Transcriptome Analysis Revealed Hub Genes Related to Tipburn Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)
Abstract
1. Introduction
2. Results
2.1. Phenotypic Observations of Tipburn After Calcium Deficiency Treatment
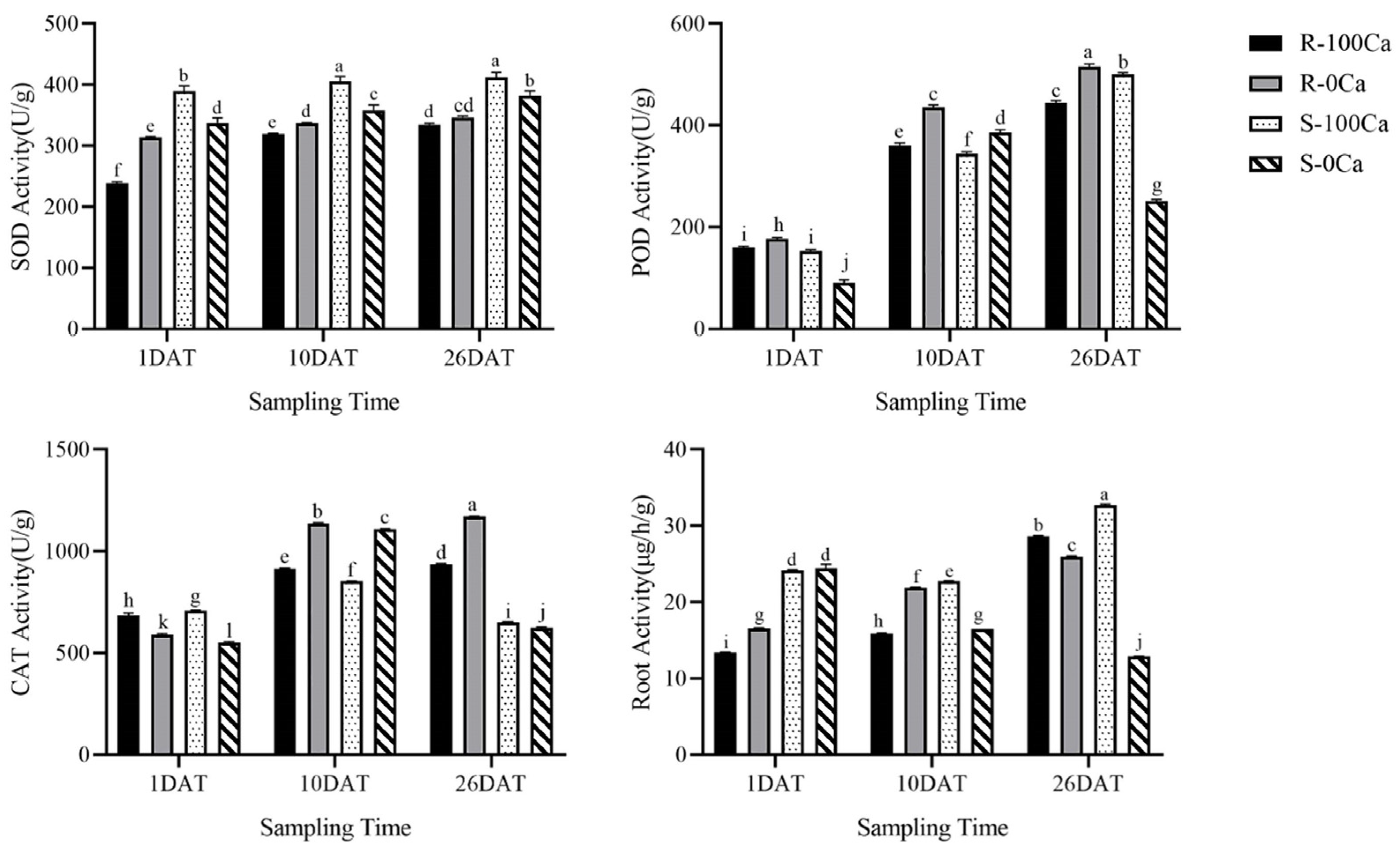
2.2. Antioxidant Enzymes Activities of Plant Leaves After Calcium Deficiency Treatment
2.3. Sequencing Data Quality Assessment
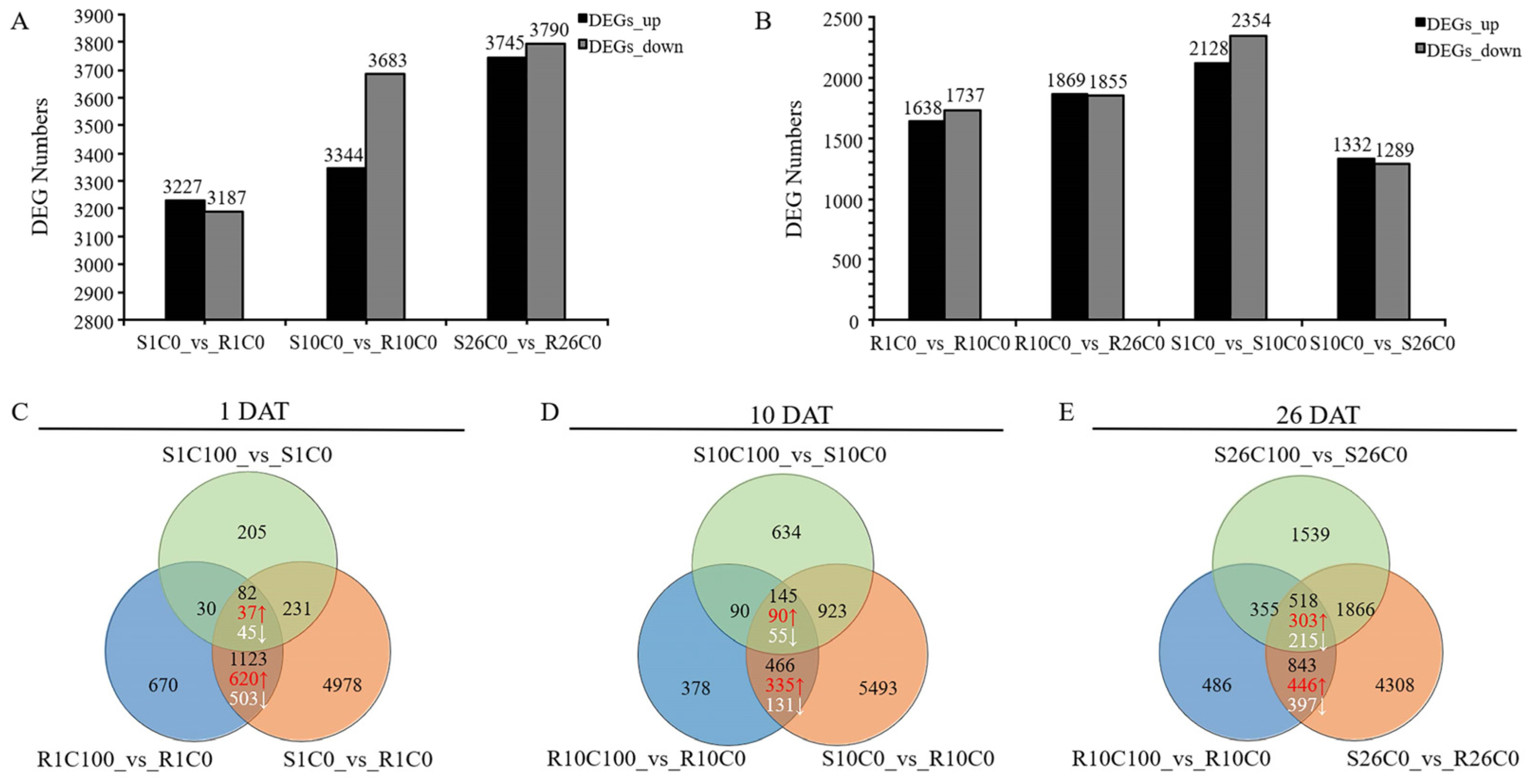
2.4. Statistics of Differentially Expressed Genes
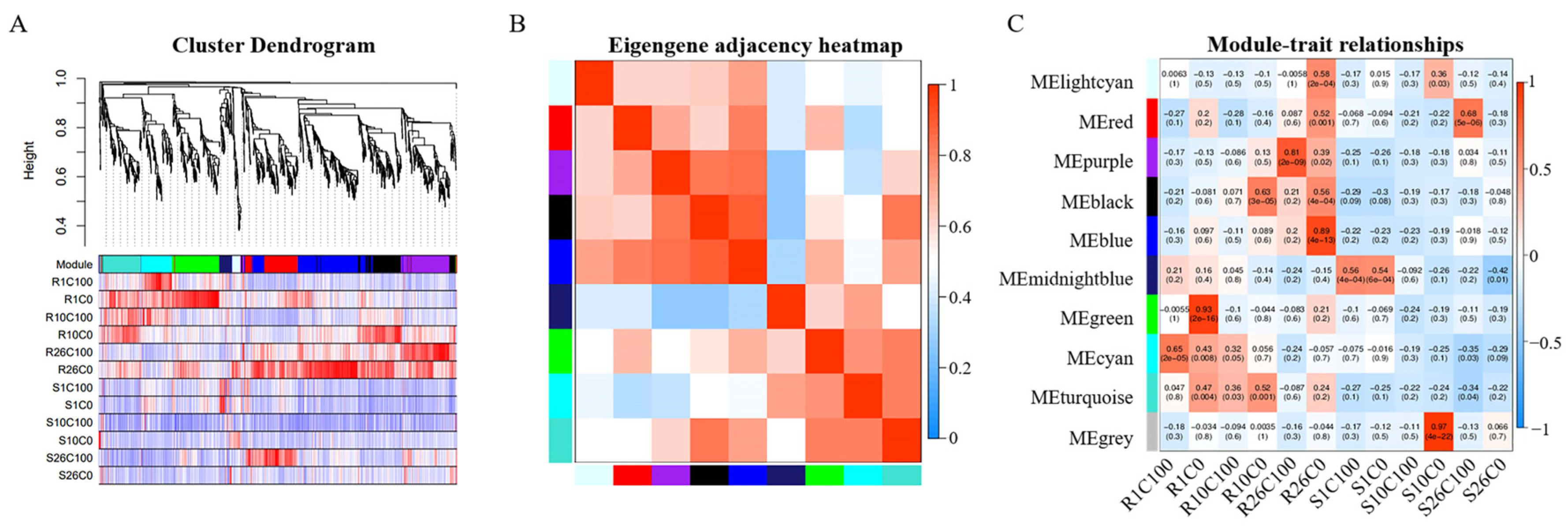
2.5. Weighted Gene Co-Expression Network Analysis of DEGs
2.6. Analysis of the Gene Expression Regulation of Key Modules
2.7. DEGs in Response to the Tipburn Process
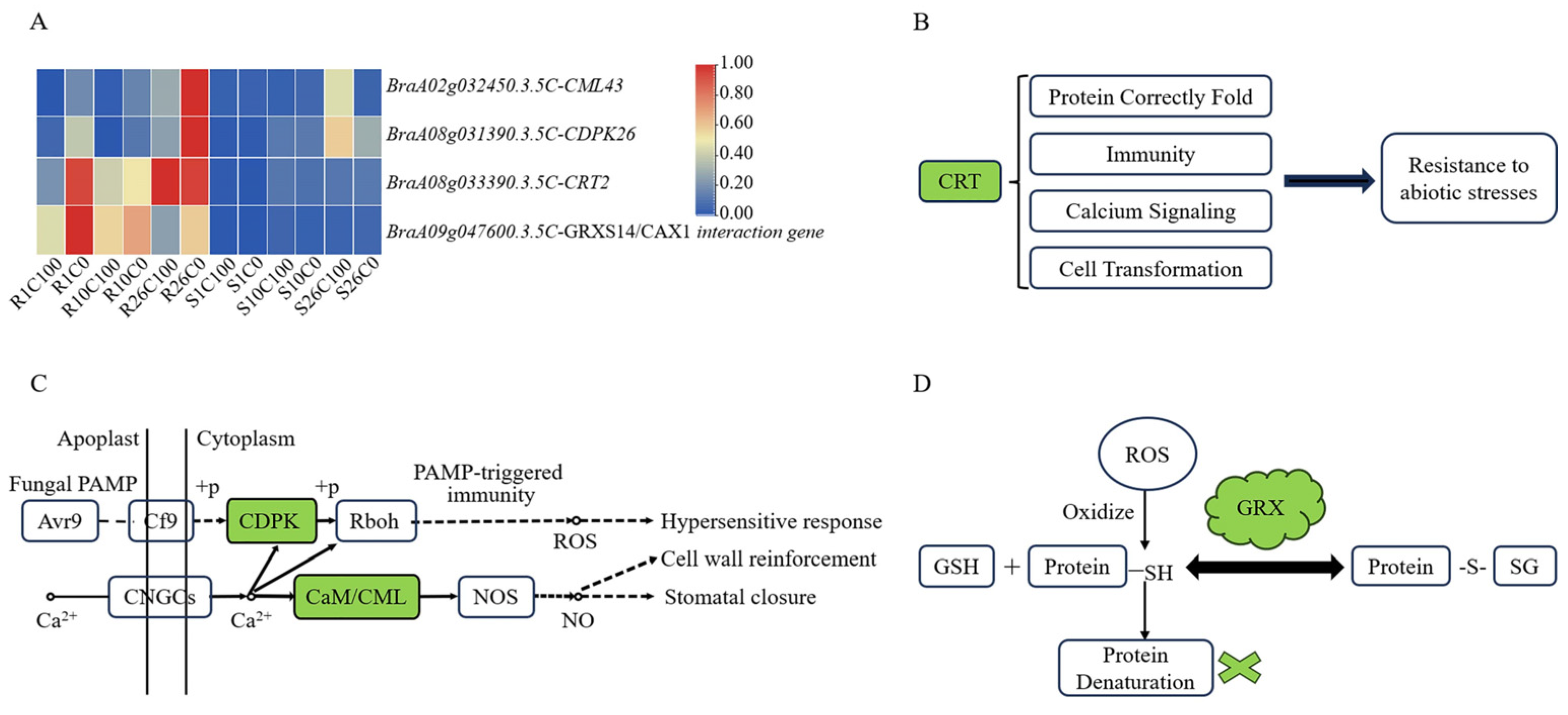
2.7.1. Calcium-Related Genes May Play an Important Role in Tipburn Resistance
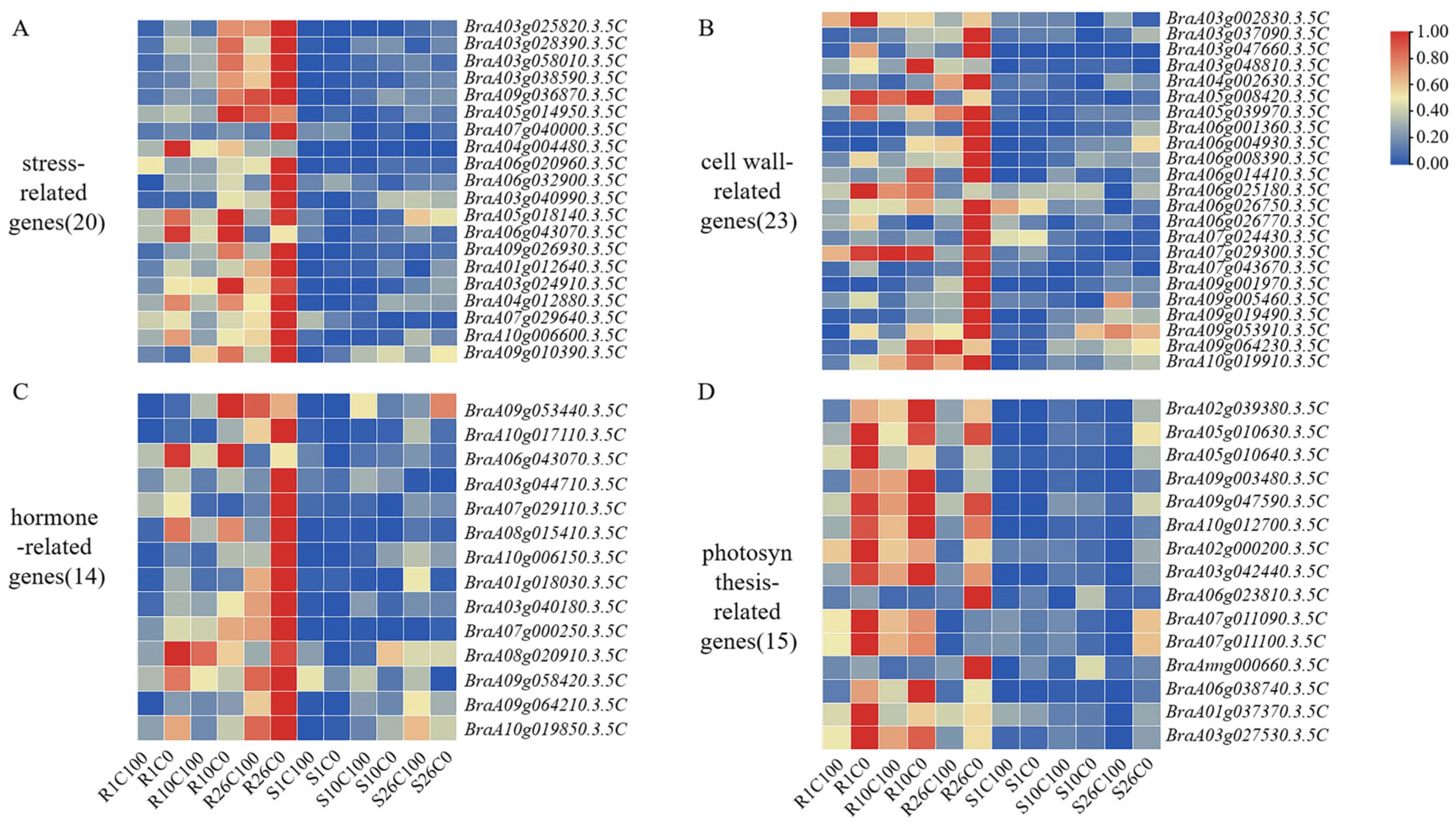
2.7.2. Stress-Related Genes May Play an Important Role in Calcium Deficiency Resistance
2.7.3. DEGs Related to Cell Walls and Hormones May Play an Important Role in the Late Stage of Calcium Deficiency Resistance
2.7.4. DEGs Associated with Photosynthesis Respond to Calcium Deficiency Stress
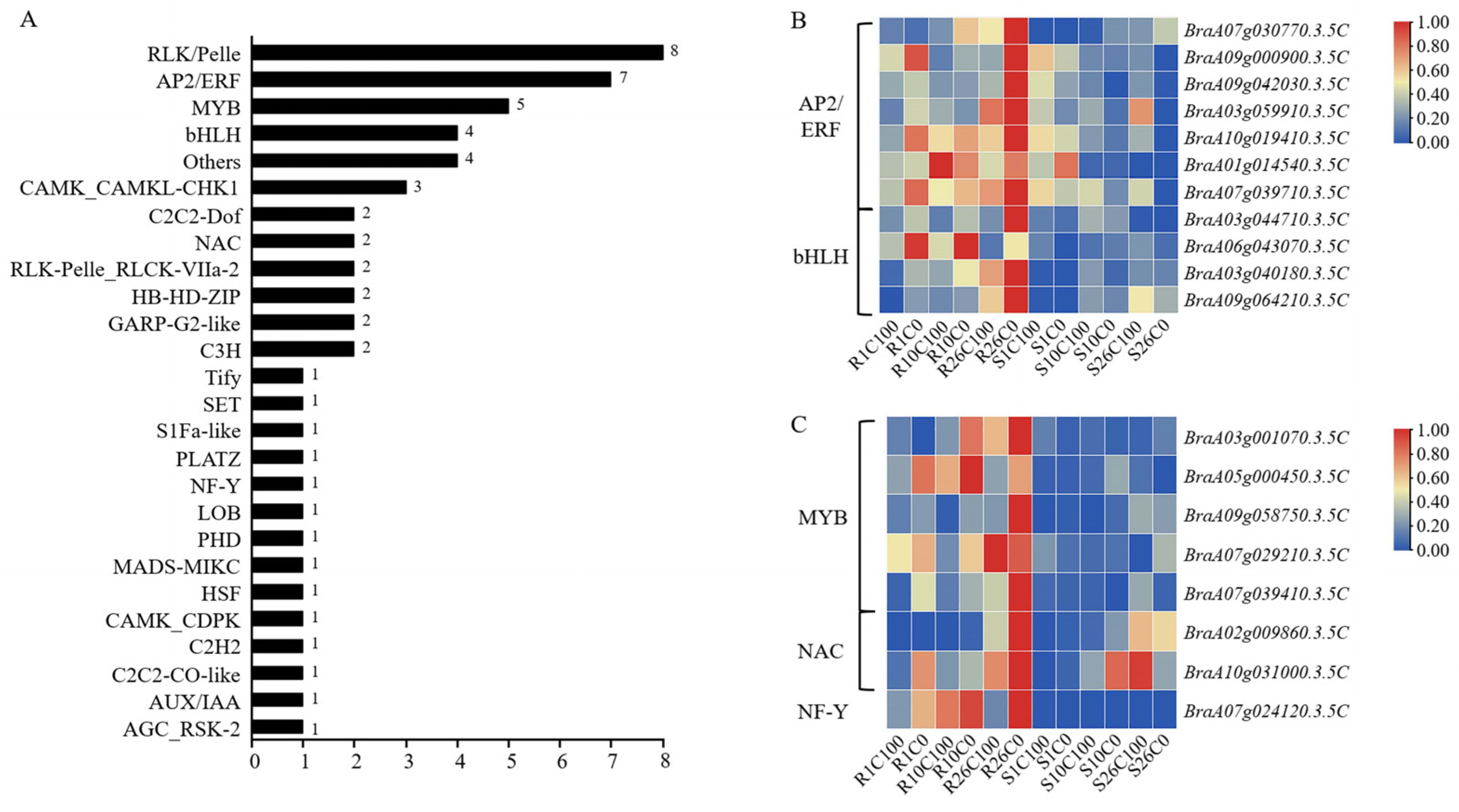
2.7.5. Transcription Factors Showed Different Expression Levels Between Treatments
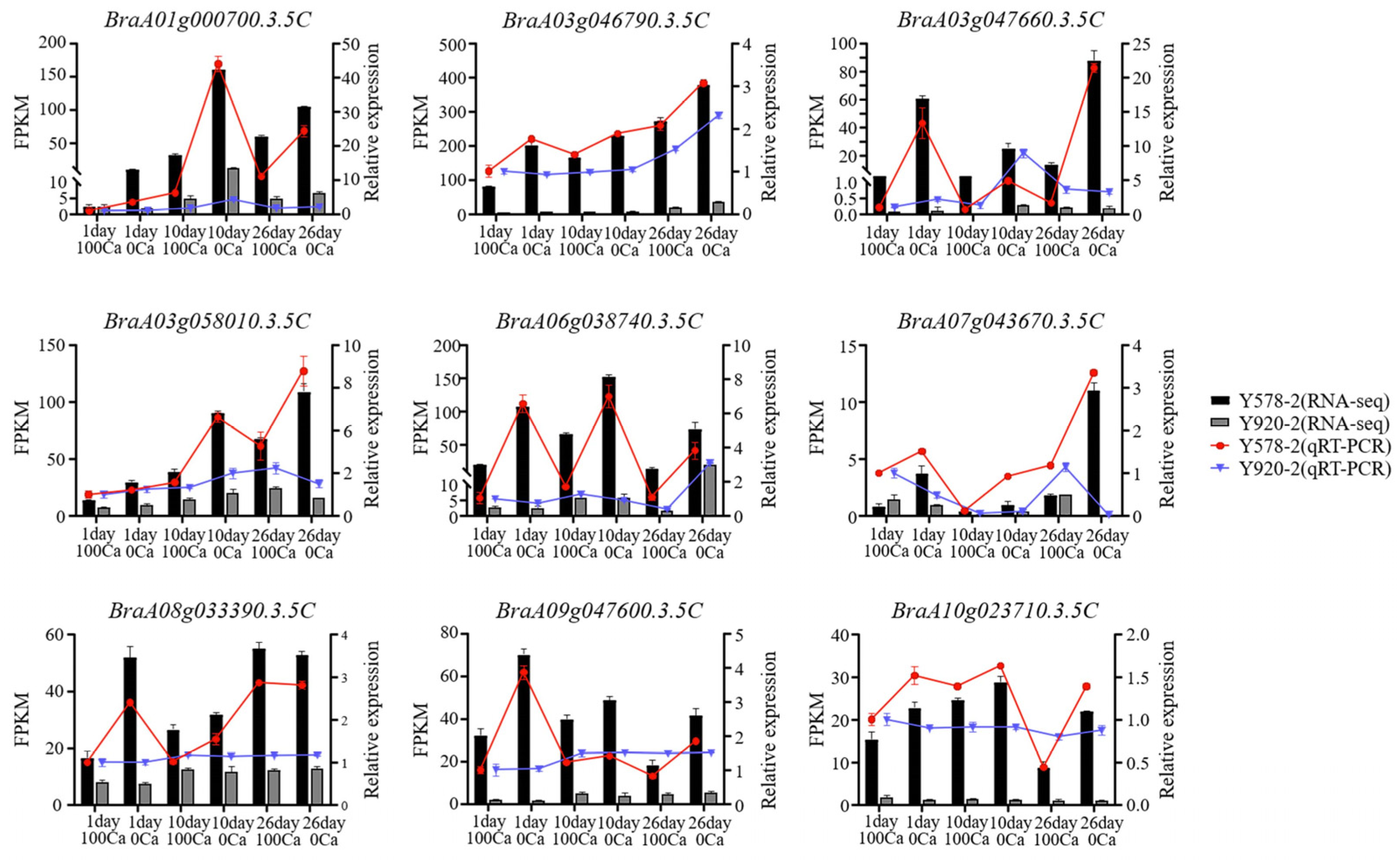
2.8. Verification of RNA-seq Accuracy Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Culture Conditions
4.2. Determination of Antioxidant Enzymes Activities
4.3. Library Construction and Sequencing
4.4. Differential Expression Analysis and Gene Annotation
4.5. Weighted Gene Co-Expression Network Analysis
4.6. qRT-PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, Y.; Tanaka, K. Transcriptional regulation of tetrapyrrole biosynthetic genes explains abscisic acid-induced heme accumulation in the unicellular red alga Cyanidioschyzon merolae. Front. Plant Sci. 2016, 7, 1300. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.C.; Wang, Y.J.; Zheng, X.Y. QTL mapping and analysis of leaf head related traits in Chinese cabbage. Sci. Agric. Sin. 2004, 37, 106–111. (In Chinese) [Google Scholar]
- Zhang, H.; Xu, Y.L.; Wang, C.N.; Huang, Z.Y.; Fan, W.Q.; Li, M.; Zhang, B. Development and validation of molecular markers for resistance to tipburn in Chinese cabbage. Acta Agric. Boreal-Sin. 2021, 36, 195–203. (In Chinese) [Google Scholar]
- Song, C.; Ye, X.Y.; Liu, G.Y.; Zhang, S.F.; Li, G.L.; Zhang, H.; Li, F.; Sun, R.F.; Wang, C.G.; Xu, D.H.; et al. Comprehensive evaluation of nutritional qualities of Chinese cabbage (Brassica rapa ssp. pekinensis) varieties based on multivariate statistical analysis. Horticulturae 2023, 9, 1264. [Google Scholar]
- Zhan, Y.D.; Tian, H.H.; Ji, X.L.; Liu, Y. Myzus persicae (Hemiptera: Aphididae) infestation increases the risk of bacterial contamination and alters nutritional content in storage Chinese cabbage. J. Sci. Food Agric. 2020, 100, 3007–3012. [Google Scholar] [CrossRef] [PubMed]
- Mcginty, R.A.; Thompson, R.C. Preliminary notes on tipburn of Lettuce. Agric. Food Sci. 1926, 27, 341–346. [Google Scholar]
- Lee, J.; Park, I.; Lee, Z.; Kim, S.W.; Baek, N.; Park, H.; Park, S.U.; Kwon, S.; Kim, H. Regulation of the major vacuolar Ca2+ transporter genes, by intercellular Ca2+ concentration and abiotic stresses, in tip-burn resistant Brassica oleracea. Mol. Biol. Rep. 2012, 40, 177–188. [Google Scholar] [CrossRef]
- Bangerth, F. Calcium-related physiological disorders of plants. Environ. Sci. Biol. 1979, 17, 97–122. [Google Scholar] [CrossRef]
- Aloni, B.; Pashkar, T.; Libel, R. The possible involvement of gibberellins and calcium in tipburn of Chinese cabbage: Study of intact plants and detached leaves. Plant Growth Regul. 1986, 4, 3–11. [Google Scholar] [CrossRef]
- HO, L.C.; ADAMS, P. Effects of diurnal changes in the salinity of the nutrient solution on the accumulation of calcium by tomato fruit. Ann. Bot. 1989, 64, 373–382. [Google Scholar] [CrossRef]
- Saure, M.C. Causes of the tipburn disorder in leaves of vegetables. Sci. Hortic. 1998, 76, 131–147. [Google Scholar] [CrossRef]
- Cui, Z.Y. The cause and control of Chinese cabbage tipburn. Mod. Rural Sci. Technol. 2019, 11, 31–32. (In Chinese) [Google Scholar]
- Su, T.B.; Yu, S.C.; Yu, R.F.; Zhang, F.L.; Yu, Y.J.; Zhang, D.S.; Zhao, X.Y.; Wang, W.H. Effects of endogenous salicylic acid during calcium deficiency-induced tipburn in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Report. 2015, 34, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.W.M.; Schorrak, L.M.; Smith, R.K.; Bent, A.F.; Sussman, M.R. A cyclic nucleotide-gated ion channel, CNGC2, is crucial for plant development and adaptation to calcium stress. Plant Physiol. 2003, 132, 728–731. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.Y.; He, M.R.; Wang, Z.L.; Zhang, J.W.; Song, Y.L.; He, Z.L.; Dong, Y.J. Application of nitric oxide and calcium nitrate enhances tolerance of wheat seedlings to salt stress. Plant Growth Regul. 2015, 77, 343–356. [Google Scholar] [CrossRef]
- White, P.J. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef]
- Dayod, M.; Tyerman, S.D.; Leigh, R.A.; Gilliham, M. Calcium storage in plants and the implications for calcium biofortification. Protoplasma 2010, 247, 215–231. [Google Scholar] [CrossRef]
- Liao, Y.; Wu, B.H.; Zeng, G.W.; Zhu, Z.J. Relation between changes in ultrastructure and composition of cell wall and calcium related physiological disorder of Chinese cabbage. Acta Phytophysiol. Sin. 2000, 2, 111–116+179. (In Chinese) [Google Scholar]
- Liu, M.L. Effects of Different Calcium Levels on the Growth and Development of Tipburn in Chinese Cabbage Seedlings. Master’s Thesis, Northwest A&F University, Yangling, China, 2014. (In Chinese). [Google Scholar]
- Su, T.B.; Li, P.R.; Wang, H.P.; Wang, W.H.; Zhao, X.Y.; Yu, Y.J.; Zhang, D.S.; Yu, S.C.; Zhang, F.L. Natural variation in a calreticulin gene causes reduced resistance to Ca2+ deficiency-induced tipburn in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Cell Environ. 2019, 42, 3044–3060. [Google Scholar] [CrossRef]
- Wang, W.H.; Wang, J.L.; Wei, Q.Z.; Li, B.Y.; Zhong, X.M.; Hu, T.H.; Hu, H.J.; Bao, C.L. Transcriptome-wide identification and characterization of circular RNAs in leaves of Chinese cabbage (Brassica rapa L. ssp. pekinensis) in response to calcium deficiency-induced tip-burn. Sci. Rep. 2019, 9, 14544. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.P.; Shen, C.W.; Yuan, R.H.; Zhang, H.X.; Xiao, Y.; Wang, X.L.; Pan, F.F.; Wu, C.H.; Li, Q.F.; Yuan, J.Y.; et al. Identification of genes related to tipburn resistance in Chinese cabbage and preliminary exploration of its molecular mechanism. BMC Plant Biol. 2021, 21, 567. [Google Scholar] [CrossRef]
- Zhang, S.; Gao, H.Z.; Wang, L.X.; Zhang, Y.H.; Zhou, D.D.; Anwar, A.; Li, J.J.; Wang, F.D.; Li, C.; Zhang, Y.; et al. Comparative transcriptome and co-expression network analyses reveal the molecular mechanism of calcium-deficiency-triggered tipburn in Chinese Cabbage (Brassica rapa L. ssp. Pekinensis). Plants 2022, 11, 3555. [Google Scholar] [CrossRef] [PubMed]
- Michael, J.B.; Martin, D.B.; Llewelyn, R.H. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar]
- Conn, S.; Gilliham, M.; Athman, A.; Schreiber, A.W.; Baumann, U.; Moller, I.; Cheng, N.H.; Stancombe, M.A.; Hirschi, K.D.; Webb, A.; et al. Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 2011, 23, 240–257. [Google Scholar] [CrossRef]
- Geisler, M.; Frangne, N.; Gomès, E.; Martinoia, E.; Palmgren, M.G. The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in Yeast. Plant Physiol. 2000, 124, 1814–1827. [Google Scholar] [CrossRef]
- Dominique, R. Modulating plant calcium for better nutrition and stress tolerance. Int. Sch. Res. Not. 2013, 2013, 952043. [Google Scholar]
- Cheng, N.; Liu, J.-Z.; Brock, A.M.; Nelson, R.S.; Hirschi, K.D. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J. Biol. Chem. 2006, 281, 26280–26288. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Li, X.F.; Su, M.K.; Shi, J.P.; Zhang, Q.L.; Liu, X.Y. LeGRXS14 reduces salt stress tolerance in Arabidopsis thaliana. Plants 2023, 12, 2320. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Sun, Y.; Wang, C.C.; Zhang, W.L.; Sun, M.H.; Hu, H.T.; Liu, J.Z.; Yang, L. A rice CPYC-type glutaredoxin OsGRX20 in protection against bacterial blight, methyl viologen and salt stresses. Front. Plant Sci. 2018, 9, 111. [Google Scholar] [CrossRef]
- Chen, K.S.; Li, F.; Zhang, S.L. Changes of glycosyltransferase mRNA level in xyloglucan during fruit ripening of kiwifruit. Acta Bot. Sin. 1999, 41, 1231–1234. (In Chinese) [Google Scholar]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [PubMed]
- Lauri, V.; Julia, S.; Thorsten, H. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932. [Google Scholar]
- Hu, K.M.; Cao, J.B.; Zhang, J.; Xia, F.; Ke, Y.G.; Zhang, H.T.; Xie, W.Y.; Liu, H.B.; Cui, Y.; Cao, Y.L.; et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 2017, 3, 17009. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Xie, D.S.; Ma, L.; Jozef, S.; Xu, C.X. Immunohistochemical analysis of cell wall hydroxyproline-rich glycoproteins in the roots of resistant and susceptible wax gourd cultivars in response to Fusarium oxysporum f. sp. Benincasae infection and fusaric acid treatment. Plant Cell Rep. 2011, 30, 1555–1569. [Google Scholar] [CrossRef]
- Niu, Y.; Hu, B.; Li, X.; Chen, H.; Takac, T.; Samaj, J.; Xu, C. Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int. J. Mol. Sci. 2018, 19, 350. [Google Scholar] [CrossRef] [PubMed]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Ma, L.; Jiang, S.; Lin, G.; Cai, J.; Ye, X.; Chen, H.; Li, M.; Li, H.; Takac, T.; Samaj, J.; et al. Wound-induced pectin methylesterases enhance banana (Musa spp. AAA) susceptibility to Fusarium oxysporum f. sp. cubense. J. Exp. Bot. 2013, 64, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.L.; Takac, T.; Li, X.Q.; Chen, H.B.; Wang, Y.Y.; Xu, E.F.; Xie, L.; Su, Z.H.; Samaj, J.; Xu, C.X. Variable content and distribution of arabinogalactan proteins in banana (Musa spp.) under low temperature stress. Front. Plant Sci. 2015, 6, 353. [Google Scholar] [CrossRef] [PubMed]
- Engelsdorf, T.; Gigli-Bisceglia, N.; Veerabagu, M.; McKenna, J.F.; Vaahtera, L.; Augstein, F.; Zipfel, C.; Hamann, T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018, 11, eaao3070. [Google Scholar] [CrossRef] [PubMed]
- Solmaz, I.; Brett, T.; Waldner, M.; Nayidu, N.; Tu, J.Y.; Kusalik, A.J.; Todd, C.D.; Wei, Y.D.; Bonham-Smith, P.C. Transcriptome analysis of response to Plasmodiophora brassicae infection in the Arabidopsis shoot and root. BMC Genom. 2018, 19, 23. [Google Scholar]
- Song, Q.M.; Li, D.Y.; Zhang, H.J.; Song, F.M. Biological function and research progress of plant NF-Y transcription factors. J. Plant Physiol. 2015, 51, 623–632. (In Chinese) [Google Scholar]
- Ten, H.C.; Bochdanovits, Z.; Jansweijer, V.M.A.; Koning, F.G.; Berke, L.; Sanchez-Perez, G.F.; Scheres, B.; Heidstra, R. Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. Report. 2011, 76, 69–83. [Google Scholar]
- Hwan, J.E.; Won, J.H.; Chul, L.S.; Wook, H.S.; Sunggi, H.; Kook, H.B. Identification of a novel pathogen-induced gene encoding a leucine-rich repeat protein expressed in phloem cells of Capsicum annuum. Biochim. Biophys. Acta 2004, 1676, 211–222. [Google Scholar]
- Xu, Z.S.; Xiong, T.F.; Ni, Z.Y.; Chen, X.P.; Chen, M.; Li, L.C.; Gao, D.Y.; Yu, X.D.; Liu, P.; Ma, Y.Z. Isolation and identification of two genes encoding leucine-rich repeat (LRR) proteins differentially responsive to pathogen attack and salt stress in tobacco. Plant Sci. 2008, 176, 38–45. [Google Scholar] [CrossRef]
- Garcia, M.E.; Lynch, T.; Peeters, J.; Snowden, C.; Finkelstein, R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 2008, 67, 643–658. [Google Scholar] [CrossRef]
- Zhang, M.J.; Zhu, L.; Xia, Q.Z. Research progress on the regulation of plant hormones on stress response. J. Hubei Univ. (Nat. Sci. Ed.) 2021, 43, 242–253+263. (In Chinese) [Google Scholar]
- Rainer, W.; Charles, A.S.; PoKai, H.; Yohei, T.; Shintaro, M.; Julian, I.S. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar]
- Yang, D.L.; Yang, Y.N.; He, Z.H. Roles of plant hormones and their interplay in rice immunity. Mol. Plant 2013, 6, 675–685. [Google Scholar] [CrossRef]
- Liu, W.C.; Song, R.F.; Zheng, S.Q.; Li, T.T.; Zhang, B.L.; Gao, X.; Lu, Y.T. Coordination of plant growth and abiotic stress responses by tryptophan synthase β subunit1 through modulating tryptophan and ABA homeostasis in Arabidopsis. Mol. Plant 2022, 15, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Y.; Wang, Y.B.; Liu, X.L.; Ma, L.; Zhang, Z.J.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.J.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Q. Research trends of photosynthesis. Science 2022, 74, 16–20+14. (In Chinese) [Google Scholar]
- Guan, S.H.; Chai, Y.Q.; Hao, Q.; Ma, Y.D.; Wan, W.L.; Liu, H.Y.; Diao, M. Transcriptomic and physiological analysis reveals crucial biological pathways associated with low-temperature stress in Tunisian soft-seed pomegranate ( Punica granatum L. ). J. Plant Interact. 2023, 18, 2152887. [Google Scholar] [CrossRef]
- Hoon, L.S.; Atikur, R.M.; Woo, K.K.; Wook, L.J.; Chung, J.H.; Jun, C.G.; Yowook, S.; Won, L.K. Screening of multiple abiotic stress-induced genes in Italian ryegrass leaves. J. Korean Soc. Grassl. Forage Sci. 2018, 38, 190–195. [Google Scholar]
- Yang, H.; Dai, L.J.; Wei, Y.X.; Deng, Z.; Li, D.J. Melatonin enhances salt stress tolerance in rubber tree (Hevea brasiliensis) seedlings. Ind. Crops Prod. 2020, 145, 111990. [Google Scholar] [CrossRef]
- Han, Y.; Zhao, J.F.; Cai, C.; Peng, Y.F.; Zhang, M.J.; Sha, W.; Ma, T.Y. Research progress of light-harvesting chlorophyll a/b binding protein gene in plant adversity stress. J. Sci. Teach. Coll. 2023, 43, 79–83. (In Chinese) [Google Scholar]
- Zhao, Q.; Ru, J.N.; Li, Y.T.; Wang, C.; Xu, Z.S.; Wang, R.H. Identification and expression pattern analysis of Lhc gene family in wheat. J. Plant Genet. Resour. 2022, 23, 1766–1781. (In Chinese) [Google Scholar]
- Zhang, Y.; Raza, A.; Huang, H.; Su, W.; Luo, D.; Zeng, L.; Ding, X.Y.; Cheng, Y.; Liu, Z.F.; Li, Q.A.; et al. Analysis of Lhcb gene family in rapeseed (Brassica napus L.) identifies a novel member “BnLhcb3.4” modulating cold tolerance. Environ. Exp. Bot. 2022, 198, 104848. [Google Scholar] [CrossRef]
- Wu, Q.Y.; Yang, J.; Cheng, N.H.; Hirschi, K.D.; White, F.F.; Park, S.H. Glutaredoxins in plant development, abiotic stress response, and iron homeostasis: From model organisms to crops. Environ. Exp. Bot. 2017, 139, 91–98. [Google Scholar] [CrossRef]
- Christine, F.H.; Graham, N. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar]
- Galant, A.; Preuss, M.L.; Cameron, J.; Jez, J.M. Plant glutathione biosynthesis: Diversity in biochemical regulation and reaction products. Front. Plant Sci. 2011, 2, 45. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.C.; Ślesak, I.; Jordá, L.a.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 2009, 150, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.; Brian, W.; Geo, P.; Ali, M.; Gordon, K.; Marijke, B.; Steven, S.; Barbara, W.; Lior, P. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar]
- Michael, I.L.; Wolfgang, H.; Simon, A. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar]
- Peter, L.; Steve, H. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar]
- Thomas, D.S.; Kenneth, J.L. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]

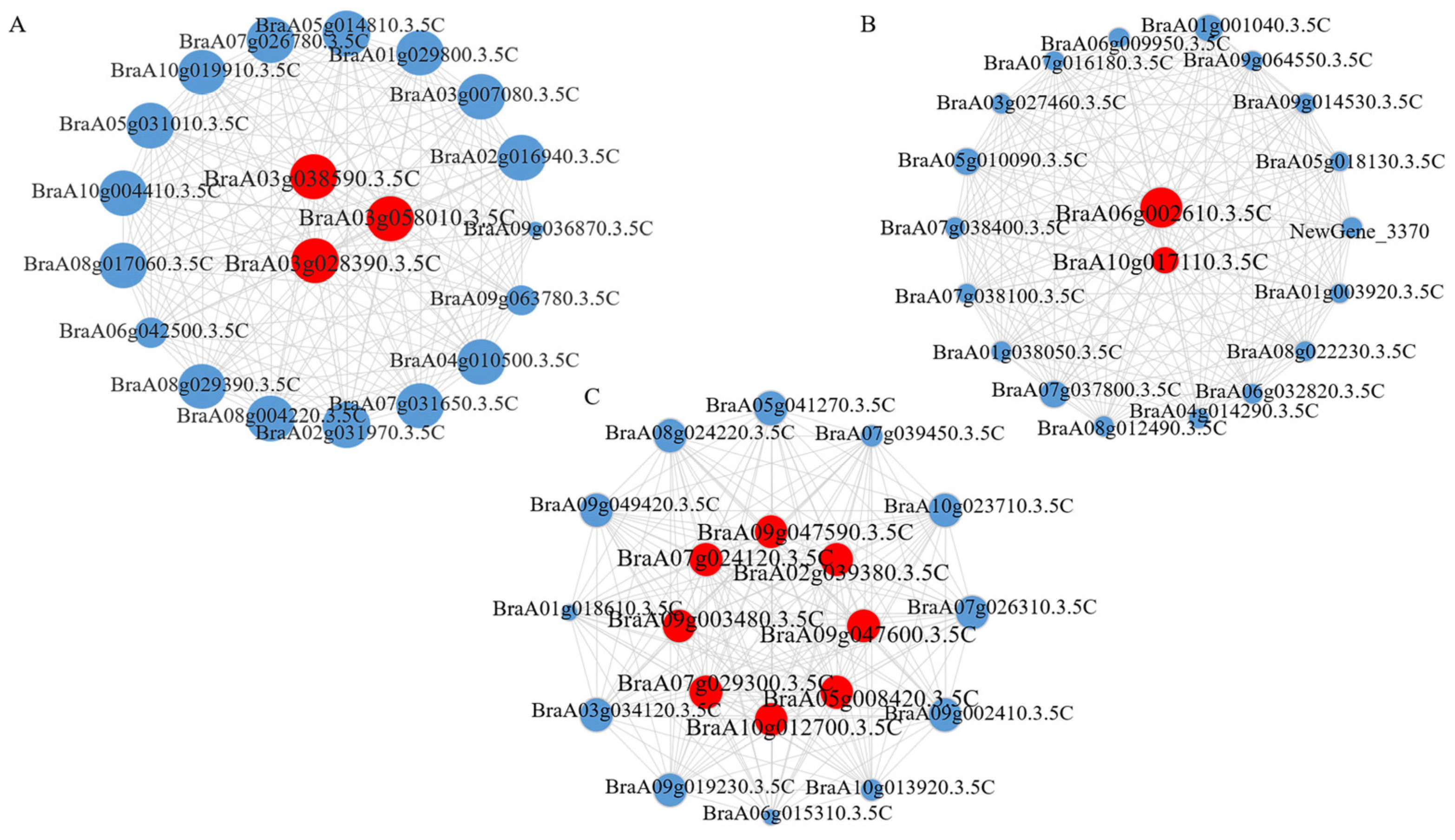
| Gene ID | Location | A. thaliana | Gene Function |
|---|---|---|---|
| BraA02g039380.3.5C | 26,487,248–26,488,449 (A02) | AT3G27690 (LHCB2) | It encodes the light-harvesting chlorophyll a/b-binding (Lhc) proteins. |
| BraA03g028390.3.5C | 14,016,000–14,017,835 (A03) | AT4G04020 (FIB) | It is regulated by abscisic acid response regulators. It is involved in abscisic acid-mediated photoprotection. |
| BraA03g038590.3.5C | 18,860,995–18,862,826 (A03) | AT3G17800 | It encodes a plant G-box-binding factor. |
| BraA03g058010.3.5C | 29,927,476–29,929,090 (A03) | AT4G31870 (GPX7) | It encodes glutathione peroxidase. |
| BraA05g008420.3.5C | 4,412,814–4,414,921 (A05) | AT2G36870 (XTH32) | It encodes a xyloglucan endotransglycosylase. |
| BraA06g002610.3.5C | 1,539,146–1,540,183 (A06) | AT1G51620 | It encodes LRR receptor-like serine/threonine-protein kinase (LRR-RLK). |
| BraA07g024120.3.5C | 18,613,864–18,614,418 (A07) | AT1G56170 (NF-YC2) | It encodes a nuclear transcription factor Y (NF-Y). |
| BraA07g029300.3.5C | 21,156,877–21,158,706 (A07) | AT1G74420 (FUT3) | It encodes xyloglucan fucosyltransferase, based on similarity to FUT1, but not functionally redundant with FUT1. |
| BraA09g003480.3.5C | 2,050,532–2,051,720 (A09) | AT3G27690 (LHCB2) | It encodes the light-harvesting chlorophyll a/b-binding (Lhc) proteins that constitute the antenna system of the photosynthetic apparatus. |
| BraA09g047590.3.5C | 34,898,886–34,900,595 (A09) | AT3G54890 (LHCA1) | It encodes the light-harvesting chlorophyll a/b-binding (Lhc) proteins. |
| BraA09g047600.3.5C | 34,901,365–34,902,146 (A09) | AT3G54900 (GRXS14) | It encodes glutaredoxin-S14/CAX1 interaction protein. It activates the CAX1 gene calcium transport activity and plays a role in plant signal transduction and response to stress. |
| BraA10g012700.3.5C | 10,248,439–10,249,537 (A10) | AT5G54270 (LHCB3) | It encodes the light-harvesting chlorophyll a/b-binding (Lhc) proteins. |
| BraA10g017110.3.5C | 12,924,138–12,926,370 (A10) | AT5G59220 (PP2C) | It is a negative regulator of osmotic stress and ABA signaling. |
| Material | Sampling Time | Calcium Treatment | Sample Number | ||
|---|---|---|---|---|---|
| Y578-2 (R) | 1 DAT | 100Ca | R1C100-1 | R1C100-2 | R1C100-3 |
| 0Ca | R1C0-1 | R1C0-2 | R1C0-3 | ||
| 10 DAT | 100Ca | R10C100-1 | R10C100-2 | R10C100-3 | |
| 0Ca | R10C0-1 | R10C0-2 | R10C0-3 | ||
| 26 DAT | 100Ca | R26C100-1 | R26C100-2 | R26C100-3 | |
| 0Ca | R26C0-1 | R26C0-2 | R26C0-3 | ||
| Y920-2 (S) | 1 DAT | 100Ca | S1C100-1 | S1C100-2 | S1C100-3 |
| 0Ca | S1C0-1 | S1C0-2 | S1C0-3 | ||
| 10 DAT | 100Ca | S10C100-1 | S10C100-2 | S10C100-3 | |
| 0Ca | S10C0-1 | S10C0-2 | S10C0-3 | ||
| 26 DAT | 100Ca | S26C100-1 | S26C100-2 | S26C100-3 | |
| 0Ca | S26C0-1 | S26C0-2 | S26C0-3 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bi, Y.; Zhang, W.; Yuan, Y.; Feng, J.; Wang, P.; Ding, C.; Zhao, Y.; Li, L.; Su, H.; Tian, B.; et al. Transcriptome Analysis Revealed Hub Genes Related to Tipburn Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plants 2025, 14, 527. https://doi.org/10.3390/plants14040527
Bi Y, Zhang W, Yuan Y, Feng J, Wang P, Ding C, Zhao Y, Li L, Su H, Tian B, et al. Transcriptome Analysis Revealed Hub Genes Related to Tipburn Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plants. 2025; 14(4):527. https://doi.org/10.3390/plants14040527
Chicago/Turabian StyleBi, Yaning, Wenjing Zhang, Yuxiang Yuan, Jianqi Feng, Peiyun Wang, Cong Ding, Yanyan Zhao, Lin Li, Henan Su, Baoming Tian, and et al. 2025. "Transcriptome Analysis Revealed Hub Genes Related to Tipburn Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis)" Plants 14, no. 4: 527. https://doi.org/10.3390/plants14040527
APA StyleBi, Y., Zhang, W., Yuan, Y., Feng, J., Wang, P., Ding, C., Zhao, Y., Li, L., Su, H., Tian, B., Wei, F., Wei, X., & Zhang, X. (2025). Transcriptome Analysis Revealed Hub Genes Related to Tipburn Resistance in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plants, 14(4), 527. https://doi.org/10.3390/plants14040527







