Genetic Basis of Seedling Root Traits in Common Wheat (Triticum aestivum L.) Identified by Genome-Wide Linkage Mapping
Abstract
1. Introduction
2. Results
2.1. Phenotypic Evaluation of Wheat Seedling Root Traits
2.2. Construction of the Linkage Map
2.3. QTL Detection for Seedling Root Traits
2.4. KASP Markers Development and Validation
2.5. Candidate Gene Identification
3. Discussion
3.1. Seven Novel Loci for Wheat Root System Traits Were Identified
3.2. Candidate Genes for Wheat Seedling Root Traits
3.3. Application of the KASP Markers for Wheat MAS Breeding
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotype Evaluation
4.3. Linkage Map Construction
4.4. Linkage Mapping and KASP Marker Development
4.5. Identification of Candidate Genes for Wheat Seedling Root Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| RL | Root length |
| RA | Root surface area |
| RV | Root volume |
| RT | Number of root tips |
| RW | Root dry weight |
| RIL | Recombinant inbred lines |
| SNP | Single-nucleotide polymorphism |
| QTL | Quantitative trait loci |
| KASP | Kompetitive allele-specific PCR |
| MAS | Marker-assisted selection |
| PVE | Phenotypic variance explained |
References
- Rogers, E.D.; Benfey, P.N. Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotech. 2015, 32, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, L.; Reynolds, M.P.; Wang, J.; Chang, X.; Mao, X.; Jing, R. Recognizing the hidden half in wheat: Root system attributes associated with drought tolerance. J. Exp. Bot. 2021, 72, 5117–5133. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Q.; Błaszkiewicz, Z.; Sun, J.; Sun, Q.; He, R.; Li, Y. Phenotyping for the dynamics of field wheat root system architecture. Sci. Rep. 2017, 7, 37649. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Roumet, P.; Salon, C.; Jeudy, C.; Lamboeuf, M.; Lafarge, S.; Dubois, F.; Habash, D.; Liu, Y.; Nadot, S. Genetic analysis of platform-phenotyped root system architecture of bread and durum wheat in relation to agronomic traits. Front. Plant Sci. 2022, 13, 853601. [Google Scholar] [CrossRef]
- Maqbool, S.; Hassan, M.A.; Xia, X.; York, L.M.; Rasheed, A.; He, Z. Root system architecture in cereals: Progress, challenges and perspective. Plant J. 2022, 110, 23–42. [Google Scholar] [CrossRef]
- Shaheen, A.; Li, Z.; Yang, Y.; Xie, J.; Zhu, L.; Li, C. Genetic regulation of wheat plant architecture and future prospects for its improvement. New Crops 2024, 14, 100048. [Google Scholar] [CrossRef]
- Ma, S.C.; Wang, T.C.; Guan, X.K.; Zhang, X. Effect of sowing time and seeding rate on yield components and water use efficiency of winter wheat by regulating the growth redundancy and physiological traits of root and shoot. Field Crops Res. 2018, 221, 166–174. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, B.C.; Turner, N.C.; Li, F.M. Grain yield, dry matter accumulation and remobilization, and root respiration in winter wheat as affected by seeding rate and root pruning. Eur. J. Agron. 2010, 33, 257–266. [Google Scholar] [CrossRef]
- Nagar, C.K.; Bharati, A.; Sinha, S.K.; Venkatesh, K.; Mandal, P.K. Nitrogen stress induced changes in root system architecture (RSA) in diverse wheat (T. aestivum L.) genotypes at seedling stage. J. Cereal Sci. 2018, 80, 10–16. [Google Scholar] [CrossRef][Green Version]
- Středa, T.; Hajzlerová, J.; Alba-Mejía, J.; Jovanović, I.; Frantová, N.; Středová, H. Quo vadis, breeding for an efficient root system, in the era of climate change? Czech. J. Genet. Plant Breed. 2024, 60, 181–211. [Google Scholar] [CrossRef]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Vyhnánek, T.; Zargar, S.M.; Kahraman, A.; Gezgin, S. Exploring strigolactones for inducing abiotic stress tolerance in plants. Czech. J. Genet. Plant Breed. 2024, 60, 55–69. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Liu, J.; He, Z.; Rasheed, A.; Wen, W.; Yan, J.; Zhang, P.; Tao, Y.; Li, B.; Zhang, M. Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 220. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wang, Y.; Liu, J.; Wang, F.; Qiu, X.; Liu, P. Genome-wide linkage mapping of root system architecture-related traits in common wheat (Triticum aestivum L.). Front. Plant Sci. 2023, 14, 1274392. [Google Scholar] [CrossRef] [PubMed]
- Ragland, D.I. The Evaluation of Root System Architecture (RSA) As a New Breeding Target for Climate-Resilient Winter Wheat (Triticum aestivum L.). Ph.D. Dissertation, Virginia Tech, Blacksburg, VA, USA, 2024. [Google Scholar]
- Wang, H.; Yang, B.; Zhao, X.; Chen, H.; Liu, F.; Ru, Y. Identification of novel QTL for seedling root architectural traits in the D genome of natural and resynthetic allohexaploid wheat. Agronomy 2024, 14, 608. [Google Scholar] [CrossRef]
- Rufo, R.; Salvi, S.; Royo, C.; Soriano, J.M. Exploring the genetic architecture of root-related traits in Mediterranean bread wheat landraces by genome-wide association analysis. Agronomy 2020, 10, 613. [Google Scholar] [CrossRef]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Canè, M.A.; Colalongo, M.C. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. Bot. 2016, 67, 1161–1178. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.M.; Alvaro, F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 2019, 9, 10537. [Google Scholar] [CrossRef]
- Liu, P.; Jin, Y.; Liu, J.; Liu, C.; Yao, H.; Luo, F.; Gao, Y.; Guo, F.; Wanjiru, B. Genome-wide association mapping of root system architecture traits in common wheat (Triticum aestivum L.). Euphytica 2019, 215, 121. [Google Scholar] [CrossRef]
- Mehrabi, A.A.; Pour-Aboughadareh, A.; Mansouri, S.; Hosseini, A. Genome-wide association analysis of root system architecture features and agronomic traits in durum wheat. Mol. Breed. 2020, 40, 55. [Google Scholar] [CrossRef]
- Alemu, A.; Feyissa, T.; Maccaferri, M.; Sciara, G.; Tuberosa, R.; Ammar, K.; Ghedini, M.; Jordi, M.; Drinić, F. Genome-wide association analysis unveils novel QTLs for seminal root system architecture traits in Ethiopian durum wheat. BMC Genom. 2021, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Xu, N.; Zhang, J.; Ren, K.; Wu, J.; Wang, C.; Luo, Y.; Hu, X.; Fan, N.; Zhang, Y. Combined genome-wide association studies (GWAS) and linkage mapping identifies genomic regions associated with seedling root system architecture (RSA) under different nitrogen conditions in wheat (Triticum aestivum L.). Agriculture 2024, 14, 1652. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Pal, N.; Chahal, A.; Srivastava, P.; Gupta, P.K. Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plant. 2021, 27, 2245–2267. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Awadalla, R.A.; Elshamy, M.M.; Börner, A.; Heikal, Y.M. Genome-wide analysis for root and leaf architecture traits associated with drought tolerance at the seedling stage in a highly ecologically diverse wheat population. Comput. Struct. Biotechnol. J. 2024, 23, 870–882. [Google Scholar] [CrossRef]
- Siddiqui, N.; Gabi, M.T.; Kamruzzaman, M.; Ambaw, A.M.; Teferi, T.J.; Dadshani, S. Genetic dissection of root architectural plasticity and identification of candidate loci in response to drought stress in bread wheat. BMC Genom. 2023, 24, 38. [Google Scholar] [CrossRef]
- Zaman, Z.; Iqbal, R.; Jabbar, A.; Zahra, N.; Saleem, B.; Kiran, A.; Ullah, S.; Akbar, T.; Khan, M.R. Genetic signature controlling root system architecture in diverse spring wheat germplasm. Physiol. Plant. 2024, 176, e14183. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.J.; Wang, C.R.; Hassan, M.A.; Wu, Y.Y.; Xia, X.C.; Shi, S.B.; Liu, Y.L.; Xie, J.Y.; He, Z.H. QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J. Integr. Agric. 2021, 20, 1180–1192. [Google Scholar] [CrossRef]
- Liu, R.; Wu, F.; Xin, Y.; Yu, L.; Wang, Z.; Liu, S.; Cheng, Y.; Wang, L.; Zhao, G. Quantitative trait loci analysis for root traits in synthetic hexaploid wheat under drought stress conditions. J. Integr. Agric. 2020, 19, 1947–1960. [Google Scholar] [CrossRef]
- Lu, Y.; Xing, L.; Xing, S.; Hu, P.; Cui, C.; Zhang, M. Characterization of a putative new semi-dominant reduced height gene, RHT_NM9, in Wheat (Triticum aestivum L.). J. Genet. Genom. 2015, 42, 685–698. [Google Scholar] [CrossRef]
- Ren, T.; Hu, Y.; Tang, Y.; Li, C.; Yan, B.; Ren, Z. Utilization of a wheat 55K SNP array for mapping of major QTL for temporal expression of the tiller number. Front. Plant Sci. 2018, 9, 333. [Google Scholar] [CrossRef]
- Gasperini, D.; Greenland, A.; Hedden, P.; Dreos, R.; Harwood, W.; Griffiths, S. Genetic and physiological analysis of RHT8 in bread wheat: An alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J. Exp. Bot. 2012, 63, 4419–4436. [Google Scholar]
- Chai, L.; Chen, Z.; Bian, R.; Zhai, H.; Cheng, X.; Peng, H.; Liu, X.; Sun, G.; Yuan, Y. Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 1815–1831. [Google Scholar] [CrossRef]
- Chai, L.; Xin, M.; Dong, C.; Chen, Z.; Zhai, H.; Zhuang, J.; Feng, H.; Zhang, Y. A natural variation in Ribonuclease H-like gene underlies RHT8 to confer Green Revolution trait in wheat. Mol. Plant 2022, 15, 377–380. [Google Scholar] [CrossRef]
- Haque, M.; Martinek, P.; Watanabe, N.; Kuboyama, T. Genetic mapping of gibberellic acid-sensitive genes for semi-dwarfism in durum wheat. Cereal Res. Commun. 2011, 39, 171–178. [Google Scholar] [CrossRef]
- Duan, S.; Cui, C.; Chen, L.; Yang, Z.; Hu, Y.G.; Ghedini, M.; Rossi, L. Fine mapping and candidate gene analysis of dwarf gene RHT14 in durum wheat (Triticum durum). Funct. Integr. Genom. 2022, 22, 141–152. [Google Scholar] [CrossRef]
- Liu, H.; Shi, Z.; Ma, F.; Xu, Y.; Han, G.; Zhang, J.; Liu, D.; An, D. Identification and validation of plant height, spike length and spike compactness loci in common wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 568. [Google Scholar] [CrossRef]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Yu, S.M. A novel class of gibberellin 2-oxidases control semi-dwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Strader, L.C. Auxin-abscisic acid interactions in plant growth and development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Kondhare, K.R.; Patil, A.B.; Giri, A.P. Auxin: An emerging regulator of tuber and storage root development. Plant Sci. 2021, 306, 110854. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Ashraf, M.; Jamil, A. Seed enhancement with cytokinins: Changes in growth and grain yield in salt stressed wheat plants. Plant Growth Regul. 2006, 50, 29–39. [Google Scholar] [CrossRef]
- Lopez-Valdivia, I.; Perkins, A.C.; Schneider, H.M.; Vallebueno-Estrada, M.; Burridge, J.D.; Vielle-Calzada, J.P. Gradual domestication of root traits in the earliest maize from Tehuacán. Proc. Natl. Acad. Sci. USA 2022, 119, e2110245119. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Hao, C.; Li, T.; Liu, Y.; Wang, X.; Ma, F.; Zhang, J.; Shi, Z. The auxin response factor TaARF15-A1 negatively regulates senescence in common wheat (Triticum aestivum L.). Plant Physiol. 2023, 191, 1254–1271. [Google Scholar] [CrossRef]
- Cho, H.; Ryu, H.; Rho, S.; Hill, K.; Smith, S.; Audenaert, D.; Peng, J.; Yang, Y.; Park, J.Y.; Jang, S.C. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014, 16, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Ober, E.S.; Alahmad, S.; Cockram, J.; Forestan, C.; Hickey, L.T.; Kant, J.; Watt, M. Wheat root systems as a breeding target for climate resilience. Theor. Appl. Genet. 2021, 134, 1645–1662. [Google Scholar] [CrossRef] [PubMed]
- Vysotskaya, L.B.; Timergalina, L.N.; Simonyan, M.V.; Veselov, S.Y.; Kudoyarova, G.R. Growth rate, IAA and cytokinin content of wheat seedling after root pruning. Plant Growth Regul. 2001, 33, 51–57. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Zhao, Y.; Li, Y.; Zhang, G.; Peng, Z.; Zhang, J. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 2018, 16, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, R.; Hu, X.; Wang, D.; Wang, Y.; Xue, R.; Duan, G.; Zhai, H.; Zhang, Y. Overexpression of wheat C2H2 zinc finger protein transcription factor TaZAT8-5B enhances drought tolerance and root growth in Arabidopsis thaliana. Planta 2024, 260, 126. [Google Scholar] [CrossRef]
- Feng, L.; Gao, Z.; Xiao, G.; Huang, R.; Zhang, H.; Li, Q. Leucine-rich repeat receptor-like kinase FON1 regulates drought stress and seed germination by activating the expression of ABA-responsive genes in rice. Plant Mol. Biol. Rep. 2014, 32, 1158–1168. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
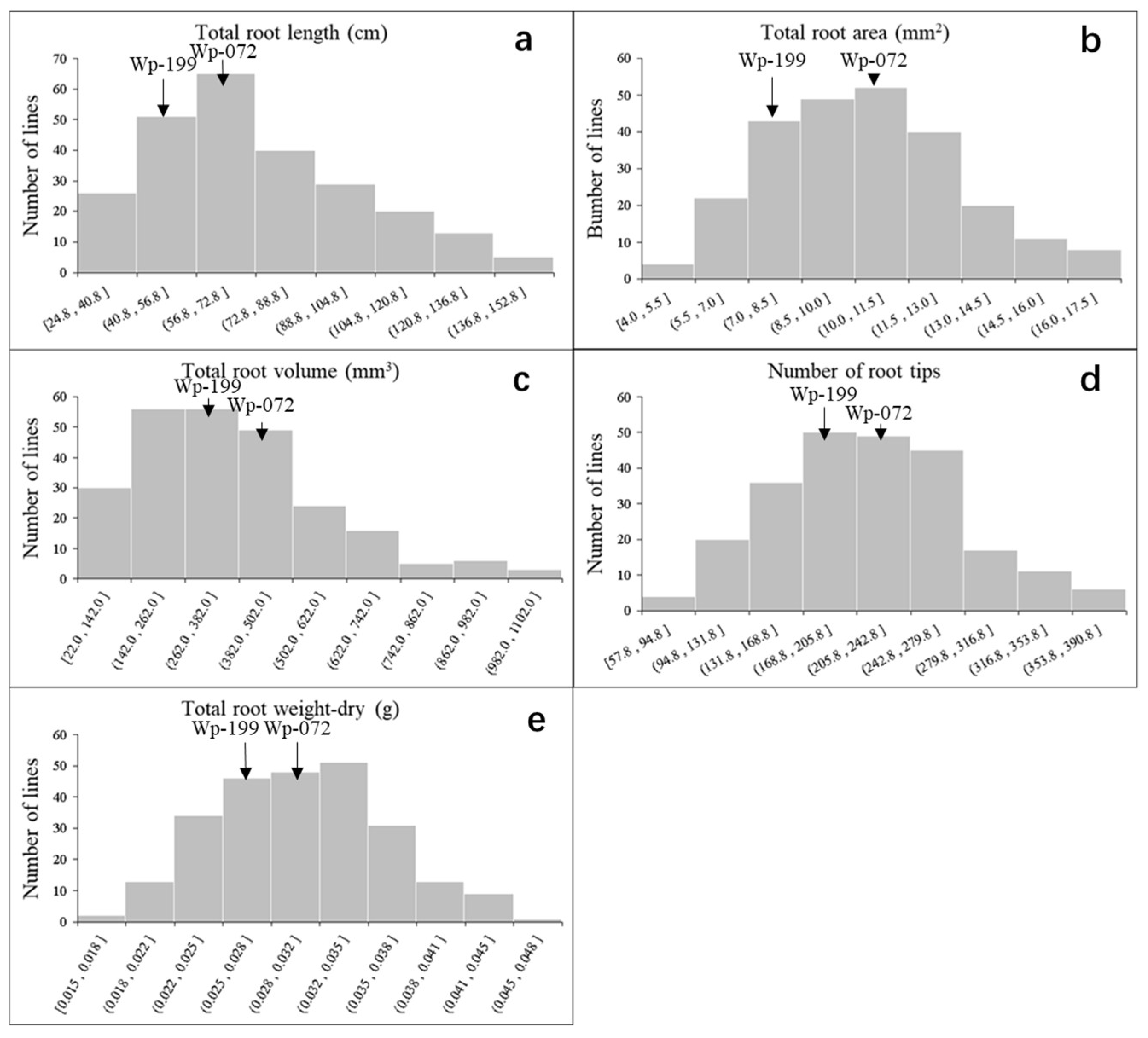
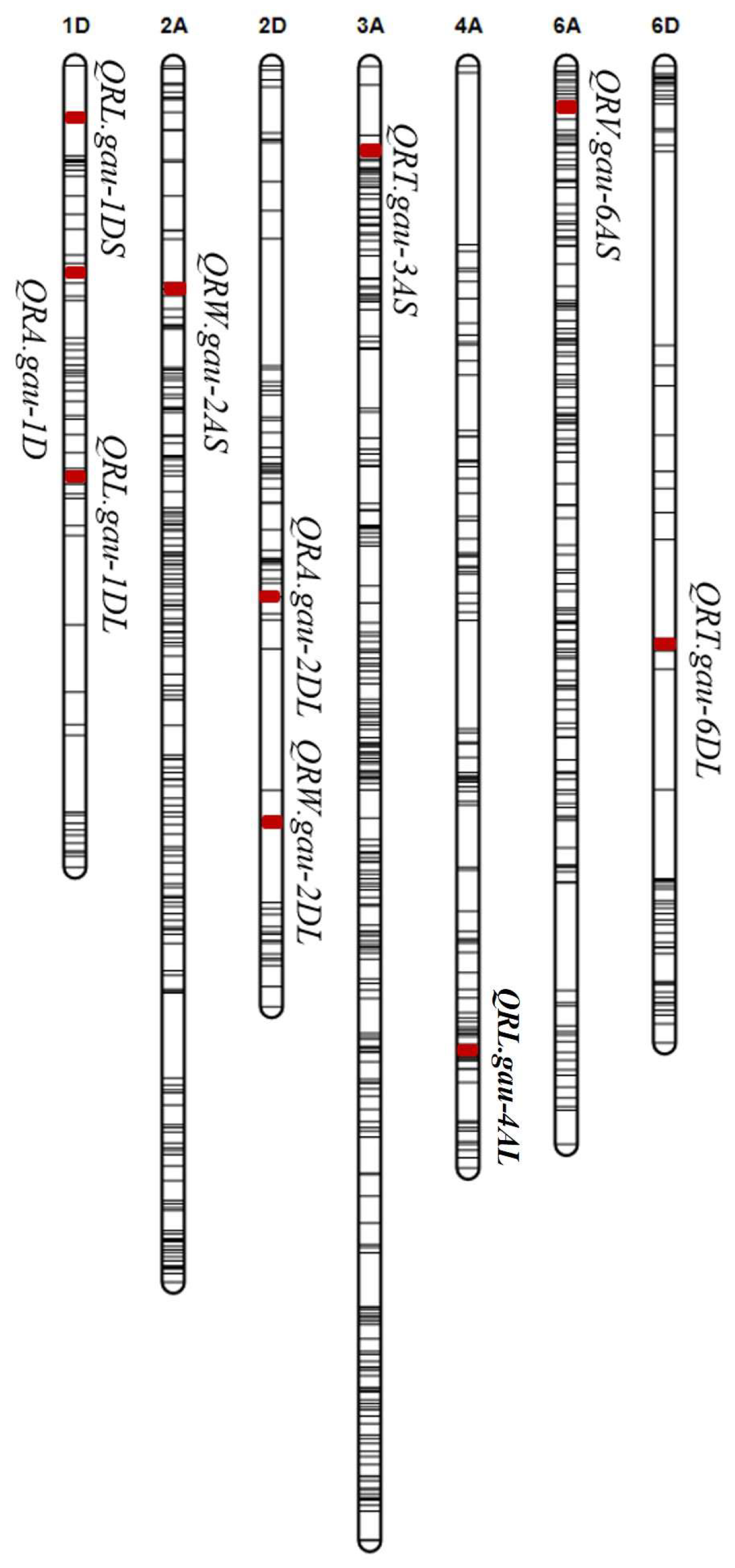
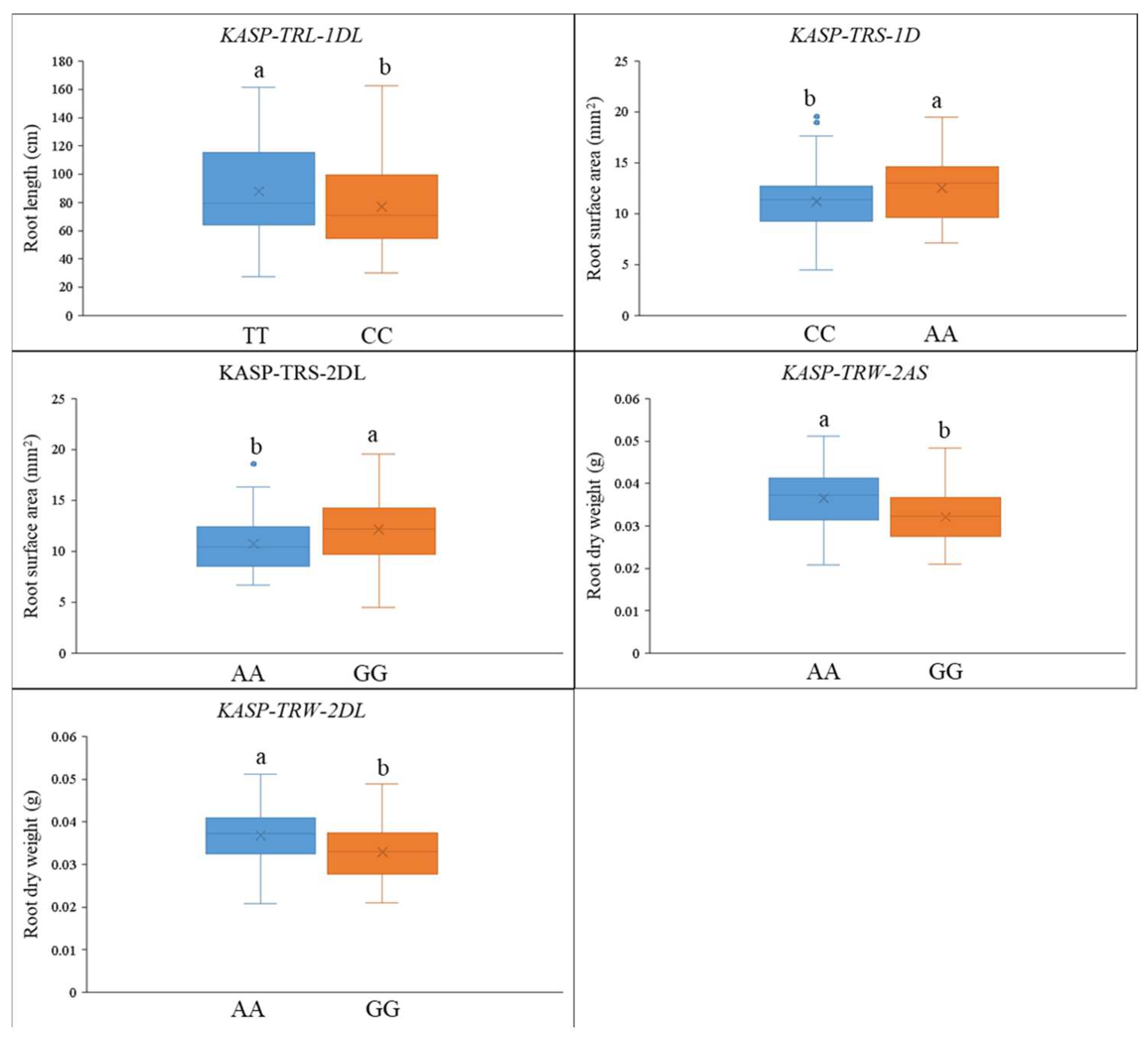
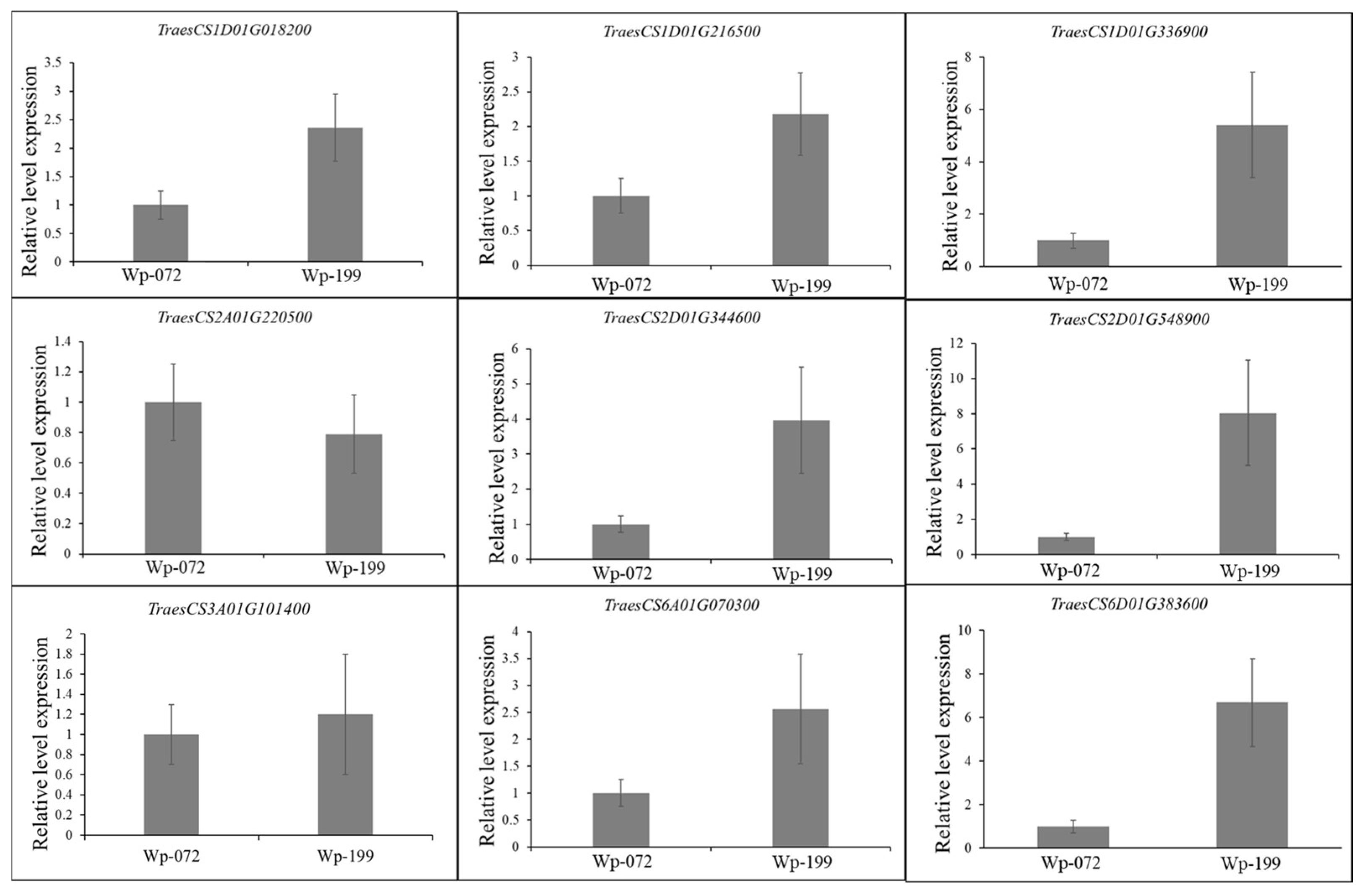
| QTL | Genetic Interval | Physical Position (Mb) | LOD | PVE | Add a | Favorable Allele | Unfavorable Allele |
|---|---|---|---|---|---|---|---|
| QRL.gau-1DS | Excalibur_c17152_454~ RAC875_c1471_566 | 7.1~7.9 | 2.7 | 5.2 | −6.1 | 75.8 cm | 72.4 cm |
| QRL.gau-1DL | RAC875_c103613_441~ Excalibur_c1236_840 | 424.7~431.8 | 4.2 | 8.4 | −9.2 | 76.0 cm | 72.7 cm |
| QRL.gau-4AL | BS00059454_51~ Kukri_c19883_816 | 731.4~732.5 | 2.7 | 5.4 | −6.1 | 75.8 cm | 72.3 cm |
| QRA.gau-1D | Kukri_c18350_151~ wsnp_Ex_rep_c70574_69491038 | 294.5~302.8 | 4.1 | 8.2 | −0.7 | 10.6 mm2 | 9.6 mm2 |
| QRA.gau-2DL | BobWhite_c6770_617~ BS00026262_51 | 439.6~446.0 | 4.3 | 8.4 | 0.7 | 10.7 mm2 | 9.8 mm2 |
| QRV.gau-6AS | CAP11_c4727_205~ Tdurum_contig11539_81 | 31.0~45.3 | 3.7 | 5.1 | −58.2 | 389.2 mm3 | 325.6 mm3 |
| QRW.gau-2AS | BS00039489_51~ Kukri_c15325_1360 | 203.0~208.5 | 4.7 | 8.2 | −0.0186 | 0.032 g | 0.027 g |
| QRW.gau-2DL | RAC875_c66820_684~ Ku_c22718_1072 | 623.3~629.2 | 4.2 | 8.1 | 0.0182 | 0.032 g | 0.027 g |
| QRT.gau-3AS | RAC875_c55313_89~ Kukri_c64788_552 | 60.1~70.2 | 2.6 | 4.5 | −6.0 | 224.6 | 196.9 |
| QRT.gau-6DL | Ex_c31468_763 tplb0055d02_624 | 462.6~468.3 | 3.8 | 7.2 | −7.5 | 218.7 | 190.2 |
| KASP Marker | QTL | Primer | Sequence |
|---|---|---|---|
| KASP-RL-1DL | QRL.gau-1DL | FAM | AGGTGGGTTCTTCAAAGGAAT |
| HEX | AGGTGGGTTCTTCAAAGGAAC | ||
| Common | GAGAATGCAAATGAATCCTCTGG | ||
| KASP-RA-1D | QRA.gau-1D | FAM | GCTGACGCATTTGAAAAAGATACA |
| HEX | GCTGACGCATTTGAAAAAGATACG | ||
| Common | CACATTTCCTGCACGGAGAA | ||
| KASP-RA-2DL | QRA.gau-2DL | FAM | TCCATGTCGTTTTATAACATTGACA |
| HEX | TCCATGTCGTTTTATAACATTGACG | ||
| Common | GTGAACACTAAGTTGTTTGTGGTTA | ||
| KASP-RW-2AS | QRW.gau-2AS | FAM | TGTTAGAATCTTACCTACCAGCATA |
| HEX | TGTTAGAATCTTACCTACCAGCATG | ||
| Common | TCCGAGGATGGGTATTTAACATG | ||
| KASP-RW-2DL | QRW.gau-2DL | FAM | ATGGTCGTCAACTCCATACAA |
| HEX | ATGGTCGTCAACTCCATACAG | ||
| Common | GCATCAGTTCAACAAGGCTG |
| Marker Name | QTL | Genotype a | Number of Lines | Phenotype | p-Value |
|---|---|---|---|---|---|
| KASP-RL-1DL | QRL.gau-1DL | TT | 56 | 276.6 cm | 0.048 * |
| CC | 77 | 230.5 cm | |||
| KASP-RA-1D | QRA.gau-1D | AA | 56 | 12.5 mm2 | 0.014 * |
| CC | 84 | 11.2 mm2 | |||
| KASP-RA-2DL | QRA.gau-2DL | AA | 53 | 10.7 mm2 | 0.007 ** |
| GG | 89 | 12.1 mm2 | |||
| KASP-RW-2AS | QRW.gau-2AS | AA | 66 | 0.037 g | 0.001 ** |
| GG | 79 | 0.032 g | |||
| KASP-RW-2DL | QRW.gau-2DL | AA | 41 | 0.037 g | 0.002 ** |
| GG | 102 | 0.033 g |
| QTL | Candidate Gene | Chr. | Start (bp) | End (bp) | Annotation |
|---|---|---|---|---|---|
| QRL.gau-1DS | TraesCS1D01G018200 | 1D | 7,913,129 | 7,917,136 | Zinc finger family protein |
| QRA.gau-1D | TraesCS1D01G216500 | 1D | 302,349,108 | 302,352,162 | Auxin canalization protein |
| QRL.gau-1DL | TraesCS1D01G336900 | 1D | 426,869,492 | 426,870,633 | Gibberellin 2-oxidase |
| QRW.gau-2AS | TraesCS2A01G220500 | 2A | 208,242,970 | 208,243,194 | Leucine-rich repeat receptor-like protein kinase |
| QRA.gau-2DL | TraesCS2D01G344600 | 2D | 440,633,947 | 440,634,603 | Leucine-rich repeat receptor-like protein kinase |
| QRW.gau-2DL | TraesCS2D01G548900 | 2D | 624,361,783 | 624,369,474 | Auxin response factor |
| QRT.gau-3AS | TraesCS3A01G101400 | 3A | 65,952,815 | 65,953,444 | E3 ubiquitin protein ligase |
| QRV.gau-6AS | TraesCS6A01G070300 | 6A | 38,453,855 | 38,454,175 | F-box family protein |
| QRT.gau-6DL | TraesCS6D01G383600 | 6D | 462,483,712 | 462,487,539 | Ethylene receptor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Wang, J.; Zhang, H.; Yao, L.; Si, E.; Li, B.; Meng, Y.; Wang, H. Genetic Basis of Seedling Root Traits in Common Wheat (Triticum aestivum L.) Identified by Genome-Wide Linkage Mapping. Plants 2025, 14, 490. https://doi.org/10.3390/plants14030490
Ma X, Wang J, Zhang H, Yao L, Si E, Li B, Meng Y, Wang H. Genetic Basis of Seedling Root Traits in Common Wheat (Triticum aestivum L.) Identified by Genome-Wide Linkage Mapping. Plants. 2025; 14(3):490. https://doi.org/10.3390/plants14030490
Chicago/Turabian StyleMa, Xiaole, Juncheng Wang, Hong Zhang, Lirong Yao, Erjing Si, Baochun Li, Yaxiong Meng, and Huajun Wang. 2025. "Genetic Basis of Seedling Root Traits in Common Wheat (Triticum aestivum L.) Identified by Genome-Wide Linkage Mapping" Plants 14, no. 3: 490. https://doi.org/10.3390/plants14030490
APA StyleMa, X., Wang, J., Zhang, H., Yao, L., Si, E., Li, B., Meng, Y., & Wang, H. (2025). Genetic Basis of Seedling Root Traits in Common Wheat (Triticum aestivum L.) Identified by Genome-Wide Linkage Mapping. Plants, 14(3), 490. https://doi.org/10.3390/plants14030490





