Systematic Analysis of the Betula platyphylla TCP Gene Family and Its Expression Profile Identifies Potential Key Candidate Genes Involved in Abiotic Stress Responses
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties of the BpTCP Gene Family
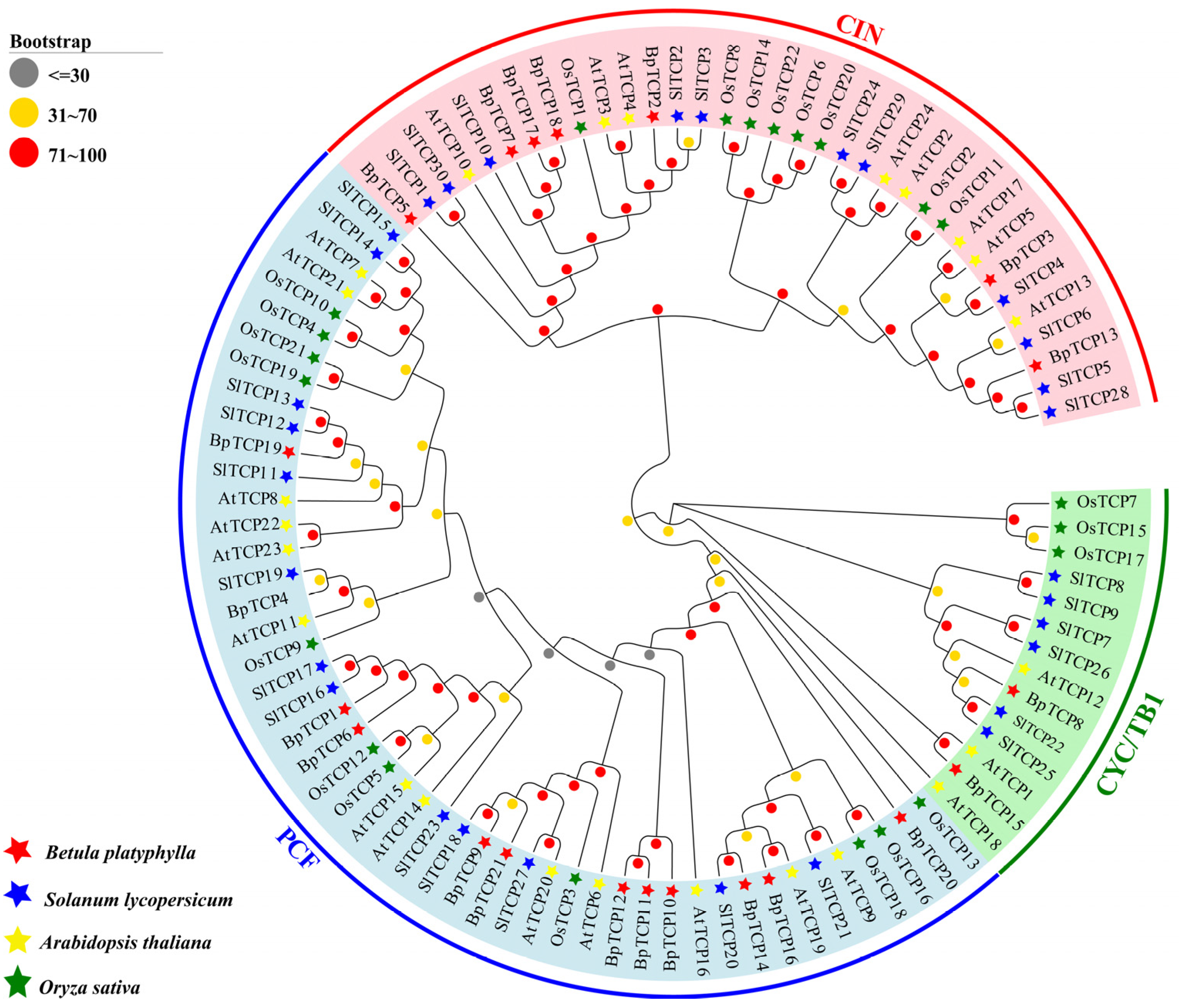
2.2. Phylogenetic Analysis of the BpTCP Gene Family
2.3. Secondary and Tertiary Structure of BpTCP Proteins
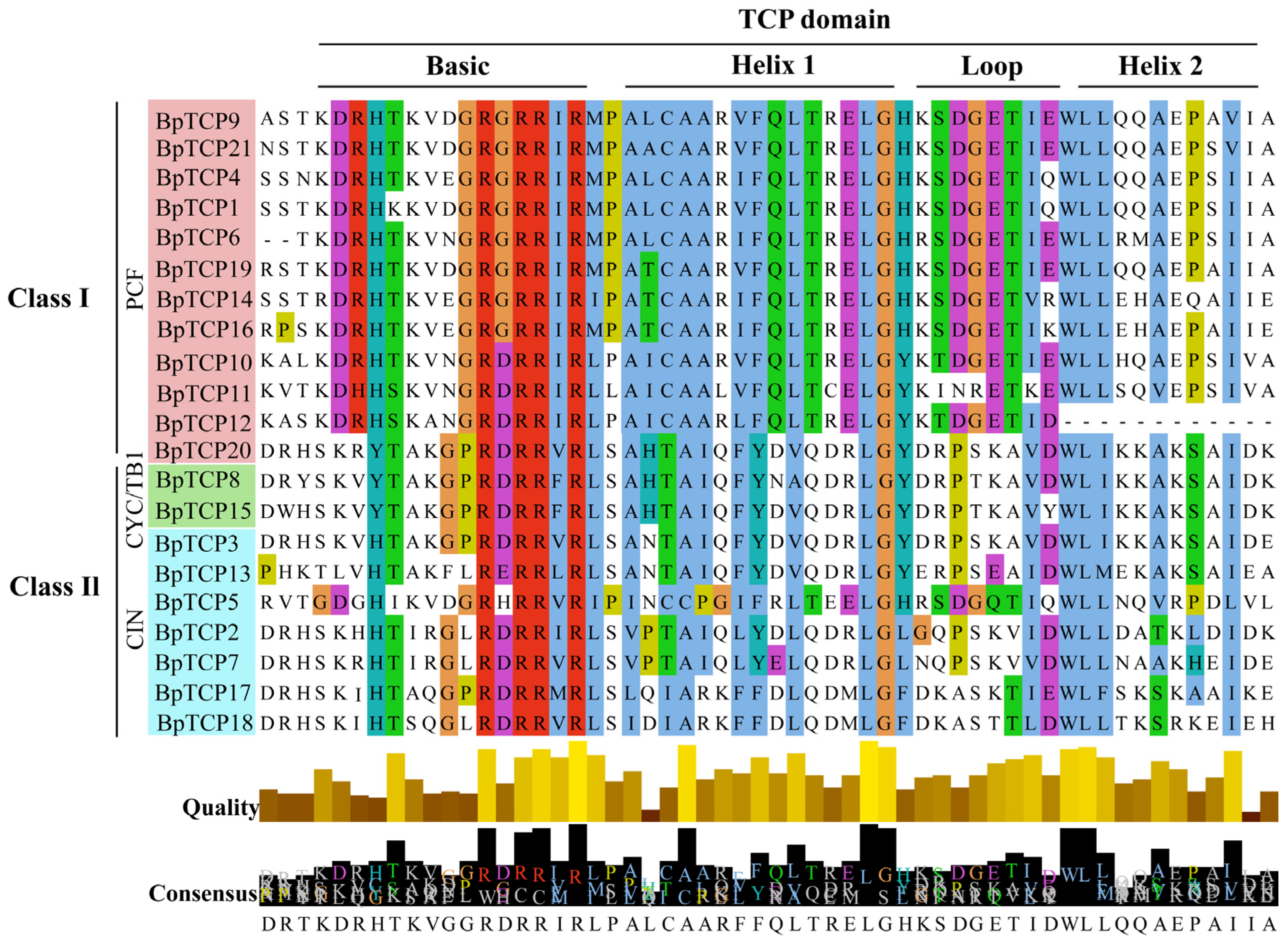
2.4. Multiple Sequence Comparison of BpTCP Proteins
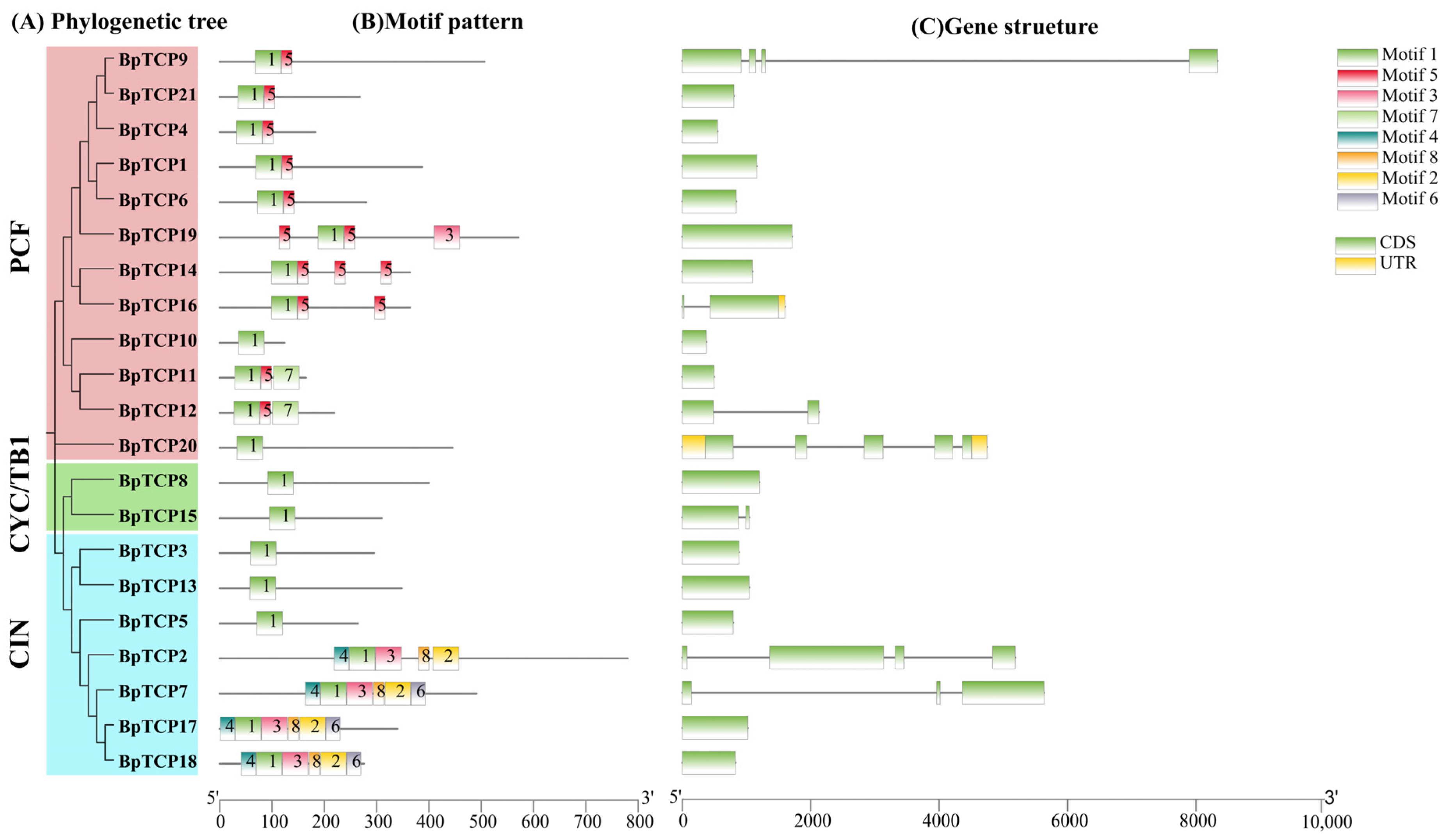
2.5. Gene Structure and Conserved Motif Analysis
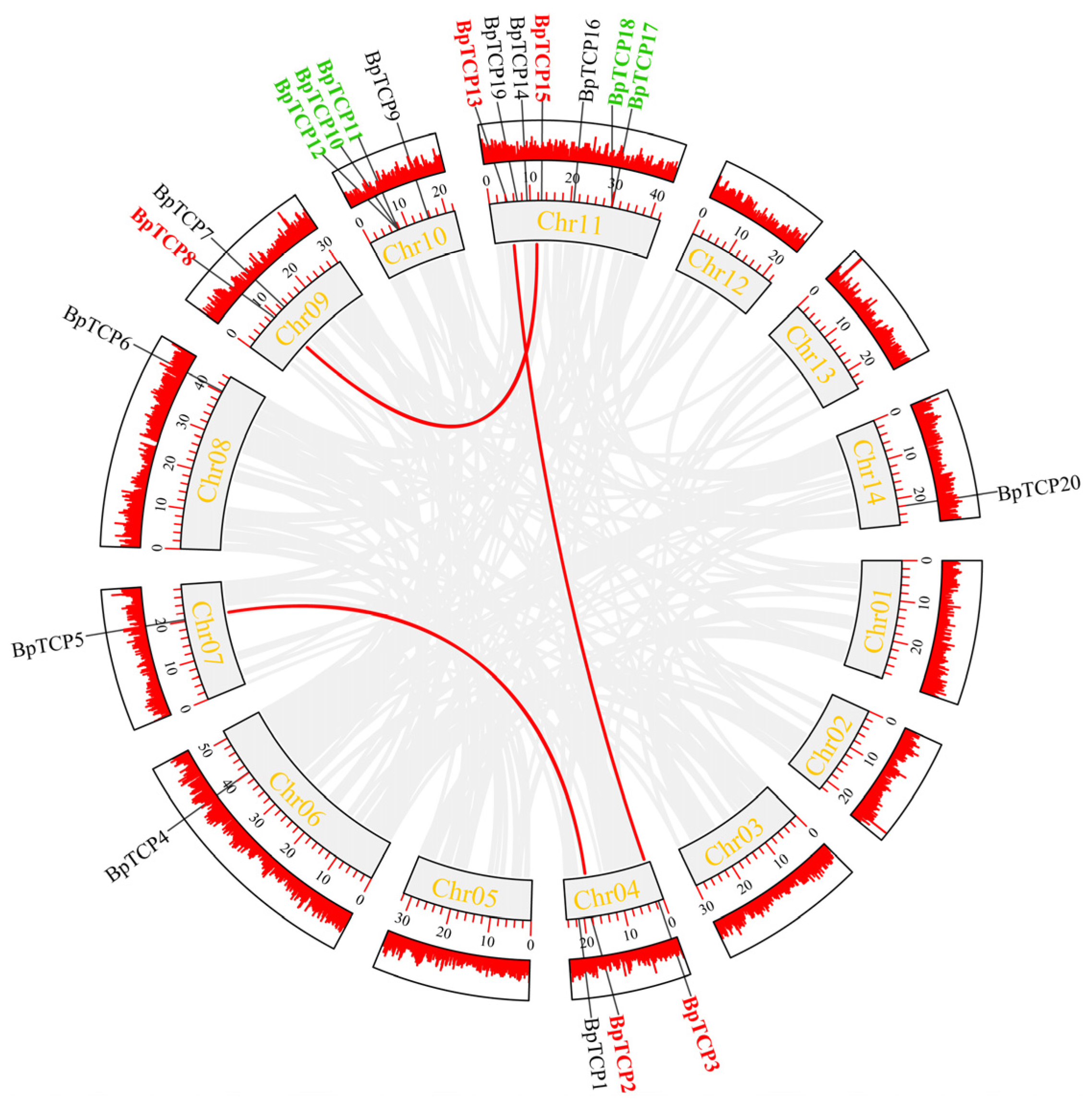
2.6. Chromosomal Localization of the BpTCP Gene Family
2.7. Intraspecies Covariance Analysis
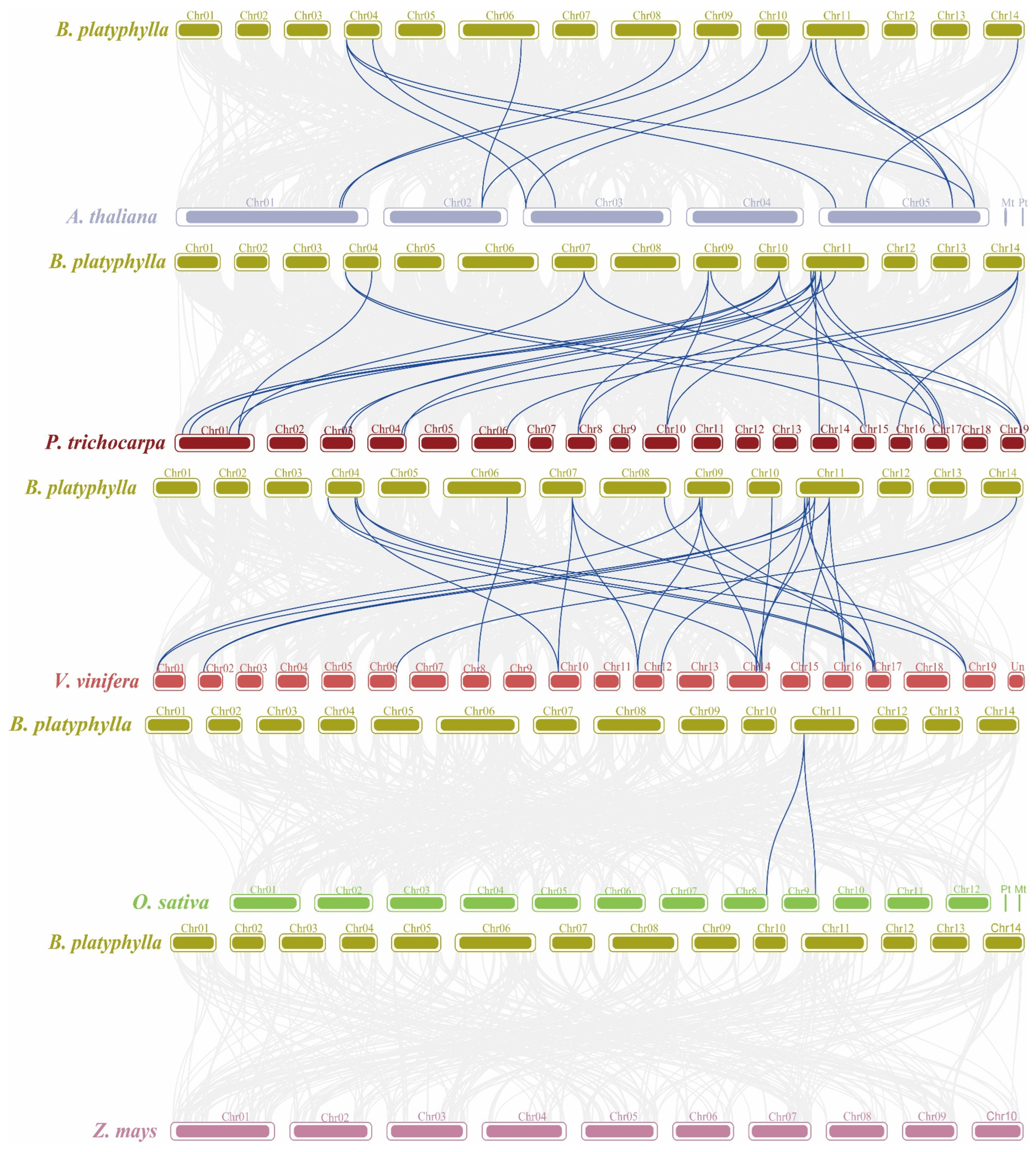
2.8. Interspecific Collinearity Analysis
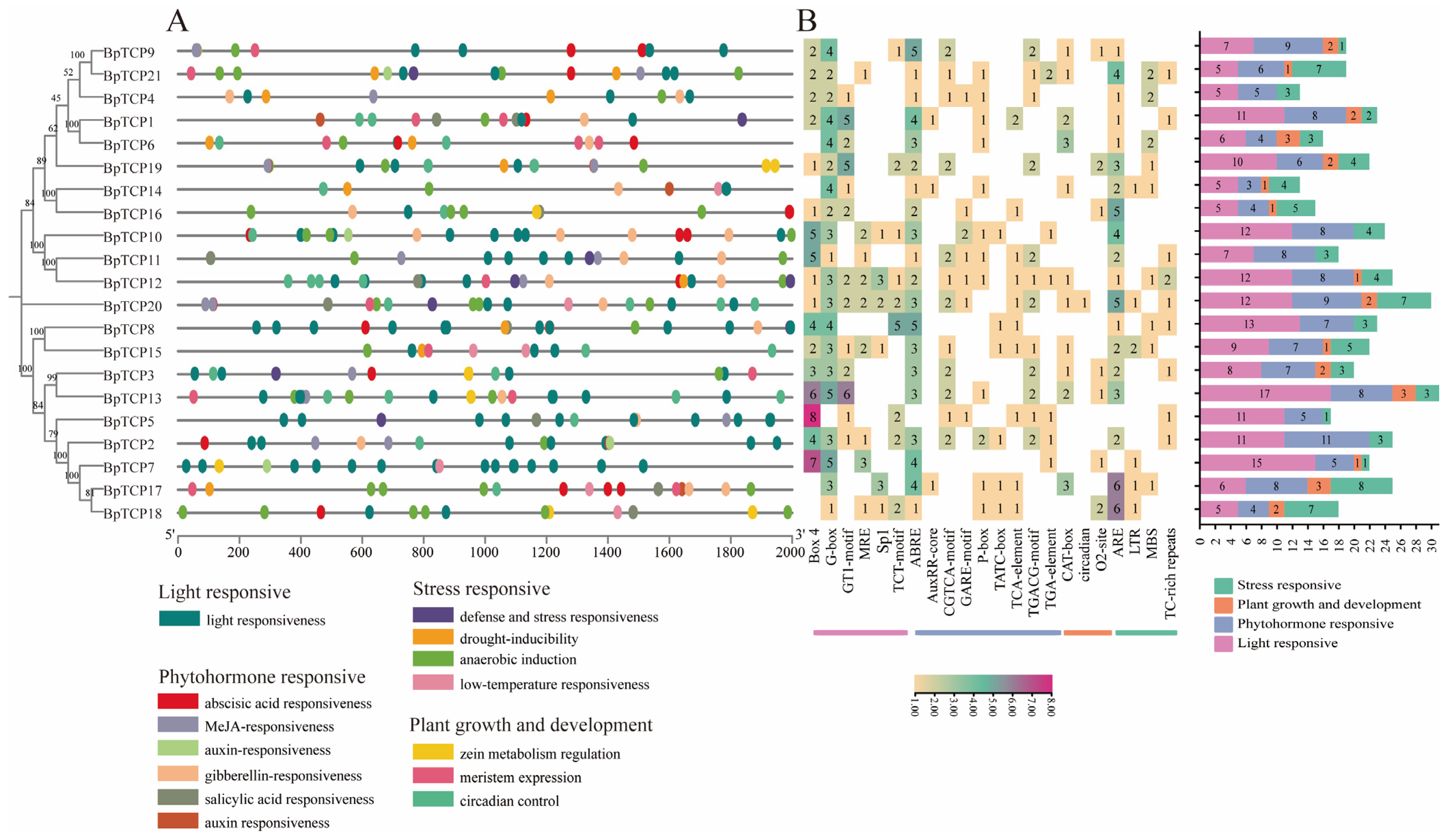
2.9. Analysis of Promoter Cis-Acting Elements of the BpTCP Gene Family
2.10. BpTCP Expression Profile in Different Tissues and During Drought
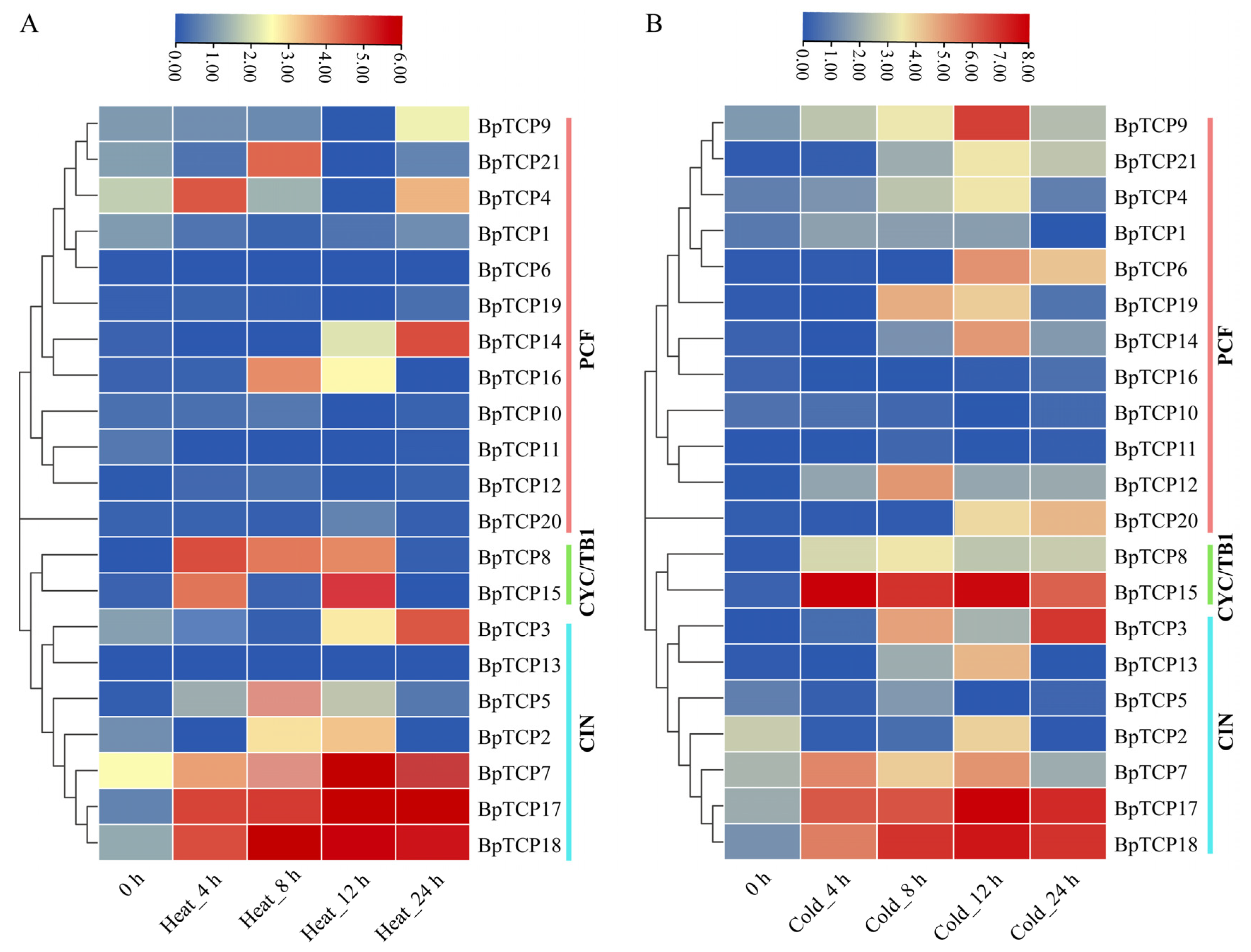
2.11. Expression Analysis of BpTCP Gene Under High- and Low-Temperature Treatments
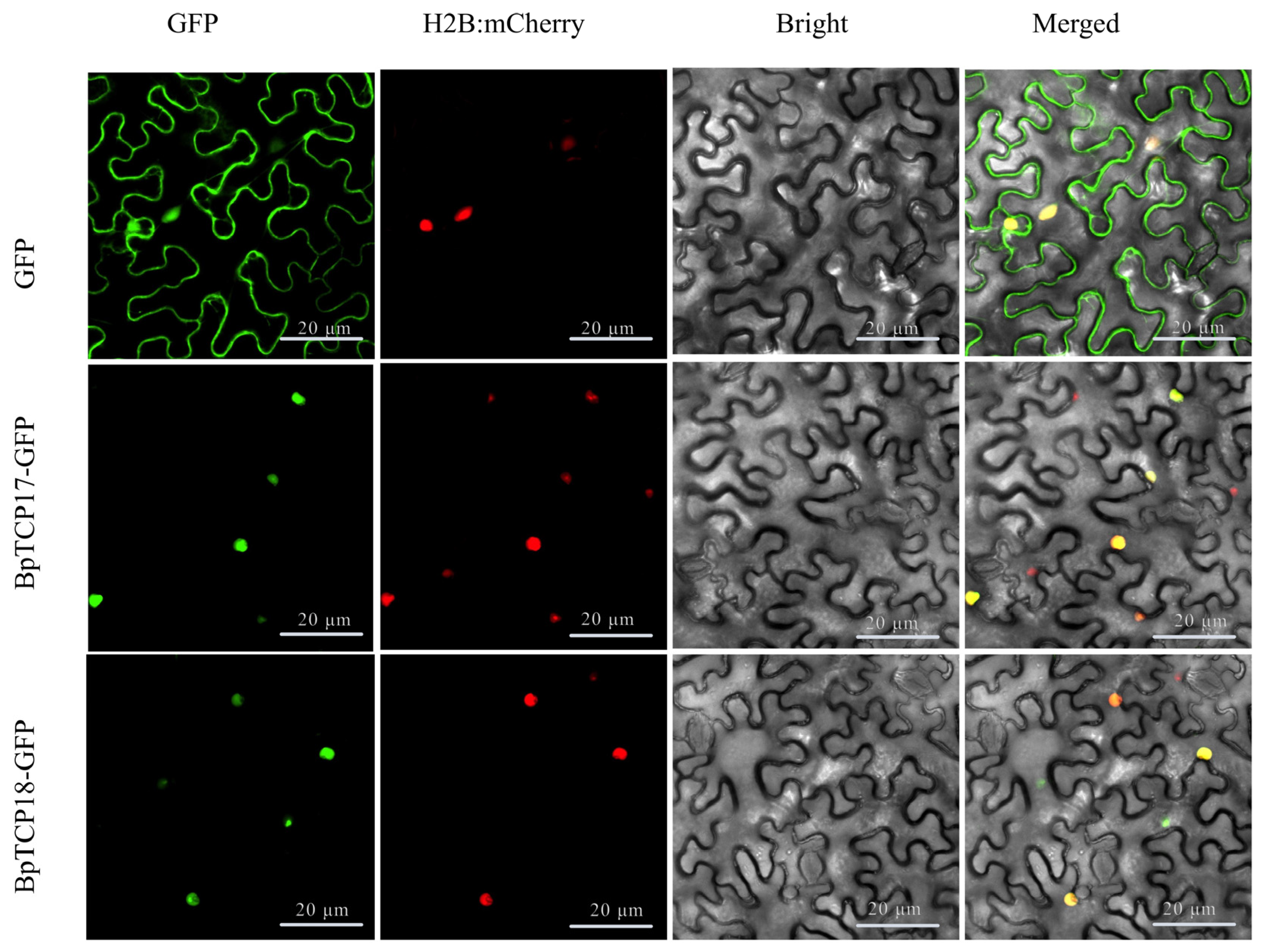
2.12. Subcellular Localization of BpTCP Proteins
3. Discussion
4. Materials and Methods
4.1. Materials and Treatments
4.2. Genome-Wide Identification of TCP Gene Family in B. platyphylla
4.3. BpTCP Protein Multiple Sequence Alignment and Evolutionary Relationship Analysis
4.4. Prediction of Secondary and Tertiary Structures of BpTCP Proteins
4.5. Gene Structure and Conserved Motif Analysis
4.6. Chromosome Localization and Covariate Covariance Analysis
4.7. Cis-Acting Element Analysis
4.8. RNA Extraction and qRT-PCR
4.9. Subcellular Localization of BpTCP Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Romani, F.; Moreno, J.E. Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytol. 2021, 230, 1345–1353. [Google Scholar] [CrossRef]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef] [PubMed]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–448. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Aggarwal, P.; Das Gupta, M.; Joseph, A.P.; Chatterjee, N.; Srinivasan, N.; Nath, U. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 2010, 22, 1174–1189. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Dianella, G.H.; Michael, J.D. Duplications in CYC-like Genes from Dipsacales Correlate with Floral Form. Int. J. Plant Sci. 2005, 166, 357–370. [Google Scholar]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J.; Lammers, M.; Karlova, R.; Bovy, A.G.; Angenent, G.C.; de Maagd, R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef]
- Huo, Y.; Xiong, W.; Su, K.; Li, Y.; Yang, Y.; Fu, C.; Wu, Z.; Sun, Z. Genome-Wide Analysis of the TCP Gene Family in Switchgrass (Panicum virgatum L.). Int. J. Genom. 2019, 2019, 8514928. [Google Scholar]
- Li, D.; Li, H.; Feng, H.; Qi, P.; Wu, Z. Unveiling kiwifruit TCP genes: Evolution, functions, and expression insights. Plant Signal Behav. 2024, 19, 2338985. [Google Scholar] [CrossRef]
- Dong, Z.; Hao, Y.; Zhao, Y.; Tang, W.; Wang, X.; Li, J.; Wang, L.; Hu, Y.; Guan, X.; Gu, F.; et al. Genome-Wide Analysis of the TCP Transcription Factor Gene Family in Pepper (Capsicum annuum L.). Plants 2024, 13, 641. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Jiang, P.; Huang, G.; Jiang, H.; Li, X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 2017, 23, 779–791. [Google Scholar] [CrossRef]
- Feng, Z.J.; Xu, S.C.; Liu, N.; Zhang, G.W.; Hu, Q.Z.; Gong, Y.M. Soybean TCP transcription factors: Evolution, classification, protein interaction and stress and hormone responsiveness. Plant Physiol. Biochem. 2018, 127, 129–142. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, Q.; Chen, F.; Wu, L.; Liu, B.; Li, F.; Zhang, J.; Bao, M.; Liu, G. Genome-Wide Identification, Characterization and Expression Analysis of TCP Transcription Factors in Petunia. Int. J. Mol. Sci. 2020, 21, 6594. [Google Scholar] [CrossRef]
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blázquez, M.A.; Alabadí, D.; Masiero, S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant. 2015, 8, 482–485. [Google Scholar] [CrossRef]
- Wei, Z.; Françoise, C.; Maharajah, P.; Sandrine, L.; Matheron, L.; Cédric, P.; Marie, B.; Francesca, R.; Stéphanie, H.; Miguel, Á.B.; et al. The MPK8-TCP14 pathway promotes seed germination in Arabidopsis. Plant J. 2019, 100, 677–692. [Google Scholar]
- Davière, J.M.; Wild, M.; Regnault, T.; Baumberger, N.; Eisler, H.; Genschik, P.; Achard, P. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 2014, 24, 1923–1928. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Luo, D. LjCYC Genes Constitute Floral Dorsoventral Asymmetry in Lotus japonicus. J. Integr. Plant Biol. 2010, 52, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Taito, T.; Kazuo, A.; Masa Aki, O.; Kenzo, N.; Shusei, S.; Tomohiko, K.; Satoshi, T.; Chiharu, U. RNA Interference of the Arabidopsis Putative Transcription Factor TCP16 Gene Results in Abortion of Early Pollen Development. Plant Mol. Biol. 2006, 61, 165–177. [Google Scholar]
- Selahattin, D.; van der Froukje, W.; Stijn, D.; Richard, W.; de Stefan, F.; Andrea, B.; Aalt, D.J.v.D.; Jose, M.M.; Lucas, C.; Marcelo Carnier, D.; et al. Arabidopsis Class I and Class II TCP Transcription Factors Regulate Jasmonic Acid Metabolism and Leaf Development Antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar]
- Edgardo, G.B.; Uciel, C.; Ramiro, E.R.; Javier, F.P.; Carla, S. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2018, 176, 1694–1708. [Google Scholar]
- Shen, J.; Zhang, Y.; Ge, D.; Wang, Z.; Song, W.; Gu, R.; Chen, G.; Cheng, Z.; Liu, R.; Zhang, X. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. USA 2019, 116, 17105–17114. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Jia, C.; Miao, H.; Zhang, J.; Liu, J.; Xu, B.; Jin, Z. Genome-Wide Identification and Transcript Analysis of TCP Gene Family in Banana (Musa acuminata L.). Biochem. Genet. 2022, 60, 204–222. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, N.; Fu, X.; Zhang, H.; Liu, S.; Pu, X.; Xiao, W.; Si, H. StTCP15 regulates potato tuber sprouting by modulating the dynamic balance between abscisic acid and gibberellic acid. Front. Plant Sci. 2022, 13, 1009552. [Google Scholar] [CrossRef]
- Ling, L.; Zhang, W.; An, Y.; Du, B.; Wang, D.; Guo, C. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 2020, 20, 537–550. [Google Scholar] [CrossRef]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive Expression of a miR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, N.; Li, S.; Liao, W.; Shen, J.; Peng, M. The regulatory effects of MeTCP4 on cold stress tolerance in Arabidopsis thaliana: A transcriptome analysis. Plant Physiol. Biochem. 2019, 138, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Pradipto, M.; Akhilesh, K.T. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 29, 9998. [Google Scholar]
- Liu, Y.J.; An, J.P.; Gao, N.; Wang, X.; Chen, X.X.; Wang, X.F.; Zhang, S.; You, C.X. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 2022, 45, 3233–3248. [Google Scholar] [CrossRef]
- Yin, Z.; Li, Y.; Zhu, W.; Fu, X.; Han, X.; Wang, J.; Lin, H.; Ye, W. Identification, Characterization, and Expression Patterns of TCP Genes and microRNA319 in Cotton. Int. J. Mol. Sci. 2018, 19, 3655. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Li, R.; Movahedi, A.; Chen, J.; Yang, L. Comprehensive Bioinformatics and Expression Analysis of TCP Transcription Factors in Liriodendron chinense Reveals Putative Abiotic Stress Regulatory Roles. Forests 2022, 13, 1401. [Google Scholar] [CrossRef]
- Chang, S.; Chao, W.; Lin, H.; Chuanping, Y.; Yucheng, W. Shotgun bisulfite sequencing of the B. platyphylla genome reveals the tree’s DNA methylation patterning. Int. J. Mol. Sci. 2014, 15, 22874. [Google Scholar]
- Geng, W.; Li, Y.; Sun, D.; Li, B.; Zhang, P.; Chang, H.; Rong, T.; Liu, Y.; Shao, J.; Liu, Z.; et al. Prediction of the potential geographical distribution of B. platyphylla Suk. in China under climate change scenarios. PLoS ONE 2022, 17, e0262540. [Google Scholar] [CrossRef]
- Elina, O. Birch as a Model Species for the Acclimation and Adaptation of Northern Forest Ecosystem to Changing Environment. Front. For. Glob. Change 2021, 4, 682512. [Google Scholar]
- Sylvie, G.; Pierre, Y.B.; Timo, K.; Shvidenko, A.S.; Dmitry, S. Boreal forest health and global change. Science 2015, 349, 819–822. [Google Scholar]
- Chen, S.; Wang, Y.; Yu, L.; Zheng, T.; Wang, S.; Yue, Z.; Jiang, J.; Kumari, S.; Zheng, C.; Tang, H.; et al. Genome sequence and evolution of B. platyphylla. Hortic. Res. 2021, 8, 37. [Google Scholar] [CrossRef]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.-J.; Tsai, W.-C. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Zhan, W.; Cui, L.; Guo, G.; Zhang, Y. Genome-wide identification and functional analysis of the TCP gene family in rye (Secale cereale L.). Gene 2023, 854, 147104. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, X.; Zheng, Q.; He, X.; Zhang, M.M.; Ke, S.; Li, Y.; Zhang, C.; Ahmad, S.; Lan, S.; et al. Genome-Wide Identification of TCP Gene Family in Dendrobium and Their Expression Patterns in Dendrobium chrysotoxum. Int. J. Mol. Sci. 2023, 24, 14320. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tang, X.; Dong, Y.; Wang, Y.; Shi, S.; Li, S.; Liu, Y.; Ge, H.; Chen, H. Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 2022, 41, 2213–2228. [Google Scholar] [CrossRef]
- Pan, J.; Ju, Z.; Ma, X.; Duan, L.; Jia, Z. Genome-wide characterization of TCP family and their potential roles in abiotic stress resistance of oat (Avena sativa L.). Front. Plant Sci. 2024, 15, 1382790. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Sun, X.L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.W.; Sun, M.Z.; Duan, X.B.; Zhu, Y.M. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef]
- Leng, X.; Wei, H.; Xu, X.; Ghuge, S.A.; Jia, D.; Liu, G.; Wang, Y.; Yuan, Y. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom. 2019, 20, 786. [Google Scholar] [CrossRef]
- Shi, P.; Guy, K.M.; Wu, W.; Fang, B.; Yang, J.; Zhang, M.; Hu, Z. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Zhang, C.; Zhao, X.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Li, M.H.; Lan, S.; Liu, Z.J. Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 2022, 13, 1068969. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, D.D.; Han, L.H.; Tao, M.; Hu, Q.Q.; Wu, W.Y.; Zhang, J.B.; Li, X.B.; Huang, G.Q. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 10118. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Chen, Y.; Chen, H.; Gong, R. Sweet cherry TCP gene family analysis reveals potential functions of PavTCP1, PavTCP2 and PavTCP3 in fruit light responses. BMC Genom. 2024, 25, 3. [Google Scholar] [CrossRef]
- Inukai, S.; Kock, K.H.; Bulyk, M.L. Transcription factor-DNA binding: Beyond binding site motifs. Curr. Opin. Genet. Dev. 2017, 43, 110–119. [Google Scholar] [CrossRef]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gómez-Cadenas, A.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012, 63, 1937–1949. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Cui, M.Y.; Han, Y.T.; Gao, K.; Feng, J.Y. Identification and Transcript Analysis of the TCP Transcription Factors in the Diploid Woodland Strawberry Fragaria vesca. Front. Plant Sci. 2016, 7, 1937. [Google Scholar] [CrossRef]
- Ptashne, M.; Gann, A. Transcriptional activation by recruitment. Nature 1997, 386, 569–577. [Google Scholar] [CrossRef]
- Voss, T.C.; Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014, 15, 69–81. [Google Scholar] [CrossRef]
- Caroline, U.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017, 18, 717–727. [Google Scholar]
- Xu, Y.; Wang, L.; Liu, H.; He, W.; Jiang, N.; Wu, M.; Xiang, Y. Identification of TCP family in moso bamboo (Phyllostachys edulis) and salt tolerance analysis of PheTCP9 in transgenic Arabidopsis. Planta 2022, 256, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Agassin, R.H.; Huang, Z.; Wang, D.; Yao, S.; Ji, K. Transcriptome-Wide Identification of TCP Transcription Factor Family Members in Pinus massoniana and Their Expression in Regulation of Development and in Response to Stress. Int. J. Mol. Sci. 2023, 24, 15938. [Google Scholar] [CrossRef] [PubMed]
- Shad, M.A.; Wu, S.; Rao, M.J.; Luo, X.; Huang, X.; Wu, Y.; Zhou, Y.; Wang, L.; Ma, C.; Hu, L. Evolution and Functional Dynamics of TCP Transcription Factor Gene Family in Passion Fruit (Passiflora edulis). Plants 2024, 13, 2568. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; He, G.; Hou, Q.; Fan, Y.; Duan, L.; Li, K.; Wei, X.; Qiu, Z.; Chen, E.; He, T. Systematic analysis and expression profiles of TCP gene family in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) revealed the potential function of FtTCP15 and FtTCP18 in response to abiotic stress. BMC Genom. 2022, 23, 415. [Google Scholar] [CrossRef]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2022, 13, 994567. [Google Scholar] [CrossRef]
- Zhou, H.; Hwarari, D.; Ma, H.; Xu, H.; Yang, L.; Luo, Y. Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front. Genet. 2022, 13, 1060546. [Google Scholar] [CrossRef]
- Liu, D.H.; Luo, Y.; Han, H.; Liu, Y.Z.; Alam, S.M.; Zhao, H.X.; Li, Y.T. Genome-wide analysis of citrus TCP transcription factors and their responses to abiotic stresses. BMC Plant Biol. 2022, 22, 325. [Google Scholar] [CrossRef]
- Zhao, M.; Peng, X.; Chen, N.; Shen, S. Genome-Wide Identification of the TCP Gene Family in Broussonetia papyrifera and Functional Analysis of BpTCP8, 14 and 19 in Shoot Branching. Plants 2020, 9, 1301. [Google Scholar] [CrossRef]
- Nie, Y.M.; Han, F.X.; Ma, J.J.; Chen, X.; Song, Y.T.; Niu, S.H.; Wu, H.X. Genome-wide TCP transcription factors analysis provides insight into their new functions in seasonal and diurnal growth rhythm in Pinus tabuliformis. BMC Plant Biol. 2022, 22, 167. [Google Scholar] [CrossRef]
- Dhaka, N.; Bhardwaj, V.; Sharma, M.K.; Sharma, R. Evolving Tale of TCPs: New Paradigms and Old Lacunae. Front. Plant Sci. 2017, 8, 479. [Google Scholar] [CrossRef]
- Nath, U.; Crawford, B.C.; Carpenter, R.; Coen, E. Genetic control of surface curvature. Science 2003, 299, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef]
- Jiao, P.; Liu, T.; Zhao, C.; Fei, J.; Guan, S.; Ma, Y. ZmTCP14, a TCP transcription factor, modulates drought stress response in Zea mays L. Environ. Exp. Bot. 2023, 208, 105232. [Google Scholar] [CrossRef]
- Fu, X.; Zhao, J.; Cao, D.; He, C.; Wang, Z.; Jiang, Y.; Liu, J.; Liu, G. Characteristics and expression of the TCP transcription factors family in Allium senescens reveal its potential roles in drought stress responses. Biocell 2023, 47, 905–917. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Mao, D.; Liu, X.; Ji, C.; Li, X.; Zhao, X.; Cheng, Z.; Chen, C.; Zhu, L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 2207–2218. [Google Scholar] [CrossRef]
- Thiebaut, F.; Rojas, C.A.; Almeida, K.L.; Grativol, C.; Domiciano, G.C.; Lamb, C.R.; Engler Jde, A.; Hemerly, A.S.; Ferreira, P.C. Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ. 2012, 35, 502–512. [Google Scholar] [CrossRef]
- Tian, C.; Zhai, L.; Zhu, W.; Qi, X.; Yu, Z.; Wang, H.; Chen, F.; Wang, L.; Chen, S. Characterization of the TCP Gene Family in Chrysanthemum nankingense and the Role of CnTCP4 in Cold Tolerance. Plants 2022, 11, 936. [Google Scholar] [CrossRef]
- Lei, N.; Yu, X.; Li, S.; Zeng, C.; Zou, L.; Liao, W.; Peng, M. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 2017, 7, 10016. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, D.; Xia, M.; Gong, M.; Li, H.; Xing, H.; Zhu, X.; Li, H.L. Genome-Wide Identification and Expression Analysis of the TCP Gene Family Related to Developmental and Abiotic Stress in Ginger. Plants 2023, 12, 3389. [Google Scholar] [CrossRef]
- Robert, C.E. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar]
- Koichiro, T.; Glen, S.; Sudhir, K. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evolution. 2021, 38, 3022–3027. [Google Scholar]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, C.; Pichel, J.C. BigSeqKit: A parallel Big Data toolkit to process FASTA and FASTQ files at scale. Gigascience. 2022, 12, giad062. [Google Scholar] [CrossRef]
- Giricz, O.; Lauer-Fields, J.L.; Fields, G.B. The normalization of gene expression data in melanoma: Investigating the use of glyceraldehyde 3-phosphate dehydrogenase and 18S ribosomal RNA as internal reference genes for quantitative real-time PCR. Anal. Biochem. 2008, 380, 137–139. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, X.; Shad, M.A.; Shu, Y.; Nong, S.; Li, X.; Wu, S.; Yang, J.; Rao, M.J.; Aslam, M.Z.; Huang, X.; et al. Genome-Wide Analysis of the Auxin/Indoleacetic Acid (Aux/IAA) Gene Family in Autopolyploid Sugarcane (Saccharum spontaneum). Int. J. Mol. Sci. 2024, 25, 7473. [Google Scholar] [CrossRef]



| Gene ID | Protein ID | AA | MW | pI | II | AI | GRAVY | SL |
|---|---|---|---|---|---|---|---|---|
| BPChr04G00604 | BpTCP1 | 386 | 40,843.18 | 6.87 | 66.17 | 56.71 | −0.597 | Nucleus |
| BPChr04G00664 | BpTCP2 | 779 | 86,635.17 | 7.16 | 56.51 | 61.90 | −0.760 | Nucleus |
| BPChr04G05342 | BpTCP3 | 294 | 33,563.17 | 6.45 | 47.85 | 62.07 | −0.914 | Nucleus |
| BPChr06G30802 | BpTCP4 | 182 | 19,700.41 | 8.45 | 58.59 | 72.47 | −0.358 | Nucleus |
| BPChr07G18922 | BpTCP5 | 263 | 30,545.66 | 5.12 | 69.49 | 47.91 | −1.108 | Nucleus |
| BPChr08G01309 | BpTCP6 | 279 | 30,575.31 | 9.44 | 73.68 | 68.89 | −0.594 | Nucleus |
| BPChr09G29718 | BpTCP7 | 490 | 54,167.47 | 8.20 | 45.87 | 68.57 | −0.533 | Chloroplast |
| BPChr09G29943 | BpTCP8 | 399 | 44,122.23 | 8.15 | 50.89 | 63.36 | −0.656 | Nucleus |
| BPChr10G03521 | BpTCP9 | 505 | 55,993.84 | 10.01 | 52.59 | 71.07 | −0.655 | Cytoplasm |
| BPChr10G17577 | BpTCP10 | 123 | 14,007.67 | 9.97 | 48.03 | 77.80 | −0.259 | Chloroplast |
| BPChr10G17579 | BpTCP11 | 164 | 18,128.67 | 5.77 | 46.78 | 73.72 | −0.423 | Nucleus |
| BPChr10G17585 | BpTCP12 | 218 | 24,210.98 | 6.18 | 34.61 | 82.75 | −0.139 | Nucleus |
| BPChr11G05694 | BpTCP13 | 347 | 38,726.77 | 6.85 | 48.58 | 62.97 | −0.842 | Nucleus |
| BPChr11G06894 | BpTCP14 | 363 | 38,410.07 | 6.24 | 51.92 | 70.72 | −0.389 | Nucleus |
| BPChr11G07004 | BpTCP15 | 309 | 35,088.50 | 7.16 | 44.20 | 64.11 | −0.784 | Nucleus |
| BPChr11G07178 | BpTCP16 | 363 | 37,925.25 | 6.07 | 56.73 | 61.68 | −0.525 | Nucleus |
| BPChr11G17722 | BpTCP17 | 339 | 37,499.35 | 9.22 | 37.38 | 63.66 | −0.493 | Nucleus |
| BPChr11G17777 | BpTCP18 | 275 | 30,761.23 | 7.13 | 30.87 | 64.33 | −0.697 | Nucleus |
| BPChr11G18700 | BpTCP19 | 570 | 60,307.68 | 6.91 | 64.31 | 52.86 | −0.791 | Nucleus |
| BPChr14G12311 | BpTCP20 | 444 | 48,868.34 | 6.19 | 46.47 | 85.86 | −0.280 | Nucleus |
| BPunChr32695 | BpTCP21 | 267 | 28,414.76 | 8.71 | 45.97 | 63.63 | −0.498 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Xu, Y.; Zhou, Y.; Liu, R.; Wang, Y.; Yao, L.; Azam, S.M.; Ma, H.; Liu, X.; Cao, S.; et al. Systematic Analysis of the Betula platyphylla TCP Gene Family and Its Expression Profile Identifies Potential Key Candidate Genes Involved in Abiotic Stress Responses. Plants 2025, 14, 880. https://doi.org/10.3390/plants14060880
Guo S, Xu Y, Zhou Y, Liu R, Wang Y, Yao L, Azam SM, Ma H, Liu X, Cao S, et al. Systematic Analysis of the Betula platyphylla TCP Gene Family and Its Expression Profile Identifies Potential Key Candidate Genes Involved in Abiotic Stress Responses. Plants. 2025; 14(6):880. https://doi.org/10.3390/plants14060880
Chicago/Turabian StyleGuo, Shengzhou, Yuan Xu, Yi Zhou, Ronglin Liu, Yongkang Wang, Ling Yao, Syed Muhammad Azam, Huanhuan Ma, Xiaomin Liu, Shijiang Cao, and et al. 2025. "Systematic Analysis of the Betula platyphylla TCP Gene Family and Its Expression Profile Identifies Potential Key Candidate Genes Involved in Abiotic Stress Responses" Plants 14, no. 6: 880. https://doi.org/10.3390/plants14060880
APA StyleGuo, S., Xu, Y., Zhou, Y., Liu, R., Wang, Y., Yao, L., Azam, S. M., Ma, H., Liu, X., Cao, S., & Wang, K. (2025). Systematic Analysis of the Betula platyphylla TCP Gene Family and Its Expression Profile Identifies Potential Key Candidate Genes Involved in Abiotic Stress Responses. Plants, 14(6), 880. https://doi.org/10.3390/plants14060880








