Dicranum motuoense (Bryophyta): A New Taxon from China, with Special References to Its Complete Organelle Genomes
Abstract
1. Introduction
2. Results
2.1. Results of Phylogenetic Analyses
2.2. Taxonomic
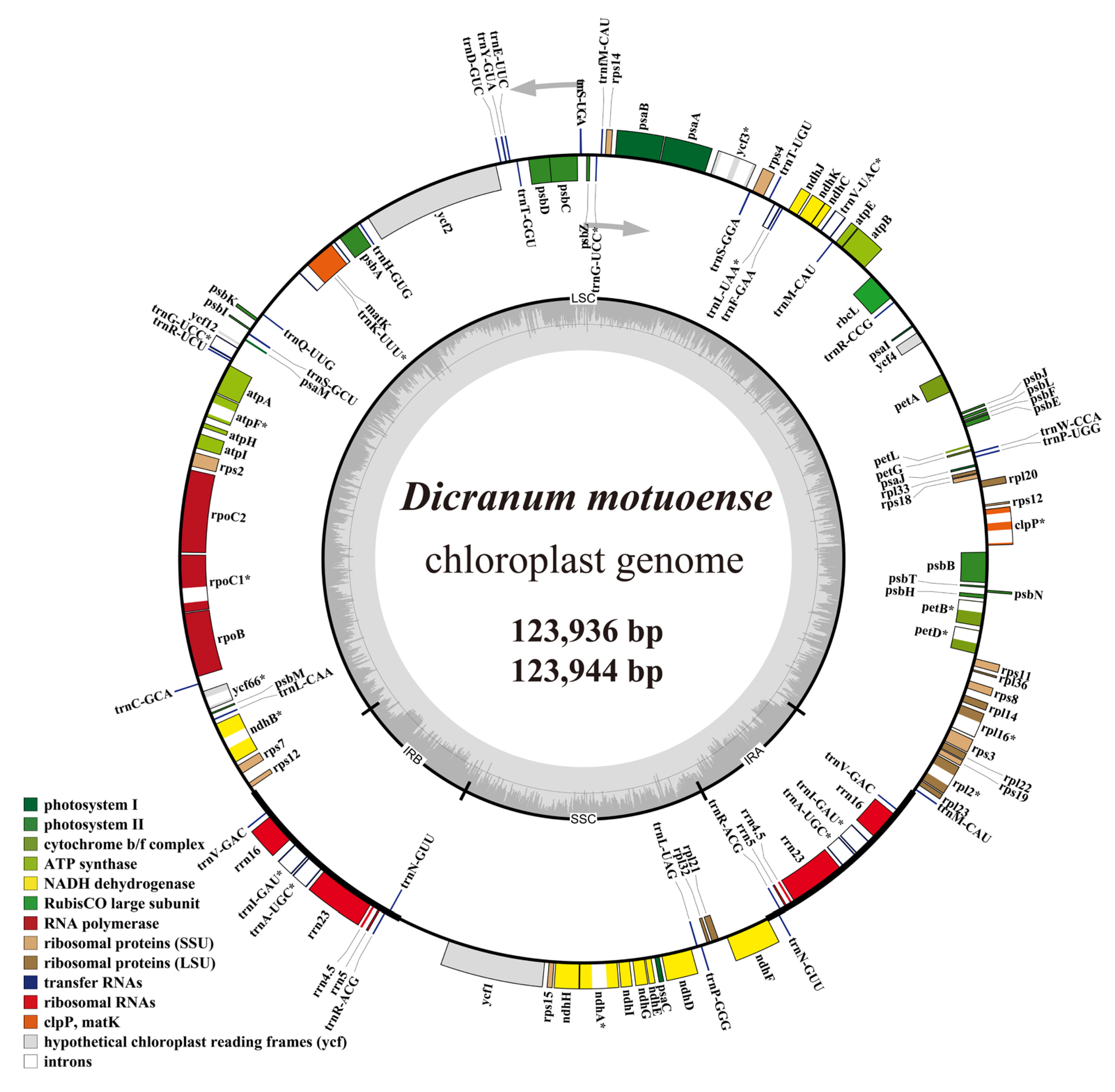
2.3. Feature of Chloroplast Genomes
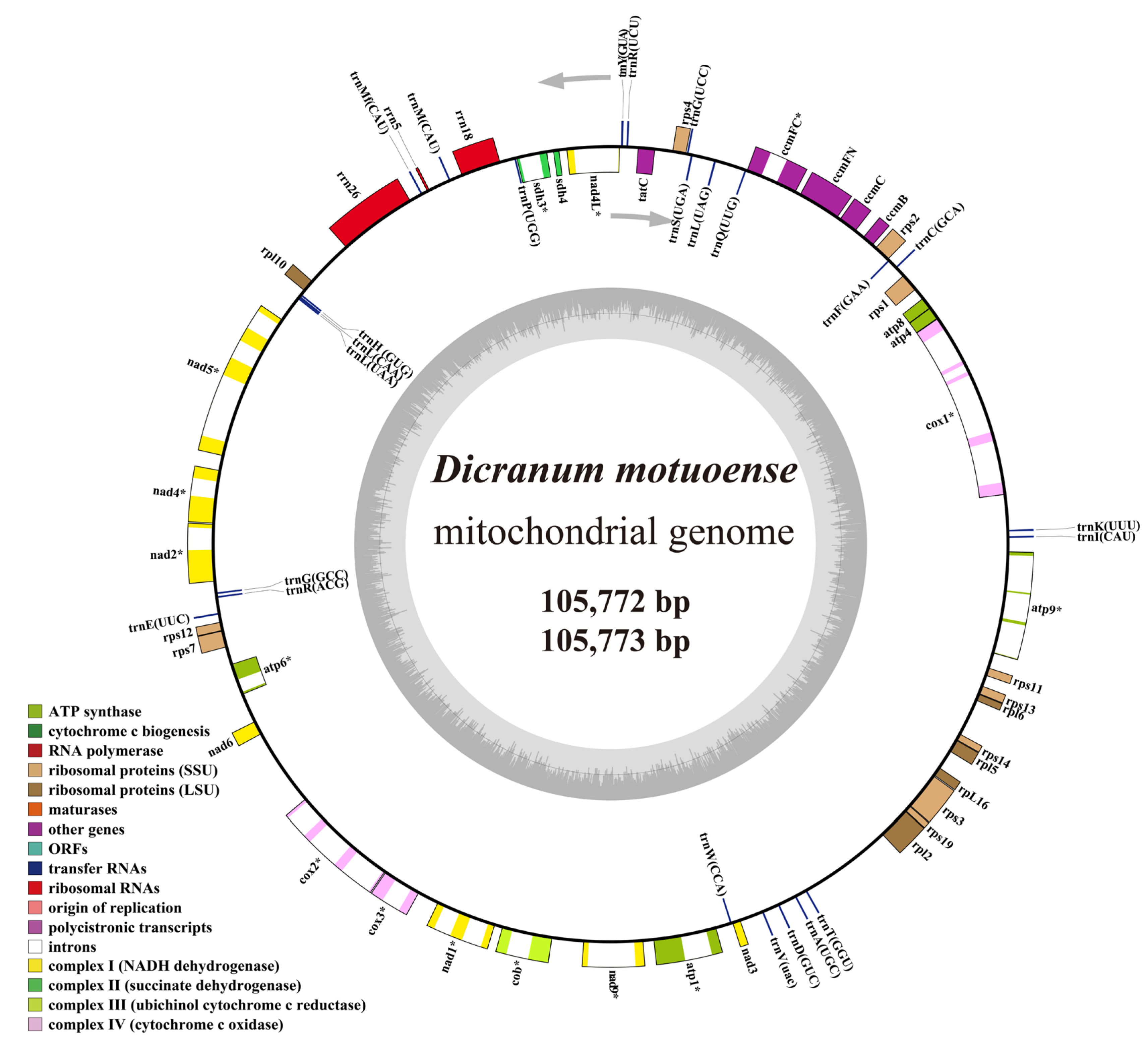
2.4. Feature of Mitochondrial Genomes
2.5. Repetitive Sequences and Codon Usage of the Organelle Genomes
3. Discussion
3.1. Differentiation with Similar Species
3.2. Organelle Genomes
3.3. Key to Species of Dicranum with Fragile Leaves in the Northern Hemisphere
| 1. Leaf apices entire or with few blunt teeth.....................................................................2 |
| 1. Leaf apices sharply denticulate or serrulate..................................................................6 |
| 2. Basal laminal cells nearly quadrate to short rectangular, 20–35(–45) μm long .......................................................................... D. viride (Sull. & Lesq.) Lindb. (in part) |
| 2. Basal laminal cells elongated and rectangular, 30–100(–120) μm long........................3 |
| 3. Costa in the lower portion of the leaf with stereid bands, sometimes weak, with 2–3 (–4) layers of cells above and below guide cells........................... D. fragilifolium Lindb. |
| 3. Costa in the lower portion of the leaf lacking stereid bands, with (0–)1–2 layers of cells above and below guide cells.....................................................................................................4 |
| 4. Alar cells unistratose............................................................. D. tauricum Sapjegin (in part) |
| 4. Alar cells bistratose or 1–2 stratose.......................................................................................5 |
| 5. Upper and middle laminal cells strongly porose; alar cells with thick walls..................... .........................................D. annapurnaense W.Z.Huang, T.X.Zheng & Y.Huan Wu |
| 5. Upper and middle laminal cells not or very slightly pitted; alar cells with thin walls....... .............................................................D. hengduanense W.Z.Huang & R.L.Zhu |
| 6. Costa in the lower portion of the leaf with stereid bands; leaf lamina cells with thick walls, sometimes with bulging.................................................................................7 |
| 6. Costa in the lower portion of the leaf without stereid bands, with substereids on both sides of guide cells; leaf lamina cells with thin or slightly thick walls.....................10 |
| 7. The epidermal layers of the costa in the lower portion of the leaf undifferentiated; upper laminal cells elongated and rectangular, 40–90 μm long, with prominent pores ........................................... D. motuoense W.Z.Huang, Tubanova & Y.Huan Wu |
| 7. The epidermal layers of the costa in the lower portion of the leaf, well differentiated on both sides, or sometimes with only the abaxial epidermis differentiated; upper laminal cells regularly quadrate to short rectangular, up to 35 μm long, not pitted.............8 |
| 8. Costa abruptly differentiated, semicircular in the transverse section, especially in the middle and proximal parts; cell walls between lamina cells without bulging or slightly bulging.................................................................. D. viride (Sull. & Lesq.) Lindb. (in part) |
| 8. Costa gradually differentiated, flattened in the transverse section along the entire length; cell walls between lamina cells slightly or strongly bulging................................9 |
| 9. Leaves weakly fragile, some leaf tips broken off; costa broader, occupying 1/3 or more of the total leaf base width.............................................................................D. fulvum Hook. |
| 9. Leaves fragile; costa narrower, occupying less than 1/3 the leaf base width ............... ................................................................................................................................................. ......................................................................D. hakkodense Cardot |
| 10. Basal laminal cells linear, 40–80(–120) μm long, with non-porose or slightly porose walls........................................................................................ D. tauricum Sapjegin (in part) |
| 10. Basal laminal cells rectangular, 20–40(–50) μm long, with smooth walls................ .............................................................................D. pacificum Ignatova & Fedoso |
4. Materials and Methods
4.1. Taxon Sampling
4.2. Morphological Study
4.3. DNA Extraction, Sequencing, Assembly and Annotation
4.4. Repetitive Sequences, and Codon Usage Preference Analyses
4.5. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinda, J.C.; Atwood, J.J. The Bryophyte Nomenclator. Available online: https://www.bryonames.org/ (accessed on 21 December 2024).
- Hedenäs, L.; Bisang, I. Key to European Dicranum species. Herzogia 2004, 17, 179–197. [Google Scholar]
- Ireland, R.R., Jr. Dicranum (Family Dicranaceae). In Flora of North America. Vol. 27, Bryophytes: Mosses, Part I; Flora of North America Editorial Committee, Ed.; Oxford University Press: New York, NY, USA, 2007; pp. 397–420. [Google Scholar]
- Lang, A.S.; Stech, M. What’s in a name? Disentangling the Dicranum scoparium species complex (Dicranaceae, Bryophyta). Syst. Bot. 2014, 39, 369–379. [Google Scholar] [CrossRef]
- Huang, W.-Z.; Shen, C.; Xu, H.; Shu, L.; Sulayman, M.; Wu, Y.-H.; Zhu, R.-L. A synopsis of Dicranum Hedw. (Dicranaceae, Bryophyta) in China, with special references to four species newly reported and re-evaluation of Dicranum psathyrum Klazenga. Plants 2024, 13, 1759. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, E.A.; Fedosov, V.E. Species of Dicranum (Dicranaceae, Bryophyta) with fragile leaves in Russia. Arctoa 2008, 17, 63–84. [Google Scholar]
- Huang, W.-Z.; Xu, H.; Ma, X.-Y.; Zhu, R.-L. Dicranum hengduanensis (Dicranaceae, Bryophyta), a new species with fragile leaves from the Hengduan Mountains in China. Bryologist 2023, 126, 226–235. [Google Scholar] [CrossRef]
- Tubanova, D.Y.; Goryunov, D.V.; Ignatova, E.A.; Ignatov, M.S. 2010. On the taxonomy of Dicranum acutifolium and D. fuscescens complexes (Dicranaceae, Bryophyta) in Russia. Arctoa 2010, 19, 151–164. [Google Scholar]
- Tubanova, D.Y.; Ignatova, E.A. A new species of Dicranum (Dicranaceae, Bryophytha) from Asiatic Russia. Arctoa 2011, 20, 183–190. [Google Scholar] [CrossRef]
- Maier, E.; Price, M.J. A lectotpye for Dicranum howellii (Dicranceae). Bryologist 2013, 116, 281–286. [Google Scholar]
- Lang, A.S.; Tubanova, D.; Stech, M. Species delimitations in the Dicranum acutifolium complex (Dicranaceae, Bryophyta) using molecular markers. J. Bryol. 2014, 36, 279–290. [Google Scholar] [CrossRef]
- Tubanova, D.Y.; Dugarova, O.D.; Kuznetsova, O.I. Dicranum afoninae (Dicranaceae, Bryophyta), a new species with the flagelliform branchlets from Asia. Arctoa 2024, 33, 31–40. [Google Scholar]
- Tubanova, D.Y.; Dugarova, O.D. Dicranum baicalense (Dicranaceae, Bryophyta), a new species from Russia. Arctoa 2022, 31, 145–154. [Google Scholar]
- Huang, W.-Z.; Tong, S.-G.; Zhu, R.-L. Dicranum shennongjiaense W.Z.Huang & R.L.Zhu (Dicranaceae, Bryophyta), a new species from Central China supported by morphological and molecular evidence. J. Bryol. 2024, 46, 1–15. [Google Scholar]
- Huang, W.-Z.; Zheng, T.-X.; Wu, Y.-H.; Yu, M.-J. Dicranum nepalense (Dicranaceae, Dicranales), a new species with fragile leaves from Nepal Himalayas. Phytotaxa 2024, 666, 51–58. [Google Scholar] [CrossRef]
- Donskov, D.G. On the leaf fragility in Dicranum (Dicranaceae, Bryophyta). Arctoa 2011, 20, 99–105. [Google Scholar] [CrossRef][Green Version]
- Hegewald, E. Zur Anatomie der Blattchenrippe von Dicranum tauricum Sap. J. Hattori Bot. Lab. 1991, 69, 117–119. [Google Scholar]
- Noguchi, A. Illustrated Moss Flora of Japan. Part 1; Hattori Botanical Laboratory: Nichinan-Shi, Japan, 1987; pp. 1–242. [Google Scholar]
- Sun, J.; Ma, Q.; Huang, Y.; Lai, C.-J.; Li, P.; Jin, X.-J.; Zhang, Y.-H. Comparative analyses of Diospyros (Ebenaceae) plastomes: Insights into genomic features, mutational hotspots, and adaptive evolution. Ecol. Evol. 2022, 13, e10301. [Google Scholar]
- Xiang, Y.-L.; Jin, X.-J.; Shen, C.; Cheng, X.-F.; Shu, L.; Zhu, R.-L. New insights into the phylogeny of the complex thalloid liverworts (Marchantiopsida) based on chloroplast genomes. Cladistics 2022, 38, 649–662. [Google Scholar] [CrossRef]
- Song, H.; Peng, H.; Fang, Z.; Zhang, B.; Zhu, Z.; Xiao, Z.; Liu, G.; Hu, Y. Koliella bifissiva sp. nov (Chlorellaceae, Chlorophyta) and analysis of its organelle genomes. Plants 2024, 13, 2604. [Google Scholar] [CrossRef]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef]
- Vanderpoorten, A.; Shaw, A.J. The application of molecular data to the phylogenetic delimitation of species in bryophytes: A note of caution. Phytotaxa 2010, 9, 229–237. [Google Scholar] [CrossRef]
- Lüth, M. Mosses of Europe—A Photographic Flora; Michael Lüth: Freiburg, Germany, 2019; Volume 1, pp. 1–1306. [Google Scholar]
- Kürschner, H.; Frey, W. Liverworts, Mosses and Hornworts of Southwest Asia (Marchantiophyta, Bryophyta, Anthocerotophyta). Nova Hedwigia 2020, 149, 1–267. [Google Scholar]
- Ren, Z.; Chen, H.; Zhang, S. The complete plastid genome of an Antarctic moss Chorisodontium aciphyllum (Hook. f. & Wilson) Broth (Dicranaceae, Dicranales). Mitochondrial DNA B 2022, 7, 683–685. [Google Scholar]
- Lubna; Asaf, S.; Jan, R.; Asif, S.; Bilal, S.; Khan, A.L.; Kim, K.-M.; Lee, I.J.; AL-Harrasi, A. Plastome diversity and evolution in mosses: Insights from structural characterization, comparative genomics, and phylogenetic analysis. Int. J. Biol. Macromol. 2024, 257, 128608. [Google Scholar]
- Mohanta, T.K.; Mishra, A.K.; Khan, A.; Hashem, A.; Abd Allah, E.F.; Al-Harrasi, A. Gene loss and evolution of the Plastome. Genes 2020, 11, 1133. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, X.X.; Li, R.Q.; Zhao, X.; Liu, Y.; Li, M.H.; Zwaenepoel, A.; Ma, H.; Goffinet, B.; Guan, Y.L.; et al. The hornwort genome and early land plant evolution. Nat. Plants 2020, 6, 107–118. [Google Scholar] [CrossRef]
- Sadamitsu, A.; Inoue, Y.; Sakakibara, K.; Tsubota, H.; Yamaguchi, T.; Deguchi, H.; Nishiyama, T.; Shimamura, M. The complete plastid genome sequence of the enigmatic moss, Takakia lepidozioides (Takakiopsida, Bryophyta): Evolutionary perspectives on the largest collection of genes in mosses and the intensive RNA editing. Plant Mol. Biol. 2021, 107, 431–449. [Google Scholar]
- Keller, J.; Rousseau-Gueutin, M.; Martin, G.E.; Morice, J.; Boutte, J.; Coissac, E.; Ourari, M.; Aïnouche, M.; Salmon, A.; Cabello-Hurtado, F. The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. DNA Res. 2017, 24, 343–358. [Google Scholar]
- Huang, W.-Z.; Ma, W.-Z.; Schneider, H.; Yu, Y.; Wu, Y.-H. Mitochondrial genome from Andreaea wangiana reveals structural conservatism and a trend of size reduction in mosses. Bryologist 2019, 122, 597–606. [Google Scholar]
- Kim, J.S.; Kim, J.H. Comparative genome analysis and phylogenetic relationship of order Liliales insight from the complete plastid genome sequences of two Lilies (Lilium longiflorum and Alstroemeria aurea). PLoS ONE 2013, 8, e68180. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, J.Y.; Wang, B.; Li, L.; Qiu, Y.L. The mitochondrial genomes of the early land plants Treubia lacunose and Anomodon rugelii: Dynamic and conservative evolution. PLoS ONE 2011, 6, e25836. [Google Scholar]
- Liu, Y.; Medina, R.; Goffinet, B. 350 Myr of mitochondrial genome stasis in mosses, an early land plant lineage. Mol. Biol. Evol. 2014, 31, 2586–2591. [Google Scholar] [PubMed]
- Wang, B.; Xue, J.Y.; Li, L.; Liu, Y.; Qiu, Y.L. The complete mitochondrial genome sequence of the liverwort Pleurozia purpurea reveals extremely conservative mitochondrial genome evolution in liverworts. Curr. Genet. 2009, 55, 601–609. [Google Scholar] [PubMed]
- Dong, S.S.; Liu, Y. The mitochondrial genomes of bryophytes. Bry. Div. Evo. 2021, 43, 112–126. [Google Scholar]
- Byun, M.Y.; Cho, S.M.; Lee, J.; Park, H.; Lee, H. The complete mitochondrial genome of an Antarctic moss Chorisodontium aciphyllum (Hook. f. & Wilson) Broth. Mitochondrial DNA B 2019, 4, 1714–1715. [Google Scholar]
- Nie, X.J.; Lv, S.Z.; Zhang, Y.X.; Du, X.H.; Wang, L.; Biradar, S.S.; Tan, X.F.; Wan, F.H.; Song, W.N. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar]
- Zheng, Y.; Zhang, H.; Wang, Q.M.; Gao, Y.; Zhang, Z.H.; Sun, Y.X. Complete chloroplast genome sequence of Clivia miniata and its characteristics. Acta Hortic. Sin. 2020, 47, 2439–2450. [Google Scholar]
- Karlim, E.F.; Boles, S.; Shaw, A.J. Resolving boundaries between species in Sphagnum section Subsecunda using microsatellite markers. Taxon 2008, 57, 1189–1200. [Google Scholar]
- Karlim, E.F.; Boles, S.; Shaw, A.J. Systematics of Sphagnum section Sphagnum in New Zealand: A microsatellite-based analysis. N. Z. J. Bot. 2008, 46, 105–118. [Google Scholar]
- Shaw, A.J.; Cao, T.; Wang, L.; Flatberg, L.I.; Flatberg, B.; Shaw, B.; Zhou, P.; Boles, S.; Terracciano, S. Genetic variation in three Chinese peat mosses (Sphagnum) based on microsatellite markers, with primer information and analysis of ascertainment bias. Bryologist 2008, 111, 271–281. [Google Scholar]
- Zhao, C.; Zhu, R.L.; Liu, Y. Simple sequence repeats in bryophyte mitochondrial genomes. Mitochondrial DNA A 2014, 27, 191–197. [Google Scholar]
- Lang, A.S.; Kruijer, J.D.; Stech, M. DNA barcoding of Arctic bryophytes: An example from the moss genus Dicranum (Dicranaceae, Bryophyta). Polar Biol. 2014, 37, 1157–1169. [Google Scholar] [CrossRef]
- Lang, A.S.; Bocksberger, G.; Stech, M. Phylogeny and species delimitations in European Dicranum (Dicranaceae, Bryophyta) inferred from nuclear and plastid DNA. Mol. Phylogenet. Evol. 2015, 92, 217–225. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; de Pamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gbaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Meth. 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.-Z.; Ma, X.-Y.; Tubanova, D.Y.; Dugarova, O.D.; Zhang, F.-Y.; Hu, J.; Zhu, R.-L.; Wu, Y.-H. Dicranum motuoense (Bryophyta): A New Taxon from China, with Special References to Its Complete Organelle Genomes. Plants 2025, 14, 650. https://doi.org/10.3390/plants14050650
Huang W-Z, Ma X-Y, Tubanova DY, Dugarova OD, Zhang F-Y, Hu J, Zhu R-L, Wu Y-H. Dicranum motuoense (Bryophyta): A New Taxon from China, with Special References to Its Complete Organelle Genomes. Plants. 2025; 14(5):650. https://doi.org/10.3390/plants14050650
Chicago/Turabian StyleHuang, Wen-Zhuan, Xin-Yin Ma, Dolgor Y. Tubanova, Oyuna D. Dugarova, Fen-Yao Zhang, Jun Hu, Rui-Liang Zhu, and Yu-Huan Wu. 2025. "Dicranum motuoense (Bryophyta): A New Taxon from China, with Special References to Its Complete Organelle Genomes" Plants 14, no. 5: 650. https://doi.org/10.3390/plants14050650
APA StyleHuang, W.-Z., Ma, X.-Y., Tubanova, D. Y., Dugarova, O. D., Zhang, F.-Y., Hu, J., Zhu, R.-L., & Wu, Y.-H. (2025). Dicranum motuoense (Bryophyta): A New Taxon from China, with Special References to Its Complete Organelle Genomes. Plants, 14(5), 650. https://doi.org/10.3390/plants14050650





