Proteome and Metabolome Analyses of Albino Bracts in Davidia involucrata
Abstract
1. Introduction
2. Results
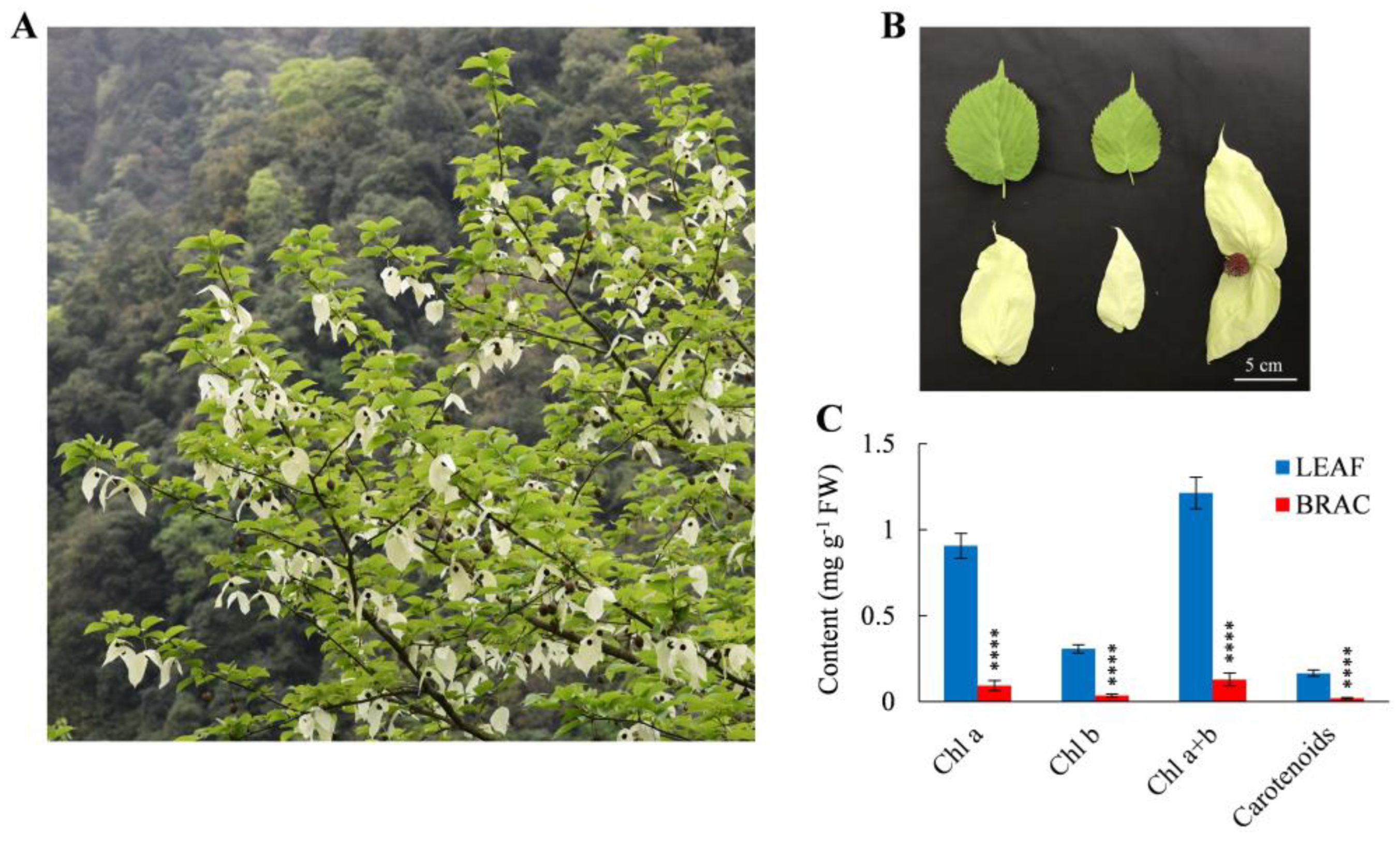
2.1. Pigment Contents and Photosynthetic Characteristics in D. involucrata Bracts
2.2. Ultrastructural Analysis of D. involucrata Bracts
2.3. Screening and GO Classification of DEPs
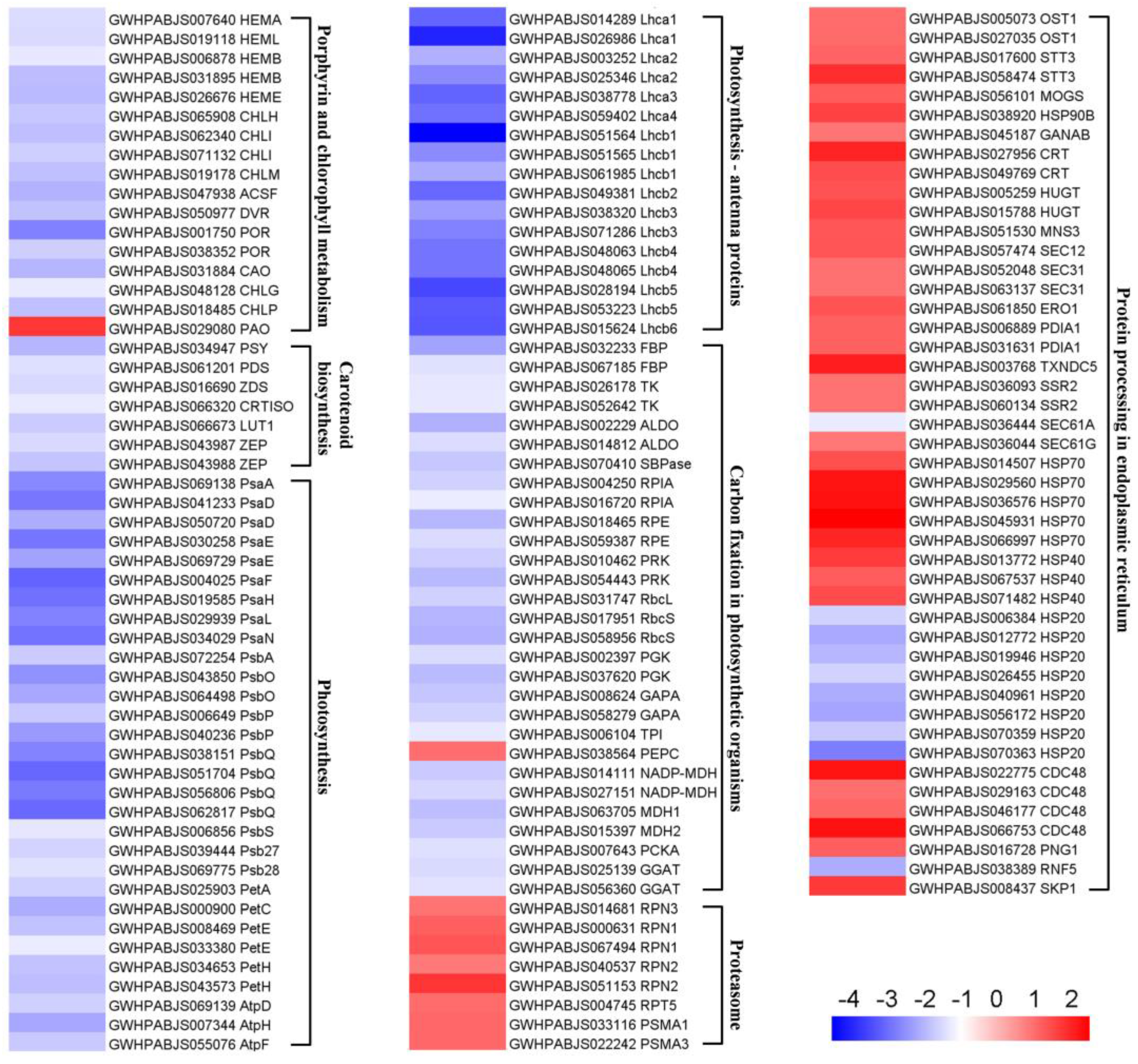
2.4. KEGG Enrichment Analysis of DEPs
2.5. Alterations in the Metabolomics of D. involucrata Bracts
2.6. Combined Proteomic and Metabolomic Analyses of Flavonoid Accumulation in D. involucrata Bracts
2.7. Evaluation of Antioxidant Status
3. Discussion
3.1. Albinism Might Relate to Chloroplast Abnormalities
3.2. Albinism Is Associated with Defective Photosynthetic Pigment Metabolism
3.3. Albino Bracts Are Subjected to Light Stress
3.4. Flavonoid Metabolism Is a Crucial Adaptive Trait for Bracts
3.5. Activation of the UPS in Albino Bracts
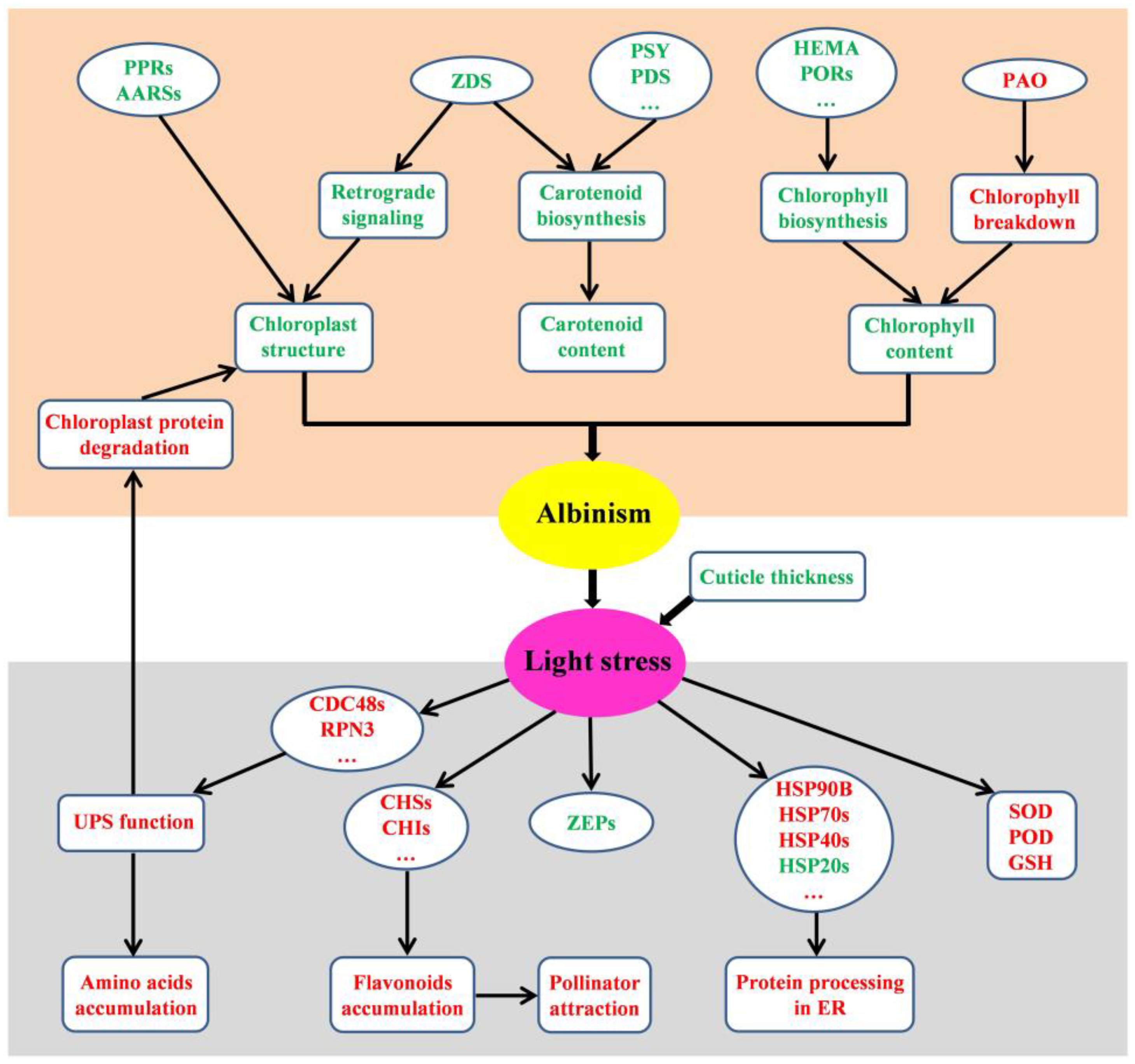
3.6. Possible Mechanisms of Albinism Induction in Bracts
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Physiological Indices
4.3. Transmission Electron Microscopy
4.4. TMT-Based Quantitative Proteomic Analysis
4.5. Database Search and Bioinformatics Analysis
4.6. Metabolite Extraction and Profiling
4.7. Analysis of Metabolomics Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 2015, 1847, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liang, F.C.; Wittmann, D.; Siegel, A.; Shan, S.O.; Grimm, B. Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. USA 2018, 115, E3588–E3596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, M.; Gao, X.; Zhou, F.; Shen, C.; Liu, Z. Multi-omics research in albino tea plants: Past, present, and future. Sci. Hortic. 2020, 261, 108943. [Google Scholar] [CrossRef]
- Yue, C.; Wang, Z.; Yang, P. Review: The effect of light on the key pigment compounds of photosensitive etiolated tea plant. Bot. Stud. 2021, 62, 21. [Google Scholar]
- Yan, C.; Peng, L.; Zhang, L.; Qiu, Z. Fine mapping of a candidate gene for cool-temperature-induced albinism in ornamental kale. BMC Plant Biol. 2020, 20, 460. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Song, P.; Peng, X.; Li, X.; Su, R.; Zhang, H.; Lin, L.; Xia, H.; Deng, Q. Metabolome and transcriptome reveal high abundance of bioactive substances in albino jujube fruit as potential function food. Food Biosci. 2024, 59, 103991. [Google Scholar] [CrossRef]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, Z.; Tian, Y.; Liu, L.; Lv, F.; Kong, W.; Bai, W.; Wang, P.; Wang, C.; Yu, X.; et al. Rice albino 1, encoding a glycyl-tRNA synthetase, is involved in chloroplast development and establishment of the plastidic ribosome system in rice. Plant Physiol. Biochem. 2019, 139, 495–503. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, W.; Wen, K.; Chen, G.; Sun, J.; Tian, Y.; Tang, W.; Yu, J.; An, H.; Wu, T.; et al. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 2017, 95, 345–357. [Google Scholar] [CrossRef]
- Wan, C.; Zhang, H.; Cheng, H.; Sowden, R.G.; Cai, W.; Jarvis, R.P.; Ling, Q. Selective autophagy regulates chloroplast protein import and promotes plant stress tolerance. EMBO J. 2023, 42, e112534. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, J.; Li, Y.; Sun, H.; Ma, T.; Huai, J.; Yang, W.; Zhang, W.; Lin, R. The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022, 39, 110664. [Google Scholar] [CrossRef] [PubMed]
- Talaat, N.B. RNAi based simultaneous silencing of all forms of light-dependent NADPH:protochlorophyllide oxidoreductase genes result in the accumulation of protochlorophyllide in tobacco (Nicotiana tabacum). Plant Physiol. Biochem. 2013, 71, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, X.; Li, Y.; Xia, Z. Transcriptome analysis of a new maize albino mutant reveals that zeta-carotene desaturase is involved in chloroplast development and retrograde signaling. Plant Physiol. Biochem. 2020, 156, 407–419. [Google Scholar] [CrossRef]
- García-Alcázar, M.; Giménez, E.; Pineda, B.; Capel, C.; García-Sogo, B.; Sánchez, S.; Yuste-Lisbona, F.J.; Angosto, T.; Capel, J.; Moreno, V.; et al. Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 2017, 7, 45333. [Google Scholar] [CrossRef]
- Yan, J.; Liu, B.; Cao, Z.; Chen, L.; Liang, Z.; Wang, M.; Liu, W.; Lin, Y.; Jiang, B. Cytological, genetic and transcriptomic characterization of a cucumber albino mutant. Front. Plant Sci. 2022, 13, 1047090. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, P.; Chen, X.; Yue, C.; Guo, Y.; Yang, J.; Sun, Y.; Ye, N. Integrated transcriptomics and metabolomics provide novel insight into changes in specialized metabolites in an albino tea cultivar (Camellia sinensis (L.) O. Kuntz). Plant Physiol. Biochem. 2021, 160, 27–36. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Serra, P.; Casati, P. Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plant. 2021, 173, 736–749. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Ruan, J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef]
- Lallemand, F.; Martin-Magniette, M.L.; Gilard, F.; Gakière, B.; Launay-Avon, A.; Delannoy, É.; Selosse, M.A. In situ transcriptomic and metabolomic study of the loss of photosynthesis in the leaves of mixotrophic plants exploiting fungi. Plant J. 2019, 98, 826–841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, H.; Wang, J.; Li, Y.; Liu, D.; Ye, Y.; Huang, R.; Li, S.; Chen, L.; Chen, J.; et al. A key mutation in magnesium chelatase I subunit leads to a chlorophyll-deficient mutant of tea (Camellia sinensis). J. Exp. Bot. 2024, 75, 935–946. [Google Scholar] [CrossRef]
- Lin, X.; Chen, X.; Wang, P.; Zheng, Y.; Guo, Y.; Hong, Y.; Yang, R.; Ye, N. Metabolite profiling in albino tea mutant Camellia sinensis ‘Fuyun 6’ using LC–ESI–MS/MS. Trees 2022, 36, 261–272. [Google Scholar] [CrossRef]
- Liu, Q.; Vetukuri, R.R.; Xu, W.; Xu, X. Transcriptomic responses of dove tree (Davidia involucrata Baill.) to heat stress at the seedling stage. Forests 2019, 10, 656. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, L.; Li, Y.; Xu, W.; Vetukuri, R.R.; Xu, X. Overexpression of an autophagy-related gene DiATG3 from Davidia involucrata improves plant thermotolerance by enhancing the accumulation of polyamines and regulating genes in calcium and MAPK signaling pathways. Environ. Exp. Bot. 2023, 208, 105235. [Google Scholar] [CrossRef]
- Liu, Q.; Feng, Z.; Xu, W.; Vetukuri, R.R.; Xu, X. Exogenous melatonin-stimulated transcriptomic alterations of Davidia involucrata seedlings under drought stress. Trees 2021, 35, 1025–1038. [Google Scholar] [CrossRef]
- Qian, S.; Tang, C.Q.; Yi, S.; Zhao, L.; Song, K.; Yang, Y. Conservation and development in conflict: Regeneration of wild Davidia involucrata (Nyssaceae) communities weakened by bamboo management in south-central China. Oryx 2018, 52, 442–451. [Google Scholar] [CrossRef]
- Sun, J.F.; Gong, Y.B.; Renner, S.S.; Huang, S.Q. Multifunctional bracts in the dove tree Davidia involucrata (Nyssaceae: Cornales): Rain protection and pollinator attraction. Am. Nat. 2008, 171, 119–124. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 2013, 82, 505–517. [Google Scholar] [CrossRef]
- García-Caparrós, P.; De Filippis, L.; Gul, A.; Hasanuzzaman, M.; Ozturk, M.; Altay, V.; Lao, M.T. Oxidative stress and antioxidant metabolism under adverse environmental conditions: A review. Bot. Rev. 2021, 87, 421–466. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Z.; Shen, W.; Zhou, H.; Li, H.; He, X.; Li, R.; Qin, B. Disruption of the rice ALS1 localized in chloroplast causes seedling-lethal albino phenotype. Plant Sci. 2024, 338, 111925. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H. Chloroplast ultrastructure in plants. New Phytol. 2019, 223, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schnell, D.J. Protein import into chloroplasts. Trends Cell Biol. 1999, 9, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Block, M.A.; Douce, R.; Joyard, J.; Rolland, N. Chloroplast envelope membranes: A dynamic interface between plastids and the cytosol. Photosynth. Res. 2007, 92, 225–244. [Google Scholar] [CrossRef]
- Nelson, N.; Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004, 5, 971–982. [Google Scholar] [CrossRef]
- Sun, Y.; Zerges, W. Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta 2015, 1847, 809–820. [Google Scholar] [CrossRef]
- Schünemann, D. Mechanisms of protein import into thylakoids of chloroplasts. Biol. Chem. 2007, 388, 907–915. [Google Scholar] [CrossRef]
- Pribil, M.; Labs, M.; Leister, D. Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 2014, 65, 1955–1972. [Google Scholar] [CrossRef]
- Sun, Y.; Jarvis, R.P. Chloroplast proteostasis: Import, sorting, ubiquitination, and proteolysis. Annu. Rev. Plant Biol. 2023, 74, 259–283. [Google Scholar] [CrossRef]
- Bykowski, M.; Mazur, R.; Wójtowicz, J.; Suski, S.; Garstka, M.; Mostowska, A.; Kowalewska, Ł. Too rigid to fold: Carotenoid-dependent decrease in thylakoid fluidity hampers the formation of chloroplast grana. Plant Physiol. 2021, 185, 210–227. [Google Scholar] [CrossRef]
- Hansson, A.; Jensen, P.E. Chlorophyll limitation in plants remodels and balances the photosynthetic apparatus by changing the accumulation of photosystems I and II through two different approaches. Physiol. Plant. 2009, 135, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Sharwood, R.E. Engineering chloroplasts to improve Rubisco catalysis: Prospects for translating improvements into food and fiber crops. New Phytol. 2017, 213, 494–510. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 6, 32741. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.P.; Willingham, N.M.; Lloyd, J.C.; Raines, C.A. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 1998, 204, 27–36. [Google Scholar] [CrossRef]
- Liu, X.L.; Yu, H.D.; Guan, Y.; Li, J.K.; Guo, F.Q. Carbonylation and loss-of-function analyses of SBPase reveal its metabolic interface role in oxidative stress, carbon assimilation, and multiple aspects of growth and development in Arabidopsis. Mol. Plant 2012, 5, 1082–1099. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.B.; Jiang, Y.; Chong, K.; Yang, Z.N. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 2009, 59, 1011–1023. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Zheng, M.; Lyu, J.; Xu, Y.; Li, X.; Niu, M.; Long, W.; Wang, D.; Wang, H.; et al. WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 2016, 170, 2110–2123. [Google Scholar] [CrossRef]
- Chu, P.; Yan, G.X.; Yang, Q.; Zhai, L.N.; Zhang, C.; Zhang, F.Q.; Guan, R.Z. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. J. Proteom. 2015, 113, 244–259. [Google Scholar] [CrossRef]
- Ezquerro, M.; Burbano-Erazo, E.; Rodriguez-Concepcion, M. Overlapping and specialized roles of tomato phytoene synthases in carotenoid and abscisic acid production. Plant Physiol. 2023, 193, 2021–2036. [Google Scholar] [CrossRef]
- Dong, H.; Deng, Y.; Mu, J.; Lu, Q.; Wang, Y.; Xu, Y.; Chu, C.; Chong, K.; Lu, C.; Zuo, J. The Arabidopsis Spontaneous Cell Death1 gene, encoding a ζ-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 2007, 17, 458–470. [Google Scholar] [CrossRef]
- Richter, A.S.; Nägele, T.; Grimm, B.; Kaufmann, K.; Schroda, M.; Leister, D.; Kleine, T. Retrograde signaling in plants: A critical review focusing on the GUN pathway and beyond. Plant Commun. 2023, 4, 100511. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.R.; Frost, E.; Vidi, P.A.; Kessler, F.; Staehelin, L.A. Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 2006, 18, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Abouelsaad, I.; Weihrauch, D.; Renault, S. Effects of salt stress on the expression of key genes related to nitrogen assimilation and transport in the roots of the cultivated tomato and its wild salt-tolerant relative. Sci. Hortic. 2016, 211, 70–78. [Google Scholar] [CrossRef]
- Bethmann, S.; Melzer, M.; Schwarz, N.; Jahns, P. The zeaxanthin epoxidase is degraded along with the D1 protein during photoinhibition of photosystem II. Plant Direct 2019, 3, e00185. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Sun, X.; Sun, C.; Li, Z.; Hu, Q.; Han, L.; Luo, H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016, 39, 1320–1337. [Google Scholar] [CrossRef]
- Sun, X.; Huang, N.; Li, X.; Zhu, J.; Bian, X.; Li, H.; Wang, L.; Hu, Q.; Luo, H. A chloroplast heat shock protein modulates growth and abiotic stress response in creeping bentgrass. Plant Cell Environ. 2021, 44, 1769–1787. [Google Scholar] [CrossRef]
- Bernado, W.P.; Rakocevic, M.; Santos, A.R.; Ruas, K.F.; Baroni, D.F.; Abraham, A.C.; Pireda, S.; Oliveira, D.D.S.; Cunha, M.D.; Ramalho, J.C.; et al. Biomass and leaf acclimations to ultraviolet solar radiation in juvenile plants of Coffea arabica and C. canephora. Plants 2021, 10, 640. [Google Scholar] [CrossRef]
- Roy, M.; Gonneau, C.; Rocheteau, A.; Berveiller, D.; Thomas, J.C.; Damesin, C.; Selosse, M.A. Why do mixotrophic plants stay green? A comparison between green and achlorophyllous orchid individuals in situ. Ecol. Monogr. 2013, 83, 95–117. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Ryan, K.G.; Markham, K.R.; Bloor, S.J.; Bradley, J.M.; Mitchell, K.A.; Jordan, B.R. UVB radiation induced increase in quercetin: Kaempferol ratio in wild-type and transgenic lines of Petunia. Photochem. Photobiol. 1998, 68, 323–330. [Google Scholar] [CrossRef]
- Gerhardt, K.E.; Lampi, M.A.; Greenberg, B.M. The effects of far-red light on plant growth and flavonoid accumulation in Brassica napus in the presence of ultraviolet B radiation. Photochem. Photobiol. 2008, 84, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Czemmel, S.; Heppel, S.C.; Bogs, J. R2R3 MYB transcription factors: Key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 2012, 249, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Pucker, B.; Kuang, T.; Song, B.; Yang, Y.; Lin, N.; Zhang, H.; Moore, M.J.; Brockington, S.F.; Wang, Q.; et al. The genome of the glasshouse plant noble rhubarb (Rheum nobile) provides a window into alpine adaptation. Commun. Biol. 2023, 6, 706. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Liu, Q.; Vain, T.; Viotti, C.; Doyle, S.M.; Tarkowská, D.; Novák, O.; Zipfel, C.; Sitbon, F.; Robert, S.; Hofius, D. Vacuole integrity maintained by DUF300 proteins is required for brassinosteroid signaling regulation. Mol. Plant 2018, 11, 553–567. [Google Scholar] [CrossRef]
- Adams, E.H.G.; Spoel, S.H. The ubiquitin-proteasome system as a transcriptional regulator of plant immunity. J. Exp. Bot. 2018, 69, 4529–4537. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, M.; Cao, K.; Xu, H.; Zhou, X. UV-B irradiation to amino acids and carbohydrate metabolism in Rhododendron chrysanthum leaves by coupling deep transcriptome and metabolome analysis. Plants 2022, 11, 2730. [Google Scholar] [CrossRef]
- Wu, X.; Gao, R.; Mao, R.; Lin, Y.; Yang, Z.; Li, J.; Cao, F.; Li, M. Inducing bract-like leaves in Arabidopsis through ectopically expressing an ASR gene from the dove tree. Ind. Crops Prod. 2022, 180, 114796. [Google Scholar] [CrossRef]
- Salazar-Iribe, A.; De-la-Peña, C. Auxins, the hidden player in chloroplast development. Plant Cell Rep. 2020, 39, 1595–1608. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, L.; Li, R.; Wang, X.; Liu, S.; Liu, X.; Jing, Y.; Xiao, J. SKL1 is essential for chloroplast development in Arabidopsis. Front. Plant Sci. 2018, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Zu, X.; Luo, L.; Wang, Z.; Gong, J.; Yang, C.; Wang, Y.; Xu, C.; Qiao, X.; Deng, X.; Song, X.; et al. A mitochondrial pentatricopeptide repeat protein enhances cold tolerance by modulating mitochondrial superoxide in rice. Nat. Commun. 2023, 14, 6789. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, X.; Jia, X.; Liu, L.; Cao, H.; Qin, W.; Li, M. Iron deficiency leads to chlorosis through impacting chlorophyll synthesis and nitrogen metabolism in Areca catechu L. Front. Plant Sci. 2021, 12, 710093. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Saito, T.; Iwata, N.; Ohmae, Y.; Iwata, R.; Tanoi, K.; Nakanishi, T.M. Leaf senescence in rice due to magnesium deficiency mediated defect in transpiration rate before sugar accumulation and chlorosis. Physiol. Plant. 2013, 148, 490–501. [Google Scholar] [CrossRef]
- Duarte-Aké, F.; Castillo-Castro, E.; Pool, F.B.; Espadas, F.; Santamaría, J.M.; Robert, M.L.; De-la-Peña, C. Physiological differences and changes in global DNA methylation levels in Agave angustifolia Haw. albino variant somaclones during the micropropagation process. Plant Cell Rep. 2016, 35, 2489–2502. [Google Scholar] [CrossRef] [PubMed]
- Us-Camas, R.; Castillo-Castro, E.; Aguilar-Espinosa, M.; Limones-Briones, V.; Rivera-Madrid, R.; Robert-Díaz, M.L.; De-la-Peña, C. Assessment of molecular and epigenetic changes in the albinism of Agave angustifolia Haw. Plant Sci. 2017, 263, 156–167. [Google Scholar] [CrossRef]
- Wei, X.; Song, X.; Wei, L.; Tang, S.; Sun, J.; Hu, P.; Cao, X. An epiallele of rice AK1 affects photosynthetic capacity. J. Integr. Plant Biol. 2017, 59, 158–163. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, T.; Chen, X.; Xu, W.; Dong, T.; Liu, Q.; Xu, X. Twigs of dove tree in high-latitude region tend to increase biomass accumulation in vegetative organs but decrease it in reproductive organs. Front. Plant Sci. 2023, 13, 1088955. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.P.; Chen, C.L.; Wang, Y.; Fu, X.Z.; Liu, J.H. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 2011, 62, 2899–2914. [Google Scholar] [CrossRef]
- Li, L.; Tong, Y.X.; Lu, J.L.; Li, Y.M.; Liu, X.; Cheng, R.F. Morphology, photosynthetic traits, and nutritional quality of lettuce plants as affected by green light substituting proportion of blue and red light. Front. Plant Sci. 2021, 12, 627311. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, B.; Geng, W.; Shen, Y.; Xuan, D.; Lai, Q.; Shen, C.; Jin, C.; Yu, C. Comparative proteomic analysis of pepper (Capsicum annuum L.) seedlings under selenium stress. PeerJ 2019, 7, e8020. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.S.; Shao, Y.J.; Yang, Z.T.; Liu, J.X. Quantitative proteomic analysis of ER stress response reveals both common and specific features in two contrasting ecotypes of Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 9741. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Guo, H.; Zhang, L.; Fan, Y.; Fan, Y.; Tang, Z.; Zeng, F. Dynamic TMT-based quantitative proteomics analysis of critical initiation process of totipotency during cotton somatic embryogenesis transdifferentiation. Int. J. Mol. Sci. 2019, 20, 1691. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, T.; Zhang, L.; Kang, M.; Zhang, Z.; Zheng, Z.; Sun, P.; Shrestha, N.; Liu, J.; Yang, Y. Genomic analyses of a “living fossil”: The endangered dove-tree. Mol. Ecol. Resour. 2020, 20, 756–769. [Google Scholar] [CrossRef]
- Pang, X.; Suo, J.; Liu, S.; Xu, J.; Yang, T.; Xiang, N.; Wu, Y.; Lu, B.; Qin, R.; Liu, H.; et al. Combined transcriptomic and metabolomic analysis reveals the potential mechanism of seed germination and young seedling growth in Tamarix hispida. BMC Genom. 2022, 23, 109. [Google Scholar] [CrossRef]
- Xie, L.; Li, H.; Zhong, Z.; Guo, J.; Hu, G.; Gao, Y.; Tong, Z.; Liu, M.; Hu, S.; Tong, H.; et al. Metabolome analysis under aluminum toxicity between aluminum-tolerant and -sensitive rice (Oryza sativa L.). Plants 2022, 11, 1717. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Wang, J.; Li, Y.; Xu, L.; Xu, W.; Vetukuri, R.R.; Xu, X. Proteome and Metabolome Analyses of Albino Bracts in Davidia involucrata. Plants 2025, 14, 549. https://doi.org/10.3390/plants14040549
Liu Q, Wang J, Li Y, Xu L, Xu W, Vetukuri RR, Xu X. Proteome and Metabolome Analyses of Albino Bracts in Davidia involucrata. Plants. 2025; 14(4):549. https://doi.org/10.3390/plants14040549
Chicago/Turabian StyleLiu, Qinsong, Jinqiu Wang, Yuying Li, Lei Xu, Wenjuan Xu, Ramesh R. Vetukuri, and Xiao Xu. 2025. "Proteome and Metabolome Analyses of Albino Bracts in Davidia involucrata" Plants 14, no. 4: 549. https://doi.org/10.3390/plants14040549
APA StyleLiu, Q., Wang, J., Li, Y., Xu, L., Xu, W., Vetukuri, R. R., & Xu, X. (2025). Proteome and Metabolome Analyses of Albino Bracts in Davidia involucrata. Plants, 14(4), 549. https://doi.org/10.3390/plants14040549





