MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger
Abstract
1. Introduction
2. Results
2.1. Expression of CYP81A and miRNAs Targeting Their Transcripts
2.2. Expression of CYP71A, CYP72A, EcGST, GSTF1, and eIF4B and of the miRNAs Targeting Their Transcripts
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Herbicide Treatment
4.2. DNA Extraction and Assessment of ACCase Gene Mutations
4.3. RNA Extraction and cDNA Synthesis
4.4. Analyzed Genes and miRNA Prediction and Validation
4.5. RT-qPCR Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ente Nazionale Risi. Available online: www.enterisi.it (accessed on 4 May 2023).
- Istat—Istituto Nazionale di Statistica. Available online: http://dati.istat.it/ (accessed on 6 May 2023).
- GIRE®. Italian Herbicide Resistance Working Group, 2023. Database of Herbicide Resistance in Italy. Available online: http://gire.mlib.cnr.it/ (accessed on 14 March 2023).
- Mascanzoni, E.; Perego, A.; Marchi, N.; Scarabel, L.; Panozzo, S.; Ferrero, A.; Acutis, M.; Sattin, M. Epidemiology and agronomic predictors of herbicide resistance in rice at a large scale. Agron. Sustain. Dev. 2018, 38, 68. [Google Scholar] [CrossRef]
- Herbicide Resistance Action Committee (HRAC). Available online: https://hracglobal.com/ (accessed on 14 March 2023).
- The European Parliament and the Council of the European Union. Regulation (EC) 1107/2009 of 21 October Concerning the Placing of Plant Protection Products on the Market and Repealing Council Directives 79/117/EEC and 91/414/EEC; OJ L309/1–50; European Union: Brussels, Belgium, 2009. [Google Scholar]
- Duke, S.O.; Dayan, F.E. The search for new herbicide mechanisms of action: Is there a ‘holy grail’? Pest Manag. Sci. 2022, 78, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Panozzo, S.; Collavo, A.; Sattin, M. Sensitivity Analysis of Italian Lolium spp. to Glyphosate in Agricultural Environments. Plants 2020, 9, 165. [Google Scholar] [CrossRef]
- International Herbicide-Resistant Weed Database. Available online: https://www.weedscience.org/ (accessed on 23 March 2023).
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C.N., Jr. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-Target-Site Resistance to Herbicides: Recent Developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Yamaguchi, T.; Fukunishi, K.; Tominaga, T.; Iwakami, S. Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in Echinochloa phyllopogon. Plant Mol. Biol. 2020, 102, 403–416. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, X.; Liu, K.; Zhang, J.; Wu, X.; Zhu, J.; Tu, J. Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol. Biol. 2006, 61, 933–943. [Google Scholar] [CrossRef]
- Dam, T.; Guida, A.D.; Hazel, C.B.; Li, B.; Williams, M.E. Polynucleotide Encoding a Maize Herbicide Resistance Gene and Methods for Use. U.S. Patent No 7,705,200, 25 October 2007. [Google Scholar]
- Li, G.; Wu, S.G.; Yu, R.X.; Cang, T.; Chen, L.P.; Zhao, X.P.; Cai, L.M.; Wu, C.X. Identification and expression pattern of a glutathione S-transferase in Echinochloa crus-galli. Weed Res. 2013, 53, 314–321. [Google Scholar] [CrossRef]
- Iwakami, S.; Kamidate, Y.; Yamaguchi, T.; Ishizaka, M.; Endo, M.; Suda, H.; Nagai, K.; Sunohara, Y.; Toki, S.; Uchino, A.; et al. CYP81A P450s are involved in concomitant cross-resistance to acetolactate synthase and acetyl-CoA carboxylase herbicides in Echinochloa phyllopogon. New Phytol. 2019, 221, 2112–2122. [Google Scholar] [CrossRef]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 Are Associated with Resistance to Two Acetolactate Synthase Inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, W.; Du, Y.; Deng, Y.; Bai, T.; Ji, M. Multiple resistance of Echinochloa phyllopogon to synthetic auxin, ALS and ACCase-inhibiting herbicides in Northeast China. Pestic. Biochem. Physiol. 2023, 193, 105450. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhao, N.; Jiang, M.; Wang, M.; Zhang, J.; Cao, H.; Liao, M. Metamifop resistance in Echinochloa glabrescens via glutathione S-transferases-involved enhanced metabolism. Pest Manag. Sci. 2013, 79, 2725–2736. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Uchino, A.; Kataoka, Y.; Shibaike, H.; Watanabe, H.; Inamura, T. Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 2014, 70, 549–558. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Dalazen, G.; Markus, C.; Merotto, A., Jr. Differential Expression of Genes Associated with Degradation Enhancement of Imzethapyr in Barnyardgrass (Echinochloa crusgalli). J. Agric. Sci. 2018, 10, 389–401. [Google Scholar]
- Markus, C.; Pecinka, A.; Karan, R.; Barney, J.N.; Merotto, A., Jr. Epigenetic regulation—Contribution to herbicide resistance in weeds? Pest Manag. Sci. 2018, 74, 275–281. [Google Scholar] [CrossRef]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A. The Function of miRNAs in Plants. Plants 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Li, W.; Li, F.; Ren, M.; Wang, W. microRNAs: Key Regulators in Plant Responses to Abiotic and Biotic Stresses via Endogenous and Cross-Kingdom Mechanisms. Int. J. Mol. Sci. 2024, 25, 1154. [Google Scholar] [CrossRef]
- Arteaga-Vázquez, M.; Caballero-Pérez, J.; Vielle-Calzada, J.P. A family of microRNAs present in plants and animals. Plant Cell 2006, 18, 3355–3369. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Profiling the Abiotic Stress Responsive microRNA Landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef]
- Zhao, W.; Xiao, W.; Sun, J.; Chen, M.; Ma, M.; Cao, Y.; Cen, W.; Li, R.; Luo, J. An Integration of MicroRNA and Transcriptome Sequencing Analysis Reveal Regulatory Roles of miRNAs in Response to Chilling Stress in Wild Rice. Plants 2022, 11, 977. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Zhu, C.; Chang, X.; Yue, C.; Wang, Z.; Lin, Y.; Lai, Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017, 17, 211. [Google Scholar] [CrossRef]
- Çakır, O.; Arıkan, B.; Karpuz, B.; Turgut-Kara, N. Expression analysis of miRNAs and their targets related to salt stress in Solanum lycopersicum H-2274. Biotechnol. Biotechnol. Equip. 2021, 35, 275–282. [Google Scholar] [CrossRef]
- Cusaro, C.M.; Grazioli, C.; Capelli, E.; Picco, A.M.; Guarise, M.; Gozio, E.; Zarpellon, P.; Brusoni, M. Involvement of miRNAs in Metabolic Herbicide Resistance to Bispyribac-Sodium in Echinochloa crus-galli (L.) P. Beauv. Plants 2022, 11, 3359. [Google Scholar] [CrossRef]
- Ye, C.Y.; Wu, D.; Mao, L.; Jia, L.; Qiu, J.; Lao, S.; Chen, M.; Jiang, B.; Tang, W.; Peng, Q.; et al. The Genomes of the Allohexaploid Echinochloa crus-galli and Its Progenitors Provide Insights into Polyploidization-Driven Adaptation. Mol. Plant 2020, 13, 1298–1310. [Google Scholar] [CrossRef]
- Kaloumenos, N.S.; Chatzilazaridou, S.L.; Mylona, P.V.; Polidoros, A.N.; Eleftherohorinos, I.G. Target-site mutation associated with cross-resistance to ALS-inhibiting herbicides in late watergrass (Echinochloa oryzicola Vasing.). Pest Manag. Sci. 2013, 9, 865–873. [Google Scholar] [CrossRef] [PubMed]
- PPDB: Pesticide Properties DataBase. Available online: https://sitem.herts.ac.uk/aeru/ppdb/en/Reports/1028.htm (accessed on 8 May 2024).
- EU Pesticides Database. Available online: https://food.ec.europa.eu/plants/pesticides/eu-pesticides-database_en (accessed on 9 May 2024).
- Ministero Della Salute. Available online: https://www.salute.gov.it/portale/home.html (accessed on 4 July 2024).
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. Computational analysis of miRNA targets in plants: Current status and challenges. Brief. Bioinform. 2011, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Pan, L.; Wang, Z.; Cai, J.; Gao, H.; Zhao, H.; Dong, L. High-throughput sequencing reveals differential regulation of miRNAs in fenoxaprop-P-ethyl-resistant Beckmannia syzigachne. Sci. Rep. 2016, 6, 28725. [Google Scholar] [CrossRef][Green Version]
- Żywicki, M.; Gracz, J.; Karłowski, W.; Twardowski, T.; Tyczewska, A. Expression of miRNAs involved in phosphate homeostasis and senescence is altered in glyphosate-treated maize. Acta Physiol. Plant 2015, 37, 265. [Google Scholar] [CrossRef]
- Cummins, I.; Cole, D.J.; Edwards, R. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. Cell Mol. Biol. 1999, 18, 285–292. [Google Scholar] [CrossRef]
- Cummins, I.; O’Hagan, D.; Jablonkai, I.; Cole, D.J.; Hehn, A.; Werck-Reichhart, D.; Edwards, R. Cloning, characterization and regulation of a family of phi class glutathione transferases from wheat. Plant Mol. Biol. 2003, 52, 591–603. [Google Scholar] [CrossRef]
- Cummins, I.; Wortley, D.J.; Sabbadin, F.; He, Z.; Coxon, C.R.; Straker, H.E.; Sellars, J.D.; Knight, K.; Edwards, L.; Hughes, D.; et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA 2013, 110, 5812–5817. [Google Scholar]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.J.; Zhang, J.J.; Xue, H.W. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009, 37, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Wu, J.; Zhang, J.; Zhao, S.; Zheng, H.; Gao, G.; Wei, L.; Li, Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011, 7, e1002176. [Google Scholar] [CrossRef]
- Radwan, O.; Liu, Y.; Clough, S.J. Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol. Plant-Microbe Interact. MPMI 2011, 24, 958–972. [Google Scholar] [CrossRef]
- Wei, L.Q.; Yan, L.F.; Wang, T. Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 2011, 12, R53. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, S.; Kong, X.; Li, Y.; Zhao, G.; He, W.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X.; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef]
- Xue, W.; Wang, Z.; Du, M.; Liu, Y.; Liu, J.Y. Genome-wide analysis of small RNAs reveals eight fiber elongation-related and 257 novel microRNAs in elongating cotton fiber cells. BMC Genom. 2013, 14, 629. [Google Scholar] [CrossRef]
- EPPO. Efficacy evaluation of herbicides: Weeds in water-seeded rice. Bulletin 2011, 41, 282–285. [Google Scholar]
- EPPO Database on PP1 Standards. Available online: https://pp1.eppo.int/standards/PP1-062-3 (accessed on 16 April 2023).
- Délye, C.; Michel, S. ‘Universal’ primers for PCR-sequencing of grass chloroplastic acetyl-CoA carboxylase domains involved in resistance to herbicides. Weed Res. 2005, 45, 323–330. [Google Scholar] [CrossRef]
- Pan, L.; Guo, Q.; Wang, J.; Shi, L.; Yang, X.; Zhou, Y.; Yu, Q.; Bai, L. CYP81A68 confers metabolic resistance to ALS and ACCase-inhibiting herbicides and its epigenetic regulation in Echinochloa crusgalli. J. Hazard. Mater. 2022, 428, 128225. [Google Scholar] [CrossRef]
- Gene Bank Database. Available online: http://www.ncbi.nlm.nih.gov/genbank (accessed on 13 January 2023).
- Grain Genes—A Database for Triticeae and Avena. Available online: https://wheat.pw.usda.gov/GG3/ (accessed on 14 March 2023).
- Primer BLAST. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi (accessed on 14 March 2023).
- Suddal, T.; Awan, M.F.; Ali, S.; Sarwar, M.F.; Iqbal, S.; Ali, Q.; Javed, M.A.; Alshahrani, M.Y. Target prediction of potential candidate miRNAs from Oryza sativa to silence the Pyricularia oryzae genome in rice blast. Sci. Rep. 2024, 14, 21813. [Google Scholar]
- Smith, T.F.; Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981, 147, 195–197. [Google Scholar] [CrossRef] [PubMed]
- miRBase: The microRNA Database. Available online: https://www.mirbase.org/ (accessed on 18 October 2023).
- Kulcheski, F.R.; de Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, X.; Chen, W.; Bai, J.; Vasseur, L.; He, W.; You, M. Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS ONE 2019, 14, e0220475. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, J.; Zhu, L.; Hu, H.; Yang, J.; Cui, J.; Xu, J. Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses. Int. J. Mol. Sci. 2021, 22, 7246. [Google Scholar] [CrossRef]
- Wrzesińska, B.; Kościelniak, K.; Frąckowiak, P.; Praczyk, T.; Obrępalska-Stęplowska, A. The analysis of reference genes expression stability in susceptible and resistant Apera spicaventi populations under herbicide treatment. Sci. Rep. 2021, 11, 22145. [Google Scholar] [CrossRef]
- Zhou, Z.; Schenke, D.; Shen, E.; Fan, L.; Cai, D. MicroRNAs constitute an additional layer in plant response to simultaneous bio- and abiotic stresses as exemplified by UV-B radiation and flg22-treatment on Arabidopsis thaliana. Plant Cell Environ. 2024, 47, 765–781. [Google Scholar] [CrossRef]
- Kassambara, A. _Ggpubr: ‘ggplot2’ Based Publication Ready Plots_. R Package Version 0.6.0. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 16 October 2023).
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 9 September 2022).
- Nakazawa, M. _fmsb: Functions for Medical Statistics Book with Some Demographic Data_. R Package Version 0.7.5. Available online: https://CRAN.R-project.org/package=fmsb (accessed on 15 September 2023).
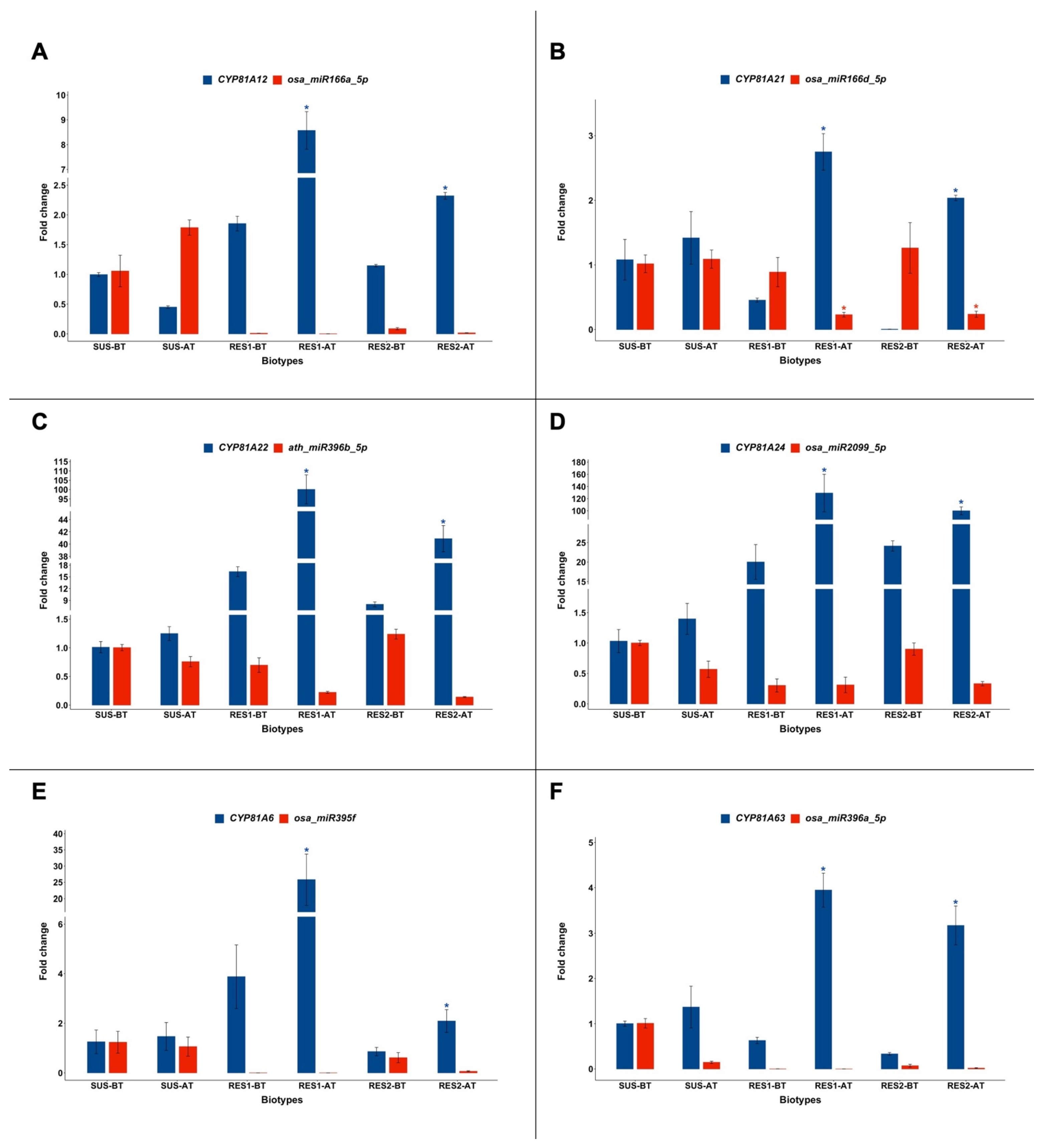
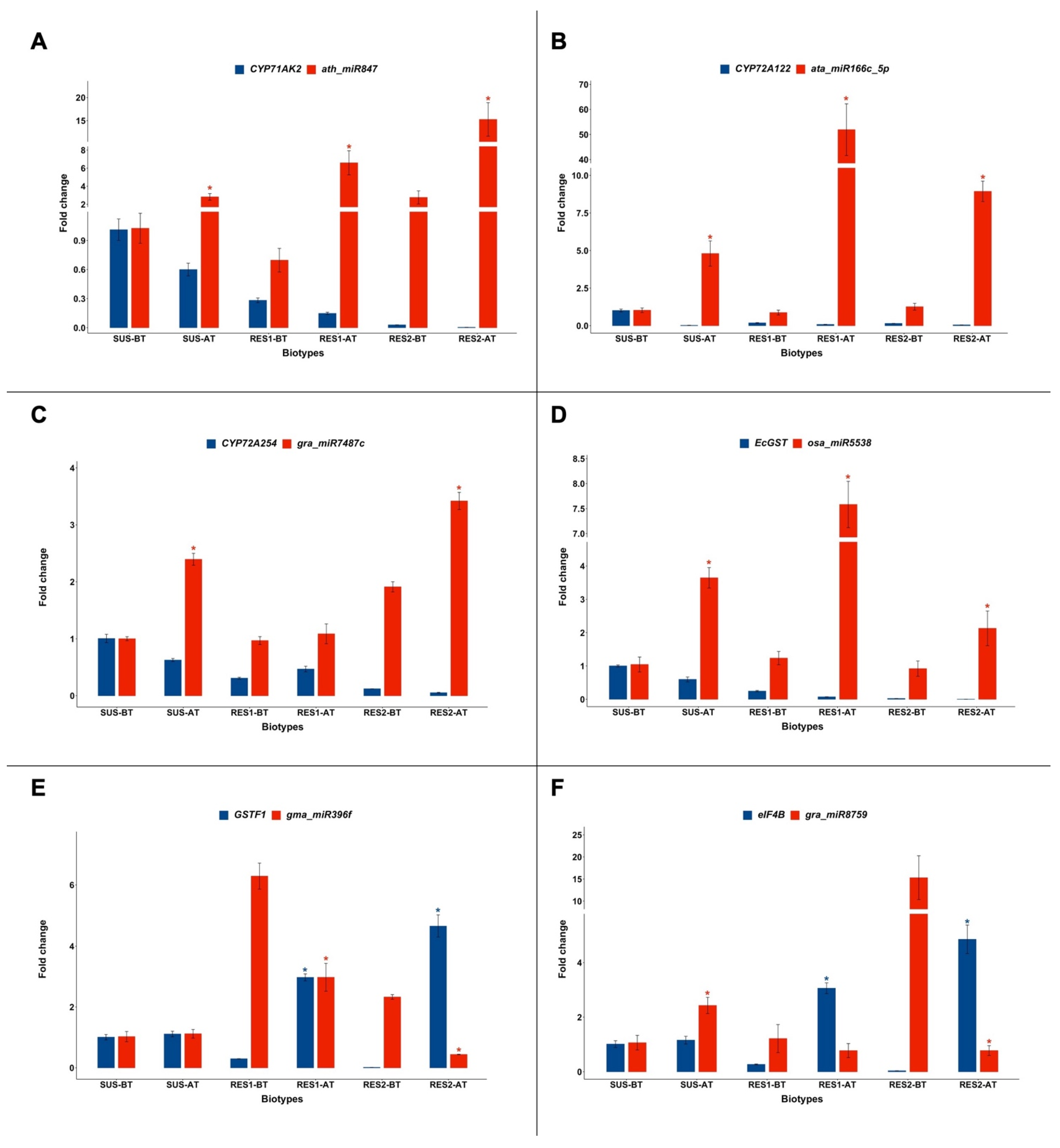
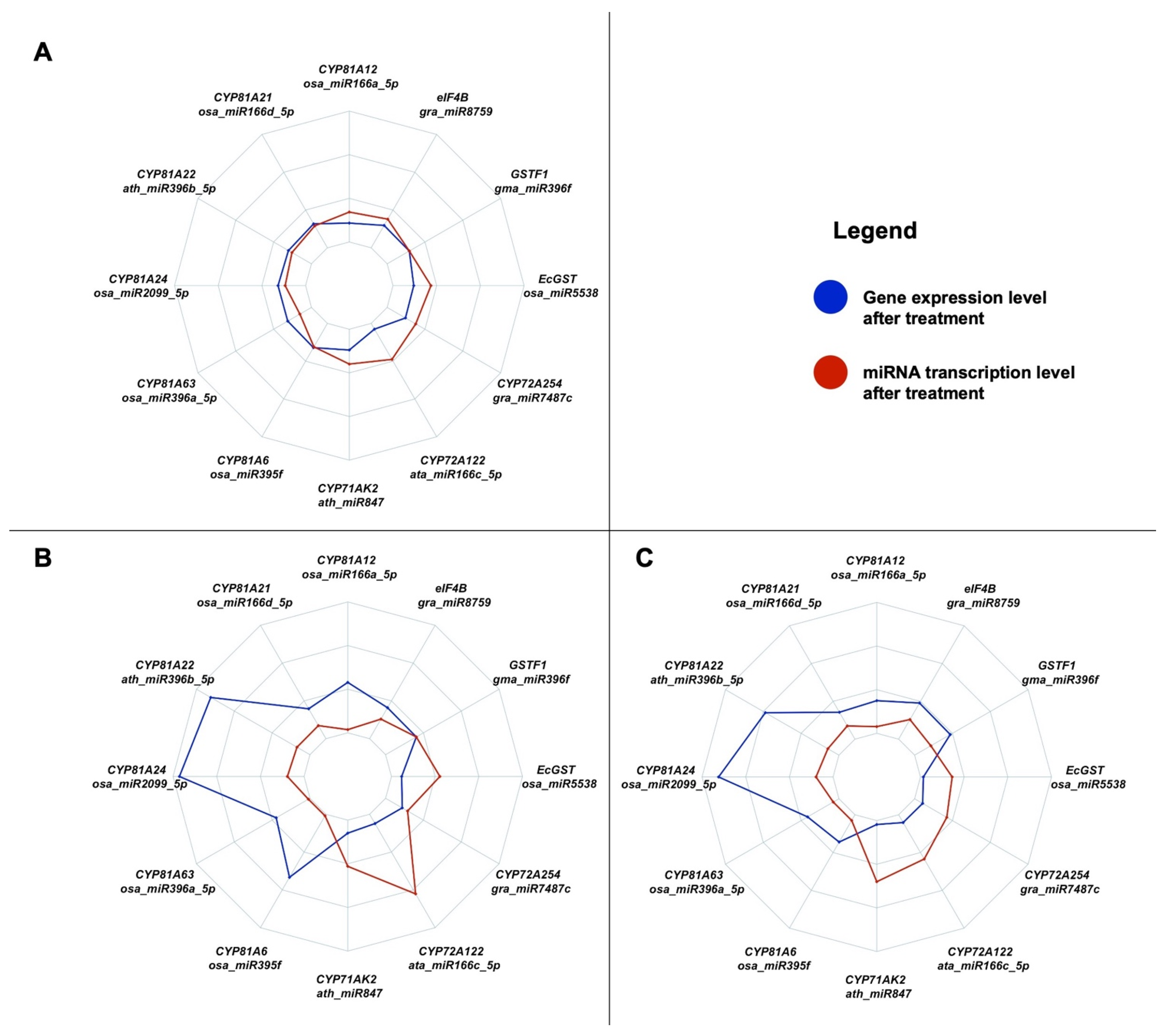
| miRNA | Target Gene |
|---|---|
| osa-miR166a-5p | CYP81A12 |
| osa-miR166d-5p | CYP81A21 |
| ath-miR396b | CYP81A22 |
| osa-miR2099-5p | CYP81A24 |
| osa-miR396a-5p | CYP81A63 |
| ath-miR847 | CYP81A6 |
| gma-miR396f | CYP71AK2 |
| ata-miR166c-5p | CYP72A122 |
| gra-miR7486c | CYP72A254 |
| gra-miR8759 | GSTF1 |
| osa-miR395f | EcGST |
| osa-miR5538 | eIF4B1 |
| Gene ID | Primer Sequence (5′-3′) | Reference |
|---|---|---|
| CYP81A12 | F: tgagctcttccatcgtcgtg R: tactttttggcgactccgct | Iwakami et al., 2014 [18] |
| CYP81A21 | F: tagcatcatccacgagacgc R: tacacgttcaccagcagcat | Iwakami et al., 2014 [18] |
| CYP81A22 | F: cggcgcgctggtccagtt R: tgacatgagcagttccatcg | Iwakami et al., 2014 [18] |
| CYP81A24 | F: gaggtctacaccgatgccac R: cttcttgagcttctccgggt | Iwakami et al., 2014 [18] |
| CYP81A63 | F: gagaccatcgctcagaccaa R: atcttgttcctcacgccgaa | Dimaano et al., 2020 [13] Iwakami et al., 2019 [17] |
| CYP81A6 | F: gactattcaacccgggcgat R: caagttctgcacggcaagag | Pan et al., 2022 [61] |
| CYP71AK2 | F: acgtgtgggacaagttcctg R: ggctttgatgcgatcgtctg | Iwakami et al., 2014 [21] |
| CYP72A122 | F: agttcaagccggagaggttc R: catcttggcttcaagcagcg | Iwakami et al., 2014 [21] |
| CYP72A254 | F: ttacgaggtactccggctgt R: gtcagggtcgtggtgaatgt | Iwakami et al., 2014 [21] |
| EcGST | F: gccgaggaggacctgaagaac R: gtgactcacagataggcttaccgt | Li et al., 2013 [16] |
| GSTF1 | F: tgcctcttcaaccccatgat R: aggtactcgtgctgggagag | Dalazen et al., 2018 [23] |
| eIF4B1 | F: cgagcagcttacaagggact R: gtggttccataccaccacga | Dalazen et al., 2018 [23] |
| Name | a.n. | miRNA Sequence (5′-3′) | Reference |
|---|---|---|---|
| ata-miR166c-5p | MIMAT0037248 | ggaacguuggcuggcucgagg | Jia et al., 2013 [56] |
| ath-miR396b | MIMAT0000945 | uuccacagcuuucuugaacuu | John-Rohades et al., 2004 [50] |
| ath-miR847 | MIMAT0004278 | ucacuccucuucuucuugaug | Rajagopalan et al., 2006 [51] |
| gma-miR396f | MIMAT0021069 | agcuuucuugaacuucuuaugccua | Radwan et al., 2011 [54] |
| gra-miR7486c | MIMAT0034235 | uuuguccacgugaacagaaaacgc | Xue et al., 2013 [57] |
| gra-miR8759 | MIMAT0034189 | ugguggaaguauugugcccgg | Xue et al., 2013 [57] |
| osa-miR395f | MIMAT0000974 | gugaauuguuugggggaacuc | John-Rohades et al., 2004 [50] |
| osa-miR5538 | MIMAT0022174 | acugaacucaaucacuugcugc | Wei et al., 2011 [55] |
| osa-miR166a-5p | MIMAT0022855 | ggaauguugucugguucaagg | Du et al., 2011 [53] |
| osa-miR166d-5p | MIMAT0022858 | ggaauguugucuggcucgagg | Du et al., 2011 [53] |
| osa-miR2099-5p | MIMAT0010062 | ugaauauguuuguacaagcuuu | Xue et al., 2009 [52] |
| osa-miR396a-5p | MIMAT0000977 | uuccacagcuuucuugaacug | Du et al., 2011 [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cusaro, C.M.; Capelli, E.; Picco, A.M.; Guarise, M.; Gozio, E.; Zarpellon, P.; Brusoni, M. MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger. Plants 2025, 14, 719. https://doi.org/10.3390/plants14050719
Cusaro CM, Capelli E, Picco AM, Guarise M, Gozio E, Zarpellon P, Brusoni M. MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger. Plants. 2025; 14(5):719. https://doi.org/10.3390/plants14050719
Chicago/Turabian StyleCusaro, Carlo Maria, Enrica Capelli, Anna Maria Picco, Marta Guarise, Enrico Gozio, Pietro Zarpellon, and Maura Brusoni. 2025. "MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger" Plants 14, no. 5: 719. https://doi.org/10.3390/plants14050719
APA StyleCusaro, C. M., Capelli, E., Picco, A. M., Guarise, M., Gozio, E., Zarpellon, P., & Brusoni, M. (2025). MicroRNA-Mediated Post-Transcriptional Regulation of Enzymes Involved in Herbicide Resistance in Echinochloa oryzicola (Vasinger) Vasinger. Plants, 14(5), 719. https://doi.org/10.3390/plants14050719







