Transcriptome and Metabolome Analyses Reveal the Mechanism of Color Differences in Pomegranate (Punica granatum L.) Red and White Petals
Abstract
1. Introduction
2. Results
2.1. Pomegranate Petal Color Phenotype
2.2. Physiological Indicators of Pomegranate Petal
2.3. RNA-Seq Data Overview
2.4. DEG Identification and KEGG Enrichment Analyses
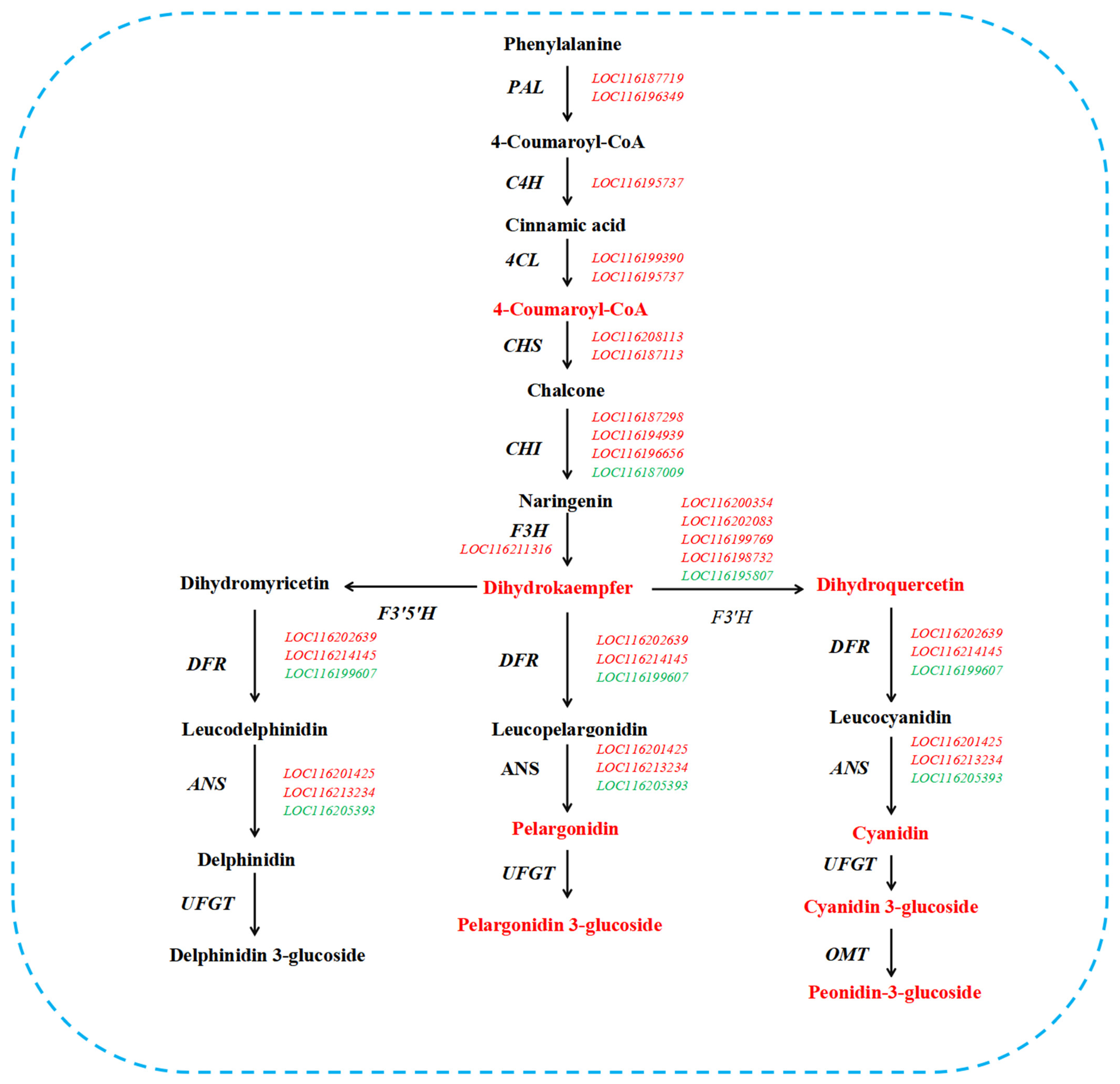
2.5. Analysis of Key Genes Associated with Pigment
2.6. Screening Pigment Metabolism-Related TFs
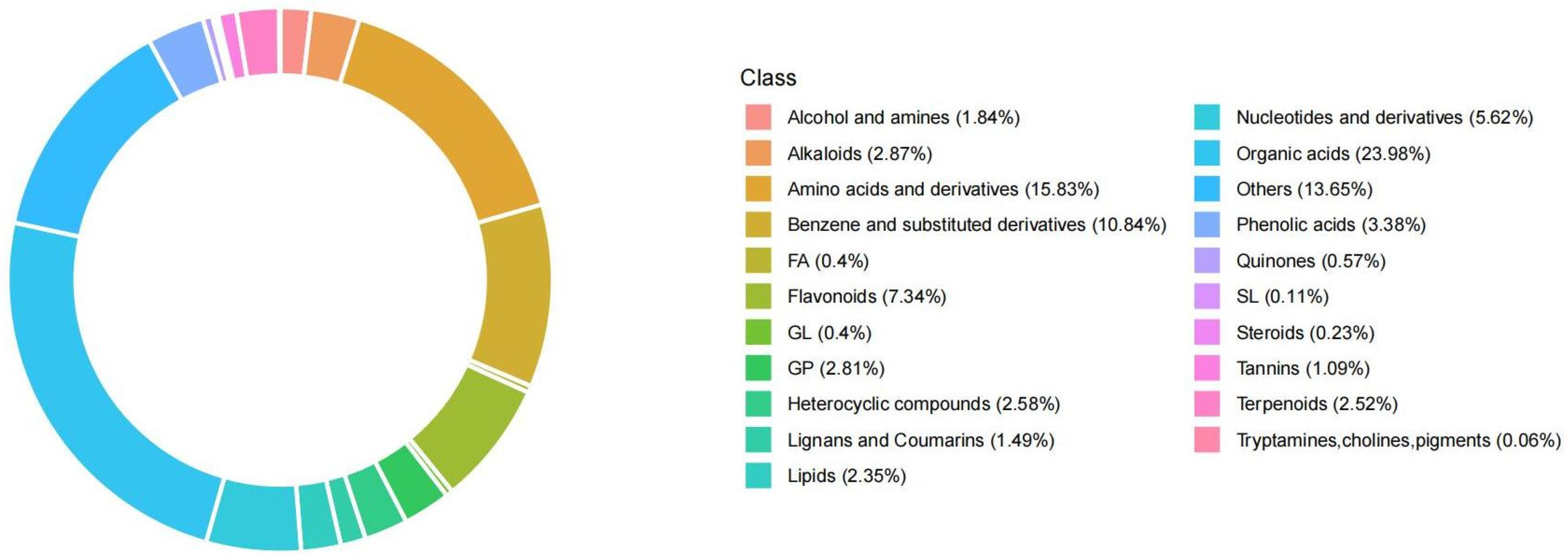
2.7. Metabolome Data Overview
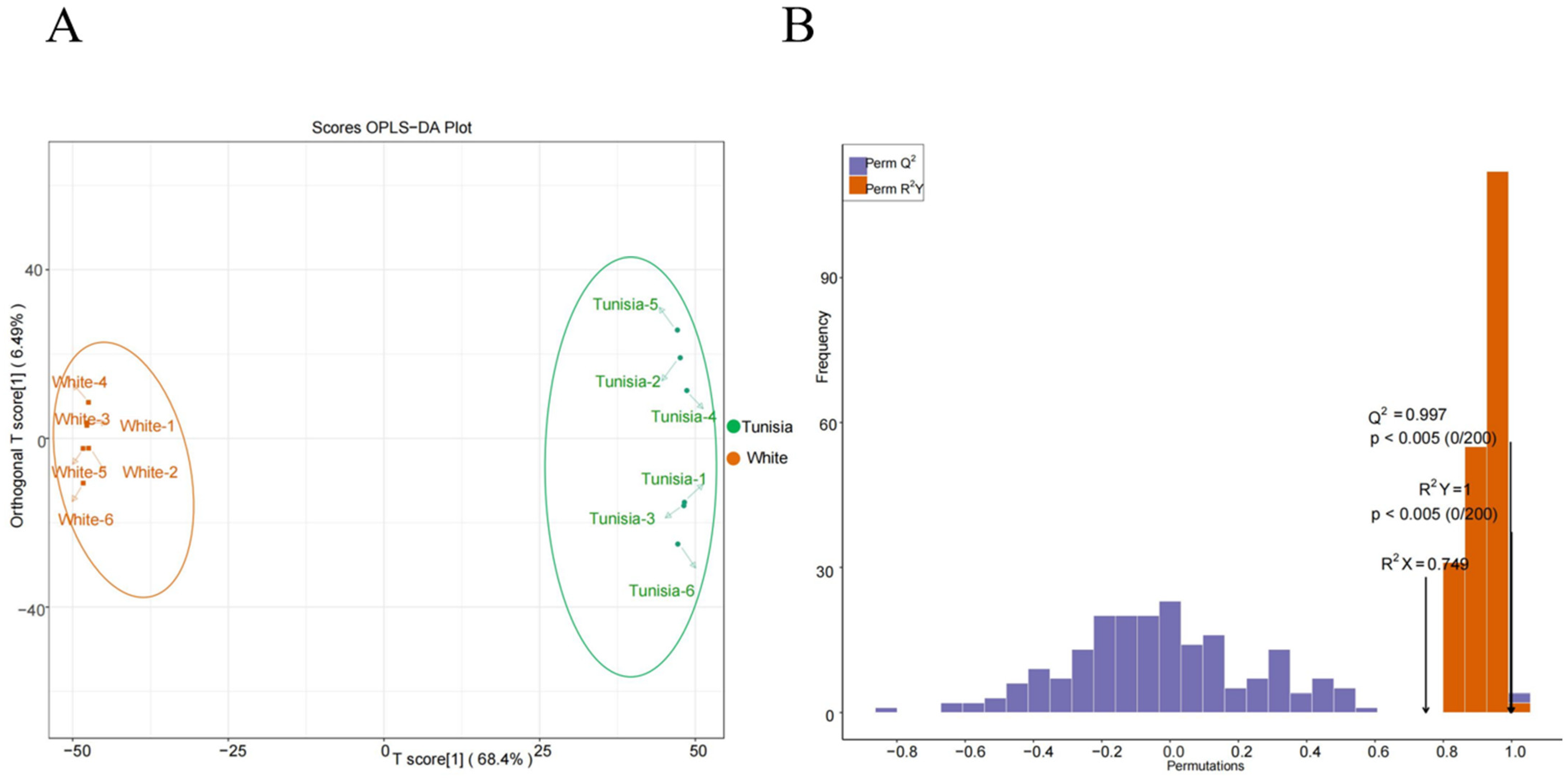
2.8. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
2.9. DEM Identification and KEGG Enrichment Analyses
2.10. Analysis of Key Metabolites Associated with Pigment
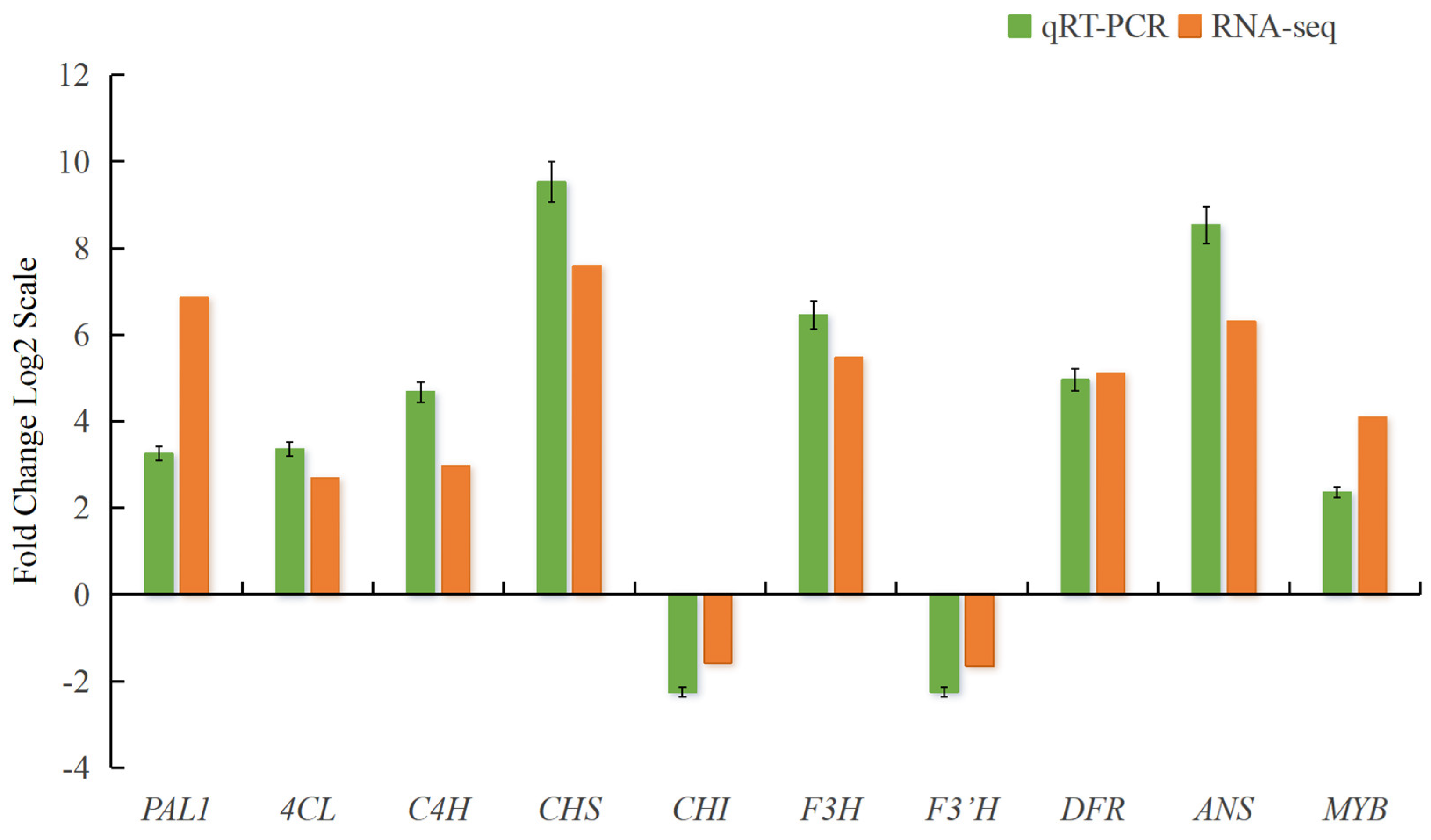
2.11. Validation of DEGs with qRT-PCR
3. Discussion
3.1. Pomegranate Petal Phenotype and Physiological Indicators
3.2. Key Genes Related to Pomegranate Petal Pigment
3.3. Key TFs Related to Pomegranate Petal Pigment
3.4. Effect of Caffeic Acid on Anthocyanin Content
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Physiological Indicators
4.2.1. Measurement of Petal Color
4.2.2. Measurement of Anthocyanin, Flavonoid, and Carotenoid Content
4.2.3. Measurement of Sucrose Content
4.3. Transcriptome Analysis
4.3.1. Library Construction and Sequencing
4.3.2. Bioinformatics Analysis
4.3.3. qRT-PCR Analysis
4.4. Metabolome Analysis
4.4.1. Dry Sample Extraction
4.4.2. HPLC Conditions
4.4.3. MS Conditions
4.4.4. Metabolomics Data Processing
4.4.5. Selected Differential Metabolites
4.4.6. KEGG Enrichment Analysis of Differential Metabolites
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Singh, B.; Singh, J.P.; Kaur, A.; Singh, N. Phenolic compounds as beneficial phytochemicals in pomegranate (Punica granatum L.) peel: A review. Food Chem. 2018, 261, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Mphahlele, R.R.; Fawole, O.A.; Makunga, N.P.; Opara, U.L. Effect of drying on the bioactive compounds, antioxidant, antibacterial and antityrosinase activities of pomegranate peel. BMC Complement. Altern. Med. 2016, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.D. Floral biology of pomegranate (Punica granatum L.). Pomegranate 2010, 4, 45–50. [Google Scholar]
- Wang, Y.; Chen, L.; Yang, Q.; Hu, Z.; Guo, P.; Xie, Q.; Chen, G. New insight into the pigment composition and molecular mechanism of flower coloration in tulip (Tulipa gesneriana L.) cultivars with various petal colors. Plant Sci. 2022, 317, 111193. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Owens, S.J.; Rørslett, B. Plants and colour: Flowers and pollination. Opt. Laser Technol. 2011, 43, 282–294. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Enaru, B.; Drețcanu, G.; Pop, T.D.; Stǎnilǎ, A.; Diaconeasa, Z. Anthocyanins: Factors affecting their stability and degradation. Antioxidants 2021, 10, 1967. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.-L.; Liang, N.-N.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Busche, M.; Pucker, B.; Weisshaar, B.; Stracke, R. Three R2R3-MYB transcription factors from banana (Musa acuminata) activate structural anthocyanin biosynthesis genes as part of an MBW complex. BMC Res. Notes 2023, 16, 103. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, H.; Wang, Q.; Khalil-Ur-Rehman, M.; Meng, L.; Tao, J. Comparison among ‘Benitaka’ grape, ABA-treated ‘Benitaka’, and its deeper-colored bud mutation revealed the coloring mechanisms on grapes. J. Plant Interact. 2019, 14, 102–109. [Google Scholar] [CrossRef]
- Bhatt, T.; Patel, K. Carotenoids: Potent to prevent diseases review. Nat. Prod. Bioprospecting 2020, 10, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Diversity and origin of carotenoid biosynthesis: Its history of coevolution towards plant photosynthesis. New Phytol. 2021, 232, 479–493. [Google Scholar] [CrossRef]
- Sun, T.; Li, L. Toward the ‘golden’ era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2020, 290, 110331. [Google Scholar] [CrossRef]
- Ma, X.; Yu, Y.-N.; Jia, J.-H.; Li, Q.-H.; Gong, Z.-H. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance. Sci. Hortic. 2022, 295, 110892. [Google Scholar] [CrossRef]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-wide identification of the Capsicum bHLH transcription factor family: Discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef]
- Liang, M.-H.; Li, X.-Y. Involvement of transcription factors and regulatory proteins in the regulation of carotenoid accumulation in plants and Algae. J. Agric. Food Chem. 2023, 71, 18660–18673. [Google Scholar] [CrossRef]
- Bar-Ya’akov, I.; Tian, L.; Amir, R.; Holland, D. Primary metabolites, anthocyanins, and hydrolyzable tannins in the pomegranate fruit. Front. Plant Sci. 2019, 10, 620. [Google Scholar] [CrossRef]
- Luo, X.; Cao, D.; Li, H.; Zhao, D.; Xue, H.; Niu, J.; Chen, L.; Zhang, F.; Cao, S. Complementary iTRAQ-based proteomic and RNA sequencing-based transcriptomic analyses reveal a complex network regulating pomegranate (Punica granatum L.) fruit peel colour. Sci. Rep. 2018, 8, 12362. [Google Scholar] [CrossRef]
- Wang, X.; Yang, C.; Zhu, W.; Weng, Z.; Li, F.; Teng, Y.; Zhou, K.; Qian, M.; Deng, Q. Transcriptomic Analysis Reveals the Mechanism of Color Formation in the Peel of an Evergreen Pomegranate Cultivar ‘Danruo No. 1’ During Fruit Development. Plants 2024, 13, 2903. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yuan, Z.; Fang, Y.; Yin, Y.; Feng, L. Characterization and evaluation of major anthocyanins in pomegranate (Punica granatum L.) peel of different cultivars and their development phases. Eur. Food Res. Technol. 2013, 236, 109–117. [Google Scholar] [CrossRef]
- Li, B.; Lin, J.; Zheng, Z.; Duan, H.; Li, D.; Wu, M. Effects of different drying methods on drying kinetics and physicochemical properties of Chrysanthemum morifolium Ramat. Int. J. Agric. Biol. Eng. 2019, 12, 187–193. [Google Scholar] [CrossRef]
- Önder, S.; Tonguç, M.; Önder, D.; Erbaş, S.; Mutlucan, M. Flower color and carbohydrate metabolism changes during the floral development of Rosa damascena. South Afr. J. Bot. 2023, 156, 234–243. [Google Scholar] [CrossRef]
- Bento, J.A.C.; Ferreira, K.C.; de Oliveira, A.L.M.; Lião, L.M.; Caliari, M.; Júnior, M.S.S. Extraction, characterization and technological properties of white garland-lily starch. Int. J. Biol. Macromol. 2019, 135, 422–428. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Dong, B.; Fu, J.; Hu, S.; Zhao, H. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans. Front. Plant Sci. 2018, 9, 1499. [Google Scholar] [CrossRef]
- Zhang, D.; Xie, A.; Yang, X.; Yang, L.; Shi, Y.; Dong, L.; Lei, F.; Sun, L.; Bao, M.; Sun, X. Analysis of physiological and biochemical factors affecting flower color of herbaceous peony in different flowering periods. Horticulturae 2023, 9, 502. [Google Scholar] [CrossRef]
- Yamagishi, M.; Ihara, H.; Arakawa, K.; Toda, S.; Suzuki, K. The origin of the LhMYB12 gene, which regulates anthocyanin pigmentation of tepals, in Oriental and Asiatic hybrid lilies (Lilium spp.). Sci. Hortic. 2014, 174, 119–125. [Google Scholar] [CrossRef]
- Télef, N.; Stammitti-Bert, L.; Mortain-Bertrand, A.; Maucourt, M.; Carde, J.P.; Rolin, D.; Gallusci, P. Sucrose deficiency delays lycopene accumulation in tomato fruit pericarp discs. Plant Mol. Biol. 2006, 62, 453–469. [Google Scholar] [CrossRef]
- Xia, H.; Zhu, L.; Zhao, C.; Li, K.; Shang, C.; Hou, L.; Wang, M.; Shi, J.; Fan, S.; Wang, X. Comparative transcriptome analysis of anthocyanin synthesis in black and pink peanut. Plant Signal. Behav. 2020, 15, 1721044. [Google Scholar] [CrossRef]
- Ju, Z.-G.; Yuan, Y.-B.; Liou, C.-L.; Xin, S.-H. Relationships among phenylalanine ammonia-Iyase activity, simple phenol concentrations and anthocyanin accumulation in apple. Sci. Hortic. 1995, 61, 215–226. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Gu, M.; Hou, B.; Zhang, C.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. Identification of PAL genes related to anthocyanin synthesis in tea plants and its correlation with anthocyanin content. Hortic. Plant J. 2022, 8, 381–394. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, C.; Yao, Y.; Mao, Z.; Sun, G.; Dai, Z. Red anthocyanins contents and the relationships with phenylalanine ammonia lyase (PAL) activity, soluble sugar and chlorophyll contents in carmine radish (Raphanus sativus L.). Hortic. Sci. 2019, 46, 17–25. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, Y.; Li, X.; Liu, Z.; Li, F.; Huang, H.; Huang, M. Analysis of Transcriptome and Expression of C4H and FLS Genes on Four Flower Colors of Impatiens uliginosa. Horticulturae 2024, 10, 415. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, H.; Fan, Y.; Yan, L. Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum). BMC Plant Biol. 2022, 22, 358. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liu, Z.; Wang, J.; Yang, B.; Chen, W.; Ou, L.; Dai, X.; Zhang, Z.; Zou, X. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 2020, 306, 125629. [Google Scholar] [CrossRef]
- Van Der Krol, A.R.; Mur, L.A.; de Lange, P.; Mol, J.N.; Stuitje, A.R. Inhibition of flower pigmentation by antisense CHS genes: Promoter and minimal sequence requirements for the antisense effect. Plant Mol. Biol. 1990, 14, 457–466. [Google Scholar] [CrossRef]
- Tai, D.; Tian, J.; Zhang, J.; Song, T.; Yao, Y. A Malus crabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS ONE 2014, 9, e110570. [Google Scholar] [CrossRef]
- Wang, L.; Albert, N.W.; Zhang, H.; Arathoon, S.; Boase, M.R.; Ngo, H.; Schwinn, K.E.; Davies, K.M.; Lewis, D.H. Temporal and spatial regulation of anthocyanin biosynthesis provide diverse flower colour intensities and patterning in Cymbidium orchid. Planta 2014, 240, 983–1002. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T.; Yamamura, S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005, 579, 6074–6078. [Google Scholar] [CrossRef]
- Yu, S.; Li, J.; Peng, T.; Ni, S.; Feng, Y.; Wang, Q.; Wang, M.; Chu, X.; Fan, Z.; Li, X. Identification of chalcone isomerase family genes and roles of cnchi4 in flavonoid metabolism in Camellia nitidissima. Biomolecules 2022, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Prescott, A.; Mackay, S.; Bartlett, J.; Vrijlandt, E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991, 1, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Zuker, A.; Tzfira, T.; Ben-Meir, H.; Ovadis, M.; Shklarman, E.; Itzhaki, H.; Forkmann, G.; Martens, S.; Neta-Sharir, I.; Weiss, D. Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene. Mol. Breed. 2002, 9, 33–41. [Google Scholar] [CrossRef]
- Nitarska, D.; Boehm, R.; Debener, T.; Lucaciu, R.C.; Halbwirth, H. First genome edited poinsettias: Targeted mutagenesis of flavonoid 3′-hydroxylase using CRISPR/Cas9 results in a colour shift. Plant Cell Tissue Organ 2021, 147, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef]
- Li, H.; Qiu, J.; Chen, F.; Lv, X.; Fu, C.; Zhao, D.; Hua, X.; Zhao, Q. Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene in Saussurea medusa. Mol. Biol. Rep. 2012, 39, 2991–2999. [Google Scholar] [CrossRef]
- Polashock, J.J.; Griesbach, R.J.; Sullivan, R.F.; Vorsa, N. Cloning of a cDNA encoding the cranberry dihydroflavonol-4-reductase (DFR) and expression in transgenic tobacco. Plant Sci. 2002, 163, 241–251. [Google Scholar] [CrossRef]
- Kim, S.; Yoo, K.S.; Pike, L.M. Development of a PCR-based marker utilizing a deletion mutation in the dihydroflavonol 4-reductase (DFR) gene responsible for the lack of anthocyanin production in yellow onions (Allium cepa). Theor. Appl. Genet. 2005, 110, 588–595. [Google Scholar] [CrossRef]
- Zhou, X.-W.; Fan, Z.-Q.; Chen, Y.; Zhu, Y.-L.; Li, J.-Y.; Yin, H.-F. Functional analyses of a flavonol synthase–like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation. J. Biosci. 2013, 38, 593–604. [Google Scholar] [CrossRef]
- Nakamura, N.; Fukuchi-Mizutani, M.; Miyazaki, K.; Suzuki, K.; Tanaka, Y. RNAi suppression of the anthocyanidin synthase gene in Torenia hybrida yields white flowers with higher frequency and better stability than antisense and sense suppression. Plant Biotechnol. 2006, 23, 13–17. [Google Scholar] [CrossRef]
- Junka, N.; Kanlayanarat, S.; Buanong, M.; Wongchaochant, S.; Wongs-Aree, C. Analysis of Anthocyanins and the Expression Patterns of Genes Involved in Biosynthesis in two Vanda Hybrids. Int. J. Agric. Biol. 2011, 13, 873–880. [Google Scholar]
- Li, J.; Lü, R.-H.; Zhao, A.-C.; Wang, X.-L.; Liu, C.-Y.; Zhang, Q.-Y.; Wang, X.-H.; Umuhoza, D.; Jin, X.-Y.; Lu, C. Isolation and expression analysis of anthocyanin biosynthetic genes in Morus alba L. Biol. Plant. 2014, 58, 618–626. [Google Scholar] [CrossRef]
- Liu, Y.C.; Yeh, C.W.; Chung, J.D.; Tsai, C.Y.; Chiou, C.Y.; Yeh, K.W. Petal-specific RNAi-mediated silencing of the phytoene synthase gene reduces xanthophyll levels to generate new Oncidium orchid varieties with white-colour blooms. Plant Biotechnol. J. 2019, 17, 2035. [Google Scholar] [CrossRef]
- Ronen, G.; Cohen, M.; Zamir, D.; Hirschberg, J. Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 1999, 17, 341–351. [Google Scholar] [CrossRef]
- Gady, A.L.; Vriezen, W.H.; Van de Wal, M.H.; Huang, P.; Bovy, A.G.; Visser, R.G.; Bachem, C.W. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breed. 2012, 29, 801–812. [Google Scholar] [CrossRef]
- Fantini, E.; Falcone, G.; Frusciante, S.; Giliberto, L.; Giuliano, G. Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 2013, 163, 986–998. [Google Scholar] [CrossRef]
- Pogson, B.; McDonald, K.A.; Truong, M.; Britton, G.; DellaPenna, D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 1996, 8, 1627–1639. [Google Scholar]
- Zhao, A.; Cui, Z.; Li, T.; Pei, H.; Sheng, Y.; Li, X.; Zhao, Y.; Zhou, Y.; Huang, W.; Song, X. mRNA and miRNA expression analysis reveal the regulation for flower spot patterning in Phalaenopsis ‘Panda’. Int. J. Mol. Sci. 2019, 20, 4250. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Fu, Z.; Wang, L.; Shang, H.; Dong, X.; Jiang, H.; Zhang, J.; Wang, H.; Li, Y.; Yuan, X.; Meng, S. An R3-MYB gene of Phalaenopsis, MYBx1, represses anthocyanin accumulation. Plant Growth Regul. 2019, 88, 129–138. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, G.; Liu, L.; Zhang, Q.; Han, Z.; Chen, X.; Li, B. A R2R3-MYB transcription factor, VvMYBC2L2, functions as a transcriptional repressor of anthocyanin biosynthesis in grapevine (Vitis vinifera L.). Molecules 2018, 24, 92. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Suzuki, T.; Harada, K.; Kobayashi, Y.; Dohra, H.; Ohno, H. Floral organ-and temperature-dependent regulation of anthocyanin biosynthesis in Cymbidium hybrid flowers. Plant Sci. 2019, 287, 110173. [Google Scholar] [CrossRef]
- Albert, N.W.; Arathoon, S.; Collette, V.E.; Schwinn, K.E.; Jameson, P.E.; Lewis, D.H.; Zhang, H.; Davies, K.M. Activation of anthocyanin synthesis in Cymbidium orchids: Variability between known regulators. Plant Cell Tissue Organ 2010, 100, 355–360. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes 2019, 10, 496. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Meng, X.; Li, G.; Gu, L.; Sun, Y.; Li, Z.; Liu, J.; Wu, X.; Dong, T.; Zhu, M. Comparative metabolomic and transcriptome analysis reveal distinct flavonoid biosynthesis regulation between petals of white and purple Phalaenopsis amabilis. J. Plant Growth Regul. 2020, 39, 823–840. [Google Scholar] [CrossRef]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-N.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Darias-Martín, J.; Martín-Luis, B.; Carrillo-López, M.; Lamuela-Raventós, R.; Díaz-Romero, C.; Boulton, R. Effect of caffeic acid on the color of red wine. J. Agr. Food Chem. 2002, 50, 2062–2067. [Google Scholar] [CrossRef]
- Qian, B.-J.; Liu, J.-H.; Zhao, S.-J.; Cai, J.-X.; Jing, P. The effects of gallic/ferulic/caffeic acids on colour intensification and anthocyanin stability. Food Chem. 2017, 228, 526–532. [Google Scholar] [CrossRef]
- Liu, J.; Zhuang, Y.; Hu, Y.; Xue, S.; Li, H.; Chen, L.; Fei, P. Improving the color stability and antioxidation activity of blueberry anthocyanins by enzymatic acylation with p-coumaric acid and caffeic acid. LWT Food Sci. Technol. 2020, 130, 109673. [Google Scholar] [CrossRef]
- Berner, M.; Krug, D.; Bihlmaier, C.; Vente, A.; Müller, R.; Bechthold, A. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J. Bacteriol. 2006, 188, 2666–2673. [Google Scholar] [CrossRef]
- Zhou, Y.; Yin, M.; Abbas, F.; Sun, Y.; Gao, T.; Yan, F.; Li, X.; Yu, Y.; Yue, Y.; Yu, R. Classification and association analysis of Gerbera (Gerbera hybrida) flower color traits. Front. Plant Sci. 2022, 12, 779288. [Google Scholar] [CrossRef]
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Liu, X.-J.; Liu, D.-D.; Xie, X.-B.; Cheng, C.-G.; Cong, P.-H.; Hao, Y.-J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef]
- Zang, H.; Guo, S.; Dong, S.; Song, Y.; Li, K.; Fan, X.; Qiu, J.; Zheng, Y.; Jiang, H.; Wu, Y. Construction of a Full-Length Transcriptome of Western Honeybee Midgut Tissue and Improved Genome Annotation. Genes 2024, 15, 728. [Google Scholar] [CrossRef]
- Wang, A.; Kang, L.; Yang, G.; Li, Z. Transcriptomic and iTRAQ-Based Quantitative Proteomic Analyses of inap CMS in Brassica napus L. Plants 2022, 11, 2460. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Ono, N.N.; Bandaranayake, P.C.; Tian, L. Establishment of pomegranate (Punica granatum) hairy root cultures for genetic interrogation of the hydrolyzable tannin biosynthetic pathway. Planta 2012, 236, 931–941. [Google Scholar] [CrossRef]
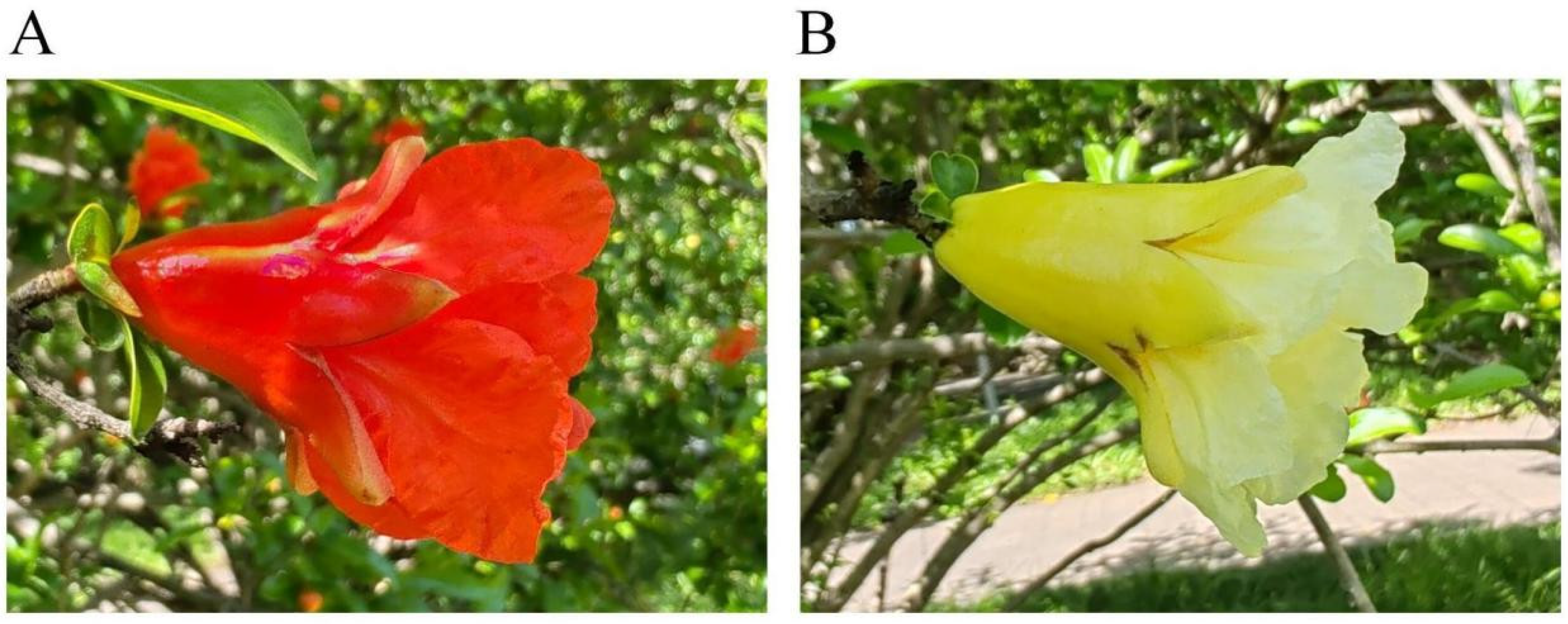
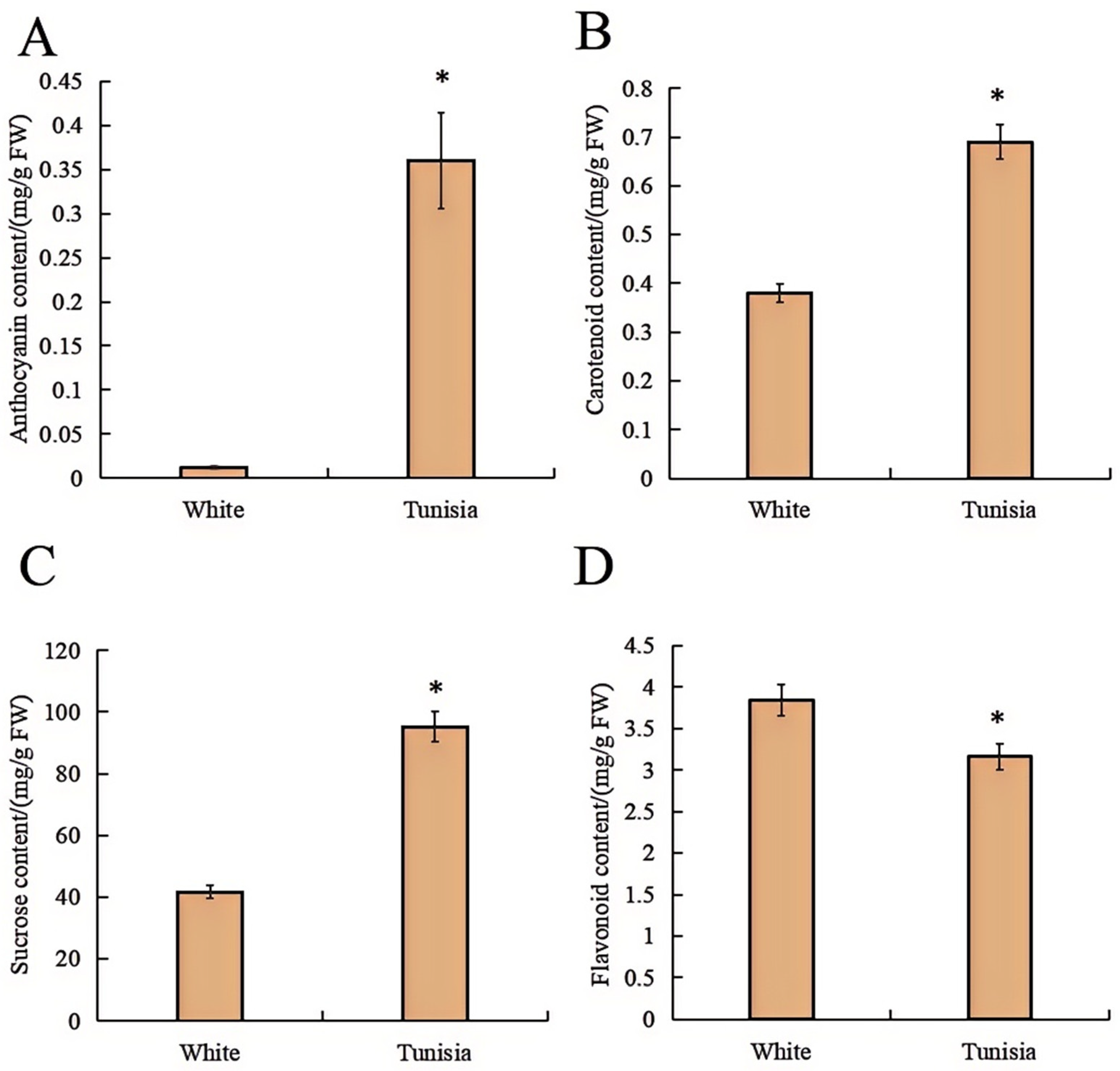
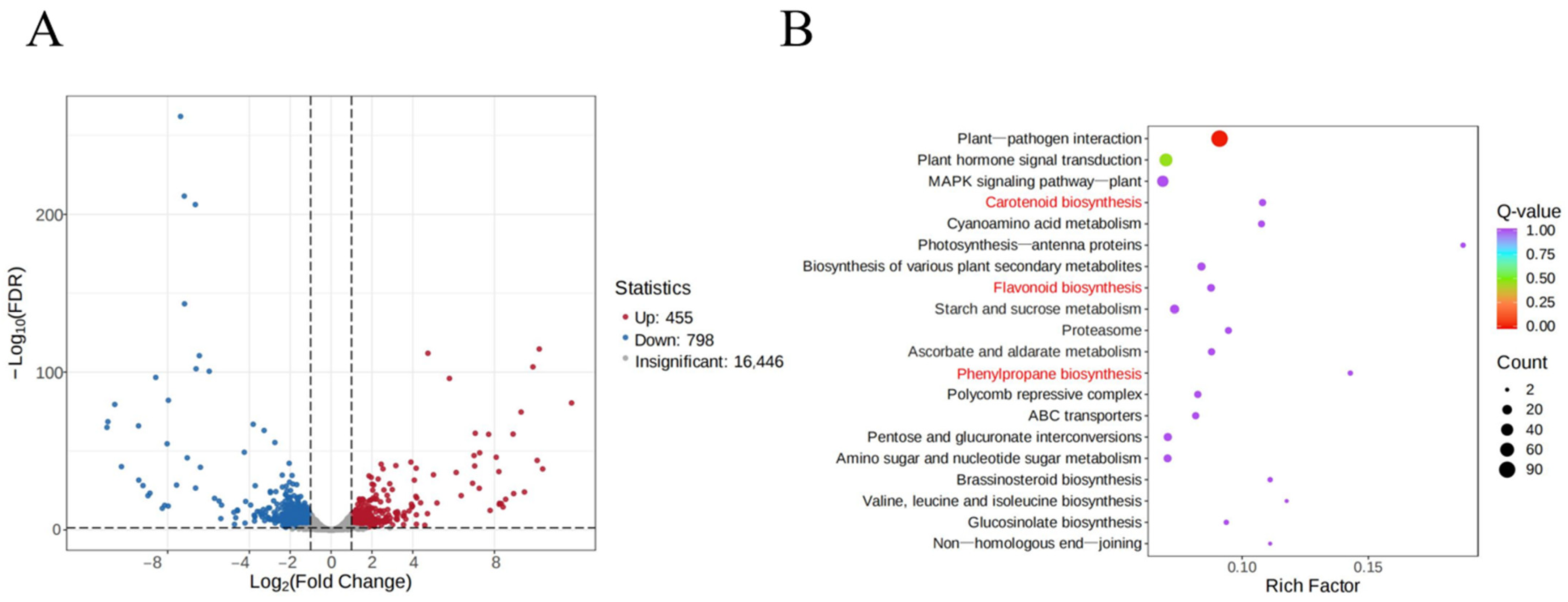
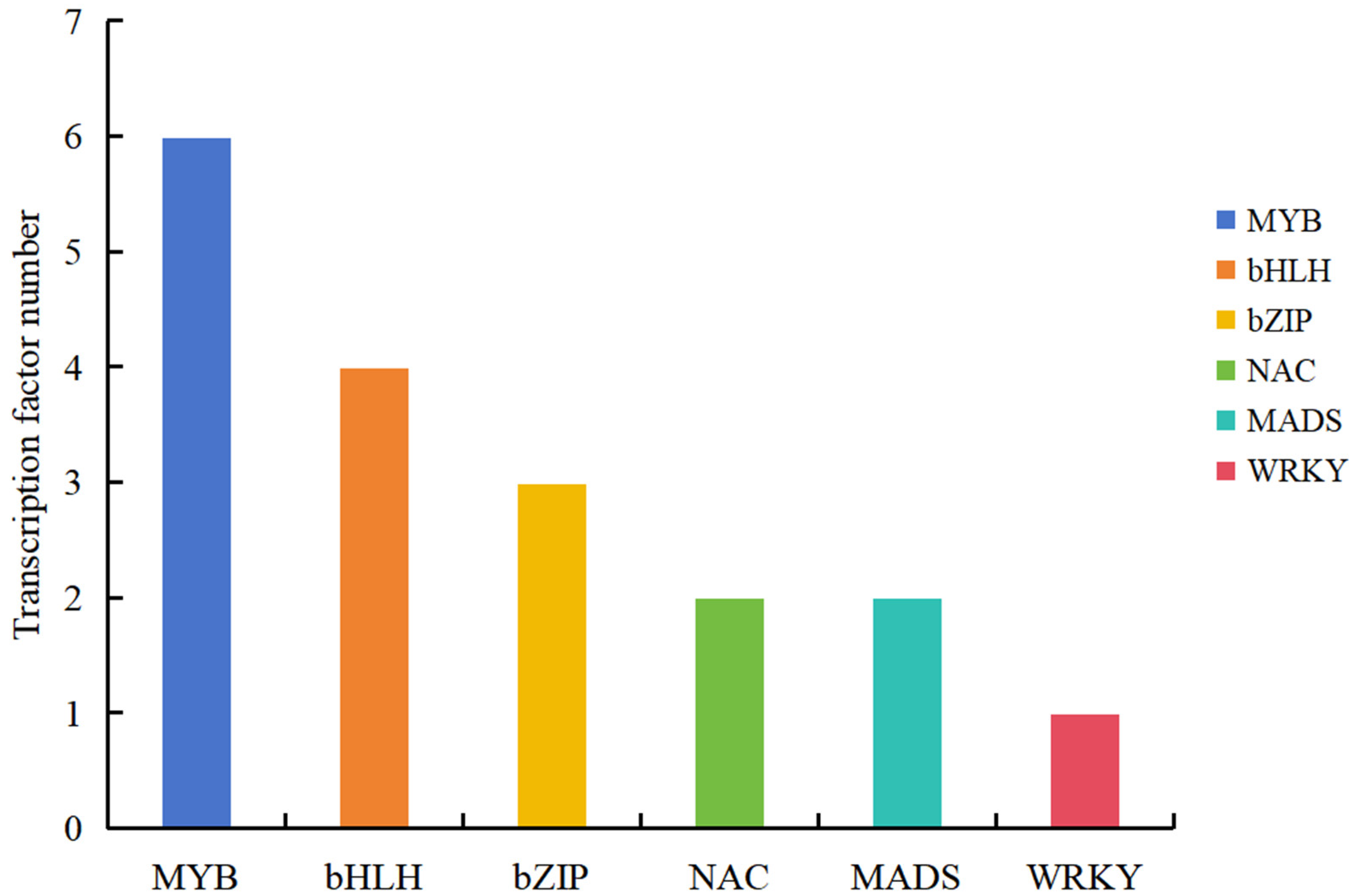

| Samples | Color | L* | a* | b* | C* |
|---|---|---|---|---|---|
| White | White | 82.72 ± 0.53 | −1.21 ± 0.15 | 8.16 ± 0.34 | 8.25 ± 0.23 |
| Tunisia | Red | 51.23 ± 0.45 # | 45.63 ± 0.88 # | 33.43 ± 1.27 # | 56.57 ± 0.89 # |
| Gene Name | Gene ID | Encoding Enzyme | Log2FC |
|---|---|---|---|
| PAL1 | LOC116187719 | phenylalanine ammonia-lyase [EC:4.3.1.24] | 6.86 |
| PAL2 | LOC116196349 | phenylalanine ammonia-lyase [EC:4.3.1.24] | 4.37 |
| C4H | LOC116195737 | trans-cinnamate 4-monooxygenase [EC:1.14.14.91] | 2.97 |
| 4CL | LOC116199390 | 4-coumarate--CoA ligase [EC:6.2.1.12] | 2.69 |
| 4CL | LOC116195737 | 4-coumarate--CoA ligase [EC:6.2.1.12] | 1.53 |
| CHS | LOC116208113 | chalcone synthase [EC:2.3.1.74] | 7.60 |
| CHS | LOC116187113 | chalcone synthase [EC:2.3.1.74] | 4.56 |
| CHI | LOC116187298 | chalcone isomerase [EC:5.5.1.6] | 5.98 |
| CHI | LOC116194939 | chalcone isomerase [EC:5.5.1.6] | 6.76 |
| CHI | LOC116196656 | chalcone isomerase [EC:5.5.1.6] | 3.25 |
| CHI | LOC116187009 | chalcone isomerase [EC:5.5.1.6] | −1.58 |
| F3H | LOC116211316 | naringenin 3-dioxygenase [EC:1.14.11.9] | 5.47 |
| F3′H | LOC116200354 | flavonoid 3′-monooxygenase [EC:1.14.14.82] | 4.73 |
| F3′H | LOC116202083 | flavonoid 3′-monooxygenase [EC:1.14.14.82] | 5.46 |
| F3′H | LOC116199769 | flavonoid 3′-monooxygenase [EC:1.14.14.82] | 3.49 |
| F3′H | LOC116198732 | flavonoid 3′-monooxygenase [EC:1.14.14.82] | 7.69 |
| F3′H | LOC116195807 | flavonoid 3′-monooxygenase [EC:1.14.14.82] | −1.65 |
| DFR | LOC116202639 | bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [EC:1.1.1.219 1.1.1.234] | 2.36 |
| DFR | LOC116214145 | bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [EC:1.1.1.219 1.1.1.234] | 5.12 |
| DFR | LOC116199607 | bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [EC:1.1.1.219 1.1.1.234] | −1.96 |
| ANS | LOC116201425 | anthocyanidin synthase [EC:1.14.20.4] | 6.32 |
| ANS | LOC116213234 | anthocyanidin synthase [EC:1.14.20.4] | 4.69 |
| ANS | LOC116205393 | anthocyanidin synthase [EC:1.14.20.4] | −1.77 |
| PSY | LOC116205576 | 15-cis-phytoene synthase [EC:2.5.1.32] | 3.63 |
| PSY | LOC116199475 | 15-cis-phytoene synthase [EC:2.5.1.32] | 4.78 |
| LCYB | LOC116201477 | lycopene beta-cyclase [EC:5.5.1.19] | 2.19 |
| Index | Metabolite | VIP | log2(FC) | Entry |
|---|---|---|---|---|
| MEDL02688 | 4-Coumaroyl-CoA | 1.59 | 6.56 | C00223 |
| MW0132689 | Dihydrokaempferol | 1.76 | 3.58 | C00974 |
| MW0130123 | Dihydroquercetin | 2.23 | 3.84 | C01617 |
| MEDL00398 | Pelargonidin | 1.35 | 8.53 | C05904 |
| MW0130498 | Cyanidin | 2.39 | 6.80 | C05905 |
| MW0165534 | Cyanidin 3-glucoside | 3.53 | 3.82 | C08604 |
| MW0145361 | Peonidin 3-glucoside | 2.49 | 4.83 | C12141 |
| MW0154969 | Pelargonidin 3-glucoside | 1.58 | 6.18 | C12137 |
| MW0139490 | Prephytoene diphosphate | 1.53 | 3.56 | C03427 |
| MW0114048 | Phytoene | 1.26 | 4.12 | C05421 |
| MW0114088 | 7,8-Dihydro-beta-carotene | 3.46 | 2.86 | C16291 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Han, P.; Zhao, R.; Yu, S.; Liu, H.; Ji, S.; Chen, W. Transcriptome and Metabolome Analyses Reveal the Mechanism of Color Differences in Pomegranate (Punica granatum L.) Red and White Petals. Plants 2025, 14, 652. https://doi.org/10.3390/plants14050652
Zhang Y, Han P, Zhao R, Yu S, Liu H, Ji S, Chen W. Transcriptome and Metabolome Analyses Reveal the Mechanism of Color Differences in Pomegranate (Punica granatum L.) Red and White Petals. Plants. 2025; 14(5):652. https://doi.org/10.3390/plants14050652
Chicago/Turabian StyleZhang, Yong, Peng Han, Ruijie Zhao, Shuhan Yu, Hang Liu, Shuren Ji, and Wei Chen. 2025. "Transcriptome and Metabolome Analyses Reveal the Mechanism of Color Differences in Pomegranate (Punica granatum L.) Red and White Petals" Plants 14, no. 5: 652. https://doi.org/10.3390/plants14050652
APA StyleZhang, Y., Han, P., Zhao, R., Yu, S., Liu, H., Ji, S., & Chen, W. (2025). Transcriptome and Metabolome Analyses Reveal the Mechanism of Color Differences in Pomegranate (Punica granatum L.) Red and White Petals. Plants, 14(5), 652. https://doi.org/10.3390/plants14050652





