GXYRT | China | ★(8) | ●(20) | E. longipetala | |||||
| GXBXS | China | ▲(1) | ★(8) | ●(20) | GXSW | China | ▲(1) | ★(8) | ●(20) |
| GXWDX | China | ▲(1) | ★(8) | ●(21) | GZLGS | China | ▲(1) | ★(8) | ●(20) |
| GZLB | China | ★(8) | ●(22) | HNQJD | China | ▲(1) | ★(8) | ●(20) | |
| GZGHX | China | ▲(1) | ★(8) | ●(15) | HNBMS | China | ★(8) | ●(24) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Fan, Q.; Xu, K.; Shi, S.; Meng, K.; Du, H.; Jin, J.; Guo, W.; Li, H.; Chen, S.; et al. Multiple Dataset-Based Insights into the Phylogeny and Phylogeography of the Genus Exbucklandia (Hamamelidaceae): Additional Evidence on the Evolutionary History of Tropical Plants. Plants 2025, 14, 1061. https://doi.org/10.3390/plants14071061
Huang C, Fan Q, Xu K, Shi S, Meng K, Du H, Jin J, Guo W, Li H, Chen S, et al. Multiple Dataset-Based Insights into the Phylogeny and Phylogeography of the Genus Exbucklandia (Hamamelidaceae): Additional Evidence on the Evolutionary History of Tropical Plants. Plants. 2025; 14(7):1061. https://doi.org/10.3390/plants14071061
Chicago/Turabian StyleHuang, Cuiying, Qiang Fan, Kewang Xu, Shi Shi, Kaikai Meng, Heying Du, Jiehao Jin, Wei Guo, Hongwei Li, Sufang Chen, and et al. 2025. "Multiple Dataset-Based Insights into the Phylogeny and Phylogeography of the Genus Exbucklandia (Hamamelidaceae): Additional Evidence on the Evolutionary History of Tropical Plants" Plants 14, no. 7: 1061. https://doi.org/10.3390/plants14071061
APA StyleHuang, C., Fan, Q., Xu, K., Shi, S., Meng, K., Du, H., Jin, J., Guo, W., Li, H., Chen, S., & Liao, W. (2025). Multiple Dataset-Based Insights into the Phylogeny and Phylogeography of the Genus Exbucklandia (Hamamelidaceae): Additional Evidence on the Evolutionary History of Tropical Plants. Plants, 14(7), 1061. https://doi.org/10.3390/plants14071061






