The Role of Reproductive Modes in Shaping Genetic Diversity in Polyploids: A Comparative Study of Selfing, Outcrossing, and Apomictic Paspalum Species
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation Within Populations
2.2. Phenotypic Variation Among Populations
2.3. Molecular and Genotypic Diversity Within Species
2.4. Genetic Differentiation and Gene Flow
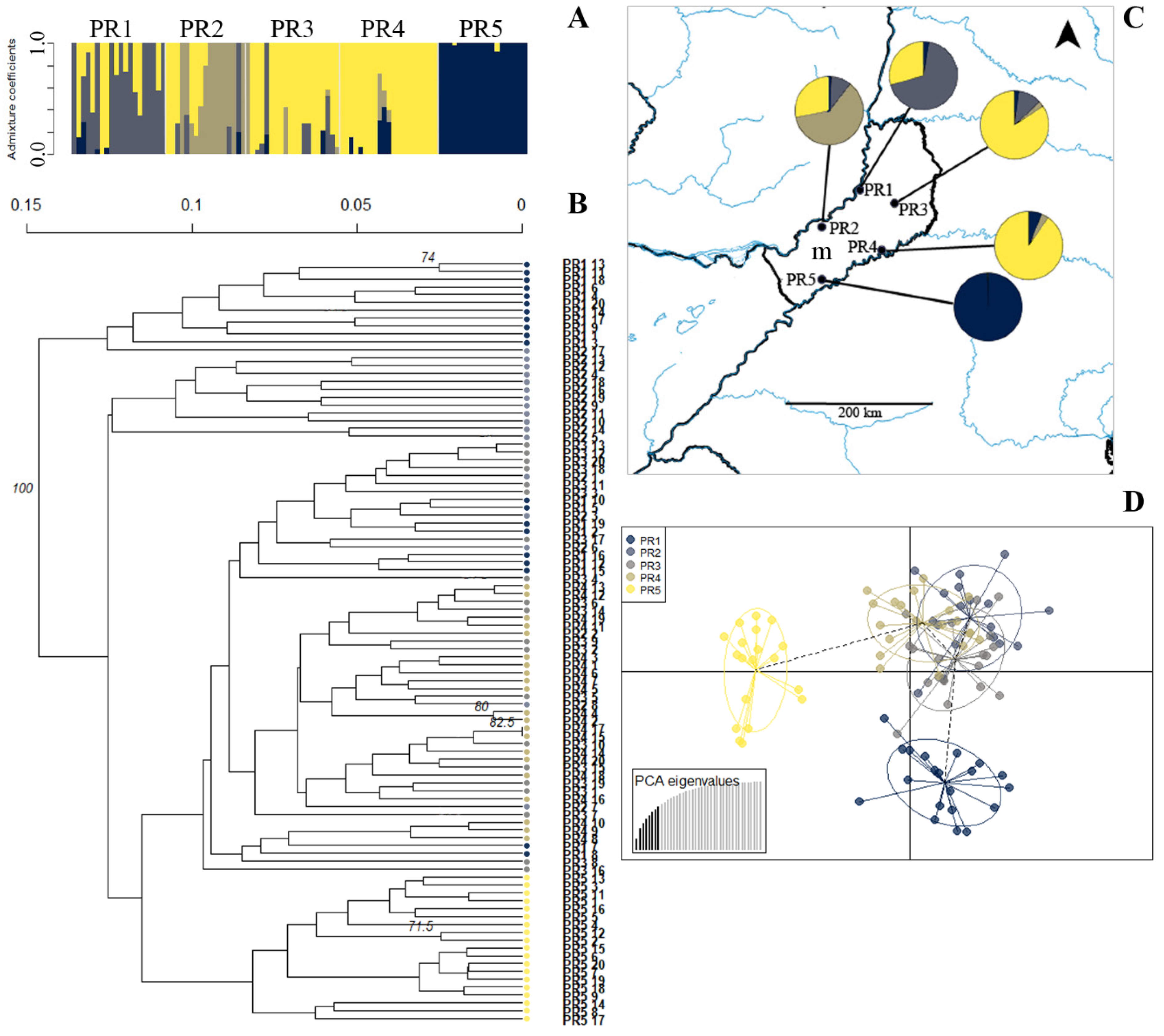
2.5. Population Structure and Cluster Analysis
3. Discussion
3.1. Opposite Mating Systems Led to Similar Patterns of Phenotypic Variation but Differed in Apomictic Species
3.2. Local Adaptation and Phenotypic Differentiation Among Populations
3.3. Apomixis Leads to Fixation of Adapted Phenotypes
3.4. Contrasting Patterns of Intrapopulation Variation Among Selfing and Outcrossing Species
3.5. Differences in Genetic Structure Among Mating Systems
3.6. Population Structure in Apomictic Species Showed No Admixture
4. Materials and Methods
4.1. Plant Material
4.2. Morpho Phenological Traits
4.3. Statistical Analyses of Morpho Phenological Traits
4.4. DNA Isolation and ISSR Protocol
4.5. Molecular Diversity Within Populations
4.6. Differentiation Between Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acuña, C.A.; Martínez, E.J.; Zilli, A.L.; Brugnoli, E.A.; Espinoza, F.; Marcón, F.; Urbani, M.H.; Quarin, C.L. Reproductive systems in Paspalum: Relevance for germplasm collection and conservation, breeding techniques, and adoption of released cultivars. Front. Plant Sci. 2019, 10, 1377. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, J.P.A.; Pupilli, F.; Acuña, C.A.; Leblanc, O.; Pessino, S.C. How to become an apomixis model: The multi-faceted case of Paspalum. Genes 2020, 11, 974. [Google Scholar] [CrossRef] [PubMed]
- Asker, S.E.; Jerling, L. Apomixis in Plants; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Holsinger, K.E. Reproductive systems and evolution in vascular plants. Proc. Natl. Acad. Sci. USA 2000, 97, 7037–7042. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The genetic structure of populations. Nature 1950, 166, 247–250. [Google Scholar] [CrossRef]
- Müller, H.J. The relation of recombination to mutational advance. Mutat. Res. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.M., Ed.; Springer: Verlag, Berlin, 1984; pp. 475–518. [Google Scholar] [CrossRef]
- Hörandl, E.; Paun, O. Patterns and sources of genetic diversity in apomictic plants: Implications for evolutionary potentials. In Apomixis: Evolutions, Mechanisms and Perspectives; Hörandl, E., Grossniklaus, U., van Dijk, P.J., Sharbel, T.F., Eds.; ARG Gantner Verlag: Rugell, Liechtenstein, 2007; pp. 170–194. [Google Scholar]
- Lynch, M. Destabilizing Hybridization, General-Purpose Genotypes and Geographic Parthenogenesis. Q. Rev. Biol. 1984, 59, 257–290. [Google Scholar] [CrossRef]
- Menken, S.B.J.; Morita, T. Uniclonal Population Structure in the Pentaploid Obligate Agamosperm Taraxacum albidum Dahlst. Plant Species Biol. 1989, 4, 29–36. [Google Scholar] [CrossRef]
- Štorchová, H.; Chrtek, J., Jr.; Bartish, I.V.; Tetera, M.; Kirschner, J.; Štěpánek, J. Genetic variation in agamospermous taxa of Hieracium sect. Alpina (Compositae) in the Tatry Mts. (Slovakia). Plant Syst. Evol. 2002, 235, 1–17. [Google Scholar] [CrossRef]
- Niissalo, M.A.; Leong-Škorničková, J.; Šída, O.; Khew, G.S. Population genomics reveal apomixis in a novel system: Uniclonal female populations dominate the tropical forest herb family, Hanguanaceae (Commelinales). AoB Plants 2020, 12, plaa053. [Google Scholar] [CrossRef]
- Houliston, G.J.; Chapman, H.M. Reproductive strategy and population variability in the facultative apomict Hieracium pilosella (Asteraceae). Am. J. Bot. 2004, 91, 37–44. [Google Scholar] [CrossRef]
- Lo, E.Y.Y.; Stefanovic, S.; Dickinson, T.A. Population genetic structure of diploid sexual and polyploid apomictic hawthorns (Crataegus; Rosaceae) in the Pacific Northwest. Mol. Ecol. 2009, 18, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Majeský, Ľ.; Vašut, R.J.; Kitner, M.; Trávníček, B. The pattern of genetic variability in apomictic clones of Taraxacum officinale indicates the alternation of asexual and sexual histories of apomicts. PLoS ONE 2012, 7, e41868. [Google Scholar] [CrossRef] [PubMed]
- Brugnoli, E.A.; Urbani, M.H.; Quarin, C.L.; Zilli, A.L.; Martínez, E.J.; Acuña, C.A. Diversity in apomictic populations of Paspalum simplex Morong. Crop Sci. 2014, 54, 1656–1664. [Google Scholar] [CrossRef]
- Toenniessen, G.H. Feeding the world in the 21st century: Plant breeding, biotechnology, and the potential role of apomixis. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; ClMMYT: Mexico, Mexico; IRD: Marseille, France; European Commission OC VI (FAIR): Brussels, Belgium, 2001; pp. 1–7. [Google Scholar]
- Loveless, M.D.; Hamrick, J.L. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Evol. Syst. 1984, 15, 65–95. [Google Scholar] [CrossRef]
- Foxe, J.P.; Slotte, T.; Stahl, E.A.; Neuffer, B.; Hurka, H.; Wrigth, S.I. Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 2009, 106, 5241–5245. [Google Scholar] [CrossRef]
- Nordborg, M. Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics 2000, 154, 923–929. [Google Scholar] [CrossRef]
- Glémin, S.; Bazin, E.; Charlesworth, D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. Biol. Sci. 2006, 273, 3011–3019. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J. Allozyme diversity in plant species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates Inc. Publishers: Sunderland, MA, USA, 1989; pp. 43–63. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W. Conservation genetics of endemic plant species. In Conservation Genetics. Case Histories from Nature; Avise, J.C., Hamrick, J.L., Eds.; Chapman and Hall: New York, NY, USA, 1996; pp. 281–304. [Google Scholar] [CrossRef]
- Nybom, H.; Bartish, I.V. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspec. Plant Ecol. Evol. Syst. 2000, 3, 93–114. [Google Scholar] [CrossRef]
- Eschmann-Grupe, G.; Neuffer, B.; Hurka, H. Extent and structure of genetic variation in two colonizing Diplotaxis species (Brassicaceae) with contrasting breeding systems. Plant Syst. Evol. 2004, 244, 31–43. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Koelling, V.A.; Hamrick, J.L.; Mauricio, R. Genetic diversity and structure in two species of Leavenworthia with self-incompatible and self-compatible populations. Heredity 2010, 106, 310–318. [Google Scholar] [CrossRef]
- Reisch, C.; Bernhardt-Römermann, M. The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecol. 2014, 215, 1493–1511. [Google Scholar] [CrossRef]
- Huang, R.; Chu, Q.H.; Lu, G.H.; Wang, Y.Q. Comparative studies on population genetic structure of two closely related selfing and outcrossing Zingiber species in Hainan Island. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Reutemann, A.V.; Honfi, A.I.; Karunarathne, P.; Eckers, F.; Hojsgaard, D.H.; Martínez, E.J. Comparative analysis of molecular and morphological diversity in two diploid Paspalum species (Poaceae) with contrasting mating systems. Plant Reprod. 2024, 37, 15–32. [Google Scholar] [CrossRef]
- Temunović, M.; Franjić, J.; Satovic, Z.; Grgurev, M.; Frascaria-Lacoste, N.; Fernández-Majarrés, J.F. Environmental heterogenity explains the genetic structure of continental and mediterranean populations of Fraxinus angustifolia Vahl. PLoS ONE 2012, 7, e42764. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Sun, G.; Wang, X.; Lu, J.; Wang, I.J.; Wang, Z. Population genetic structure is shaped by historical, geographic, and environmental factors in the leguminous shrub Caragana microphylla on the Inner Mongolia Plateau of China. BMC Plant Biol. 2017, 17, 200. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.J.; Bradburd, G.S. Isolation by environment. Mol. Ecol. 2014, 23, 5649–5662. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution 2014, 68, 1–15. [Google Scholar] [CrossRef]
- Mijangos, J.L.; Pacioni, C.; Spencer, P.B.; Craig, M.D. Contribution of genetics to ecological restoration. Mol. Ecol. 2015, 24, 22–37. [Google Scholar] [CrossRef]
- Allen, W.J.; Bufford, J.L.; Barnes, A.D.; Barratt, B.I.; Deslippe, J.R.; Dickie, I.A.; Goldson, S.L.; Howlett, B.G.; Hulme, P.E.; Lavrel, S.; et al. A network perspective for sustainable agroecosystems. Trends Plant Sci. 2022, 27, 769–780. [Google Scholar] [CrossRef]
- Quarin, C.L. The nature of apomixis and its origin in Panicoid grasses. Apomixis Newsl. 1992, 5, 8–15. [Google Scholar]
- Zuloaga, F.O.; Morrone, O.; Davidse, G.; Filgueiras, T.S.; Peterson, P.M.; Soreng, R.J.; Judziewicz, E.J. Catalogue of New World grasses (Poaceae): III. subfamilies Panicoideae, Aristidoideae, Arundinoideae, and Danthonioideae. Contr. US Natl. Herb. 2003, 46, 1–662. [Google Scholar]
- Ortiz, J.P.A.; Quarin, C.L.; Pessino, S.C.; Acuna, C.A.; Martínez, E.J.; Espinoza, F.; Hojsgaard, D.H.; Sartor, M.E.; Cáceres, M.E.; Pupilli, F. Harnessing apomictic reproduction in grasses: What we have learned from Paspalum. Ann. Bot. 2013, 112, 767–787. [Google Scholar] [CrossRef] [PubMed]
- D’Aurelio, L.D.; Espinoza, F.; Quarin, C.L.; Pessino, S.C. Genetic diversity in sexual diploid and apomictic tetraploid popultions of Paspalum notatum situated in sympatry or allopatry. Plant Syst. Evol. 2004, 244, 189–199. [Google Scholar] [CrossRef]
- Brugnoli, E.A.; Urbani, M.H.; Quarin, C.L.; Martínez, E.J.; Acuña, C.A. Diversity in diploid, tetraploid, and mixed diploid-tetraploid populations of Paspalum simplex. Crop Sci. 2013, 53, 1509–1516. [Google Scholar] [CrossRef]
- Sartor, M.E.; Rebozzio, R.N.; Quarin, C.L.; Espinoza, F. Patterns of genetic diversity in natural populations of Paspalum agamic complexes. Plant Syst. Evol. 2013, 299, 1295–1306. [Google Scholar] [CrossRef]
- Karunarathne, P.; Hojsgaard, D. Single independent autopolyploidization events from distinct diploid gene pools and residual sexuality support range expansion of locally adapted tetraploid genotypes in a South American grass. Front. Genet. 2021, 12, 736088. [Google Scholar] [CrossRef]
- Burson, B.L.; Bennett, H.W. Cytology and reproduction of three Paspalum species. J. Hered. 1970, 61, 129–132. [Google Scholar] [CrossRef]
- Quarin, C.L. A tetraploid cytotype of Paspalum durifolium: Cytology, reproductive behavior and its relationship to diploid P. intermedium. Hereditas 1994, 121, 115–118. [Google Scholar] [CrossRef]
- Burson, B.L. Cytology of Paspalum chacoense and P. durifolium and their relationship to P. dilatatum. Bot. Gaz. 1985, 146, 124–129. [Google Scholar] [CrossRef]
- Martínez, E.J.; Quarin, C.L.; Hayward, M.D. Genetic control of apospory in apomictic Paspalum species. Cytologia 1999, 64, 425–433. [Google Scholar] [CrossRef]
- Schedler, M.; Reutemann, A.V.; Hojsgaard, D.H.; Zilli, A.L.; Brugnoli, E.A.; Galdeano, F.; Acuña, C.A.; Honfi, A.I.; Martínez, E.J. Alternative evolutionary pathways in Paspalum involving allotetraploidy, sexuality, and varied mating systems. Genes 2023, 14, 1137. [Google Scholar] [CrossRef] [PubMed]
- Norrmann, G.A. Citología y modo de reproducción en dos especies de Paspalum (Gramineae). Bonplandia 1981, 17, 149–158. [Google Scholar]
- Brown, W.V.; Emery, H.P. Apomixis in the Gramineae: Panicoideae. Am. J. Bot. 1958, 45, 253–263. [Google Scholar] [CrossRef]
- Bashaw, E.C.; Hovin, A.W.; Holt, E.C. Apomixis, its evolutionary significance and utilization in plant breeding. In Proceedings of the 11th International Grasslands Congress; Norman, M.J.T., Ed.; Surfers Paradise: Australia University of Queensland Press: Brisbane, Australian, 1970; pp. 245–248. [Google Scholar]
- Karunarathne, P.; Reutemann, A.V.; Schedler, M.; Glücksberg, A.; Martínez, E.J.; Honfi, A.I.; Hojsgaard, D.H. Sexual modulation in a polyploid grass: A reproductive contest between environmentally inducible sexual and genetically dominant apomictic pathways. Sci. Rep. 2020, 10, 8319. [Google Scholar] [CrossRef]
- Jain, S.K. Population structure and the effects of breeding system. In Crop Genetic Re-Sources for Today and Tomorrow; Frankel, O.H., Hawkes, J.G., Eds.; Cambridge University Press: Cambridge, CA, USA, 1975; pp. 15–36. [Google Scholar]
- Hamrick, J.L.; Nason, J.D. Consequences of dispersal in plants. In Population Dynamics in Ecological Space and Time; Rhodes, O.E., Chesser, R.K., Smith, M.H., Eds.; The University of Chicago Press: Chicago, IL, USA, 1996; pp. 203–235. [Google Scholar]
- Ellstrand, N.C.; Roose, M.L. Patterns of genotypic diversity in clonal plant species. Am. J. Bot. 1987, 74, 123–131. [Google Scholar] [CrossRef]
- Stebbins, G.L. Variation and Evolution in Plants; Columbia University Press: New York, NY, USA, 1950. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B. Quantitative genetics in plants: The effect of the breeding system on genetic variability. Evolution 1995, 49, 911–920. [Google Scholar] [CrossRef]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef]
- Hoban, S.; Schlarbaum, S. Optimal sampling of seeds from plant populations for ex-situ conservation of genetic biodiversity, considering realistic population structure. Biol. Cons. 2014, 177, 90–99. [Google Scholar] [CrossRef]
- Coleman, S.W.; Moore, J.E.; Wilson, J.W. Quality and utilization. In Warm Season Grasses; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; ASA, CSSA, SSSA: Madison, WI, USA, 2004; pp. 267–308. [Google Scholar] [CrossRef]
- Mullet, J.E. High-biomass C4 grasses Filling the yield gap. Plant Sci. 2017, 261, 10–17. [Google Scholar] [CrossRef]
- Rathcke, B.; Lacey, E.P. Phenological patterns of terrestrial plants. Annu. Rev. Ecol. Syst. 1985, 16, 179–214. [Google Scholar] [CrossRef]
- Mozdzer, T.J.; Caplan, J.S.; Hager, R.N.; Proffitt, C.E.; Meyerson, L.A. Contrasting trait responses to latitudinal climate varia-tion in two lineages of an invasive grass. Biol. Invas. 2016, 18, 2649–2660. [Google Scholar] [CrossRef]
- Primack, R.B. Variation in the phenology of natural populations of montane shrubs in New Zealand. J. Ecol. 1980, 68, 849–862. [Google Scholar] [CrossRef]
- Takebayashi, N.; Morrell, P.L. Is self-fertilization an evolutionary dead end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am. J. Bot. 2001, 88, 1143–1150. [Google Scholar] [CrossRef]
- Merilä, J.; Crnokrak, P. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 2001, 14, 892–903. [Google Scholar] [CrossRef]
- Wright, S.I.; Kalisz, S.; Slotte, T. Evolutionary consequences of self-fertilization in plants. Proc. Royal. Soc. B 2013, 280, 20130133. [Google Scholar] [CrossRef]
- Chung, M.Y.; Merilä, J.; Li, J.; Mao, K.; López-Pujol, J.; Tsumura, Y.; Chung, M.G. Neutral and adaptive genetic diversity in plants: An overview. Front. Ecol. Evol. 2023, 11, 1116814. [Google Scholar] [CrossRef]
- Lenormand, T. Gene flow and the limits to natural selection. Trends Ecol. Evol. 2002, 17, 183–189. [Google Scholar] [CrossRef]
- Dufresne, F.; Stift, M.; Vergilino, R.; Mable, B.K. Recent progress and challenges in population genetics of polyploid organisms: An overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 2014, 23, 40–69. [Google Scholar] [CrossRef]
- Baker, H.G. Support for Baker’s law-as a rule. Evolution 1967, 21, 853–856. [Google Scholar] [CrossRef]
- Ingvarsson, P. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution 2002, 56, 2368–2373. [Google Scholar] [CrossRef] [PubMed]
- Goodwillie, C.; Kalisz, S.; Eckert, C.G. The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 47–79. [Google Scholar] [CrossRef]
- Hörandl, E. The complex causality of geographical parthenogenesis. New Phytol. 2006, 171, 525–538. [Google Scholar] [CrossRef] [PubMed]
- García, M.V.; Balatti, P.A.; Arturi, M.J. Genetic variability in natural populations of Paspalum dilatatum Poir. analyzed by means of morphological traits and molecular markers. Genet. Resour. Crop Evol. 2007, 54, 935–946. [Google Scholar] [CrossRef]
- Reis, C.A.D.O.D.; Dall’Agnol, M.; Nabinger, C.; Schifino-Wittmann, M.T. Morphological variation in Paspalum nicorae Parodi accessions, a promising forage. Sci. Agric. 2010, 67, 143–150. [Google Scholar] [CrossRef]
- Lynch, M.; Bürger, R.; Butcher, D.; Gabriel, W. The mutational meltdown in asexual populations. J. Hered. 1993, 84, 339–344. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Hörandl, E. A little bit of sex matters for genome evolution in asexual plants. Front. Plant Sci. 2015, 6, 82. [Google Scholar] [CrossRef]
- Hodač, L.; Klatt, S.; Hojsgaard, D.; Sharbel, T.F.; Hörandl, E. A little bit of sex prevents mutation accumulation even in apo-mictic polyploid plants. BMC Evol. Biol. 2019, 19, 170. [Google Scholar] [CrossRef]
- Pettengill, J.B.; Briscoe Runquist, R.D.; Moeller, D.A. Mating system divergence affects the distribution of sequence diversity within and among populations of recently diverged subspecies of Clarkia xantiana (Onagraceae). Am. J. Bot. 2016, 103, 99–109. [Google Scholar] [CrossRef]
- Dawson, I.K.; Chalmers, K.J.; Waugh, R.; Powell, W. Detection and analysis of genetic variation in Hordeum spontaneum populations from Israel using RAPD markers. Mol. Ecol. 1993, 2, 151–159. [Google Scholar] [CrossRef]
- Chirinos-Arias, M.C.; Jiménez, L.E.; Vilca-Machaca, L.S. Análisis de la Variabilidad Genética entre treinta accesiones de tarwi (Lupinus mutabilis Sweet) usando marcadores moleculares ISSR. Sci. Agropecu. 2015, 6, 17–30. [Google Scholar] [CrossRef]
- Vidal, R.; González, A.; Gutiérrez, L.; Umaña, R.; Speranza, P. Distribución de la diversidad genética y sistema reproductivo de Stipa neesiana Trin. et. Rupr. Agrocienc. Urug. 2011, 15, 1–12. [Google Scholar] [CrossRef]
- Blambert, L.; Mallet, B.; Humeau, L.; Pailler, T. Reproductive patterns, genetic diversity and inbreeding depression in two closely related Jumellea species with contrasting patterns of commonness and distribution. Ann. Bot. 2015, 118, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Zhao, N.X.; Gai, Y.B.; Lin, F.; Ren, A.Z.; Ruan, W.B.; Chen, L. RAPD analysis of genetic diversity and population genetic structure of Stipa krylovii Reshov. Inner Mongolia steppe. Russ. J. Genet. 2006, 42, 468–475. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance under diverse systems of mating. Genetics 1946, 31, 39–59. [Google Scholar] [CrossRef]
- Allard, R.W.; Jain, S.K.; Workman, P.L. The genetics of inbreeding populations. In Advances in Genetics; Caspari, E.W., Ed.; NY Academic: New York, NY, USA, 1968; pp. 55–131. [Google Scholar] [CrossRef]
- Slatkin, M. The rate of spread of an advantageous allele in a subdivided population. In Population Genetics and Ecology; Karlin, S., Nevo, E., Eds.; NY Academic: New York, NY, USA, 1976; pp. 767–780. [Google Scholar] [CrossRef]
- Duminil, J.; Hardy, O.J.; Petit, R.J. Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evol. Biol. 2009, 9, 177. [Google Scholar] [CrossRef]
- Charlesworth, D. Effects of inbreeding on the genetic diversity of populations. Philos. Trans. R. Soc. Lond. 2003, 358, 1051–1070. [Google Scholar] [CrossRef]
- Gornall, R.J. Population genetic structure in agamospermous plants. In Molecular Systematic and Plant Evolution; Hollingsworth, P.M., Bateman, R.M., Gornall, R.J., Eds.; Taylor & Francis: London, UK, 1999; pp. 118–138. [Google Scholar] [CrossRef]
- Paun, O.; Greilhuber, J.; Temsch, E.M.; Hörandl, E. Patterns, sources and ecological implications of clonal diversity in apomictic Ranunculus carpaticola (Ranunculus auricomus complex, Ranunculaceae). Mol. Ecol. 2006, 15, 897–910. [Google Scholar] [CrossRef]
- Widén, B.; Cronberg, N.; Widén, M. Genotypic diversity, molecular markers and spatial distribution of genets in clonal plants, a literature survey. Folia Geobot. 1994, 29, 245–263. [Google Scholar] [CrossRef]
- Richards, A.J. Apomixis in flowering plants: An overview. Philos. Trans. R. Soc. B 2003, 358, 1085–1093. [Google Scholar] [CrossRef]
- Paun, O.; Stuessy, T.F.; Hörandl, E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytol. 2006, 171, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Ozuna, R.; Eguiarte, L.E.; Molina-Freaner, F. Genotypic diversity among pasture and roadside populations of the invasive buffelgrass (Pennisetum ciliare L.) in north-western Mexico. J. Arid Environ. 2009, 73, 26–32. [Google Scholar] [CrossRef]
- Cosendai, A.C.; Wagner, J.; Ladinig, U.; Rosche, C.; Hörandl, E. Geographical Parthenogenesis and Population Genetic Structure in the alpine Species Ranunculus kuepferi (Ranunculaceae). Heredity 2013, 110, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Bogunić, F.; Siljak-Yakovlev, S.; Mahmutović-Dizdarević, I.; Hajrudinović-Bogunić, A.; Bourge, M.; Brown, S.C.; Muratović, E. Genome size, cytotype diversity and reproductive mode variation of Cotoneaster integerrimus (Rosaceae) from the Balkans. Plants 2021, 10, 2798. [Google Scholar] [CrossRef]
- Palacios, C.; Kresovich, S.; Gonzalez-Candelas, F. A population genetic study of the endangered plant species Limonium dufourii (Plumbaginaceae) based on amplified fragment length polymorphism (AFLP). Mol. Ecol. 1999, 8, 645–657. [Google Scholar] [CrossRef]
- Levin, D.A.; Kerster, H.W. Neighborhood structure in plants under diverse reproductive methods. Am. Nat. 1971, 105, 345–354. [Google Scholar] [CrossRef]
- Moura, Y.A.; Alves-Pereira, A.; da Silva, C.C.; Souza, L.M.; de Souza, A.P.; Koehler, S. Secondary origin, hybridization and sexual reproduction in a diploid-tetraploid contact zone of the facultatively apomictic orchid Zygopetalum mackayi. Plant Biol. 2020, 22, 939–948. [Google Scholar] [CrossRef]
- Zuloaga, F.O.; Morrone, O. Revisión de las especies de Paspalum para América del Sur austral: (Argentina, Bolivia, sur del Brasil, Chile, Paraguay y Uruguay); Missouri Botanical Garden: St. Louis, MI, USA, 2005; p. 297. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 27 July 2023).
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Van Tienderen, P.H. GENOTYPE and GENODIVE: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Winter, D.J. MMOD: An R library for the calculation of population differentiation statistics. Mol. Ecol. Resour. 2012, 12, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. Hierfstat, a package for R to compute and test variance components and F-statistics. Mol. Ecol. Notes. 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Frichot, E.; Francois, O. LEA: An R package for Landscape and Ecological Association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Thioulouse, J.; Dray, S.; Dufour, A.; Siberchicot, A.; Jombart, T.; Pavoine, S. Multivariate Analysis of Ecological Data with ade4; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Matesanz, S.; Gimeno, T.E.; de la Cruz, M.; Escudero, A.; Valladares, F. Competition may explain the fine-scale spatial patterns and genetic structure of two co-occurring plant congeners. J. Ecol. 2011, 99, 838–848. [Google Scholar] [CrossRef]
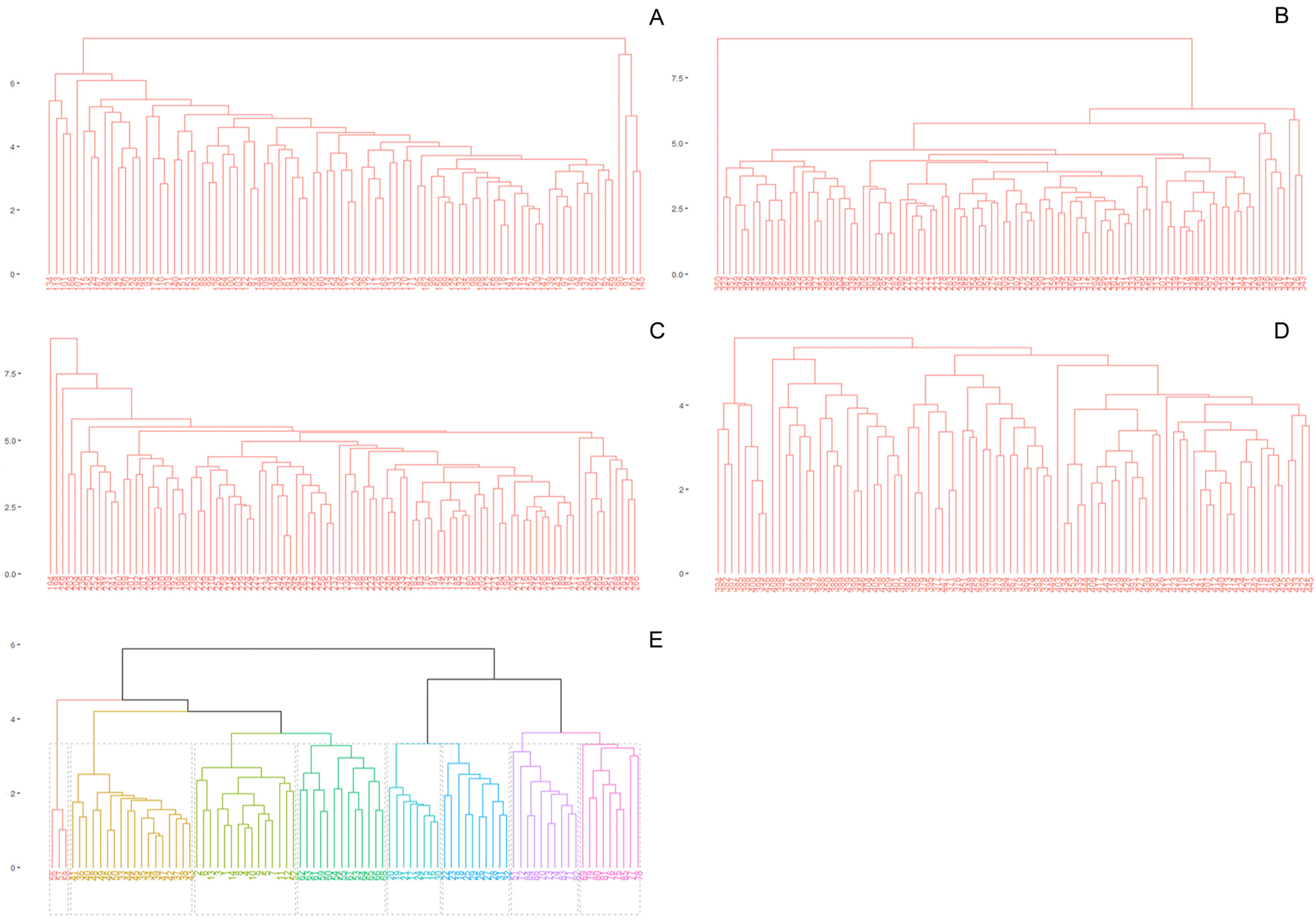
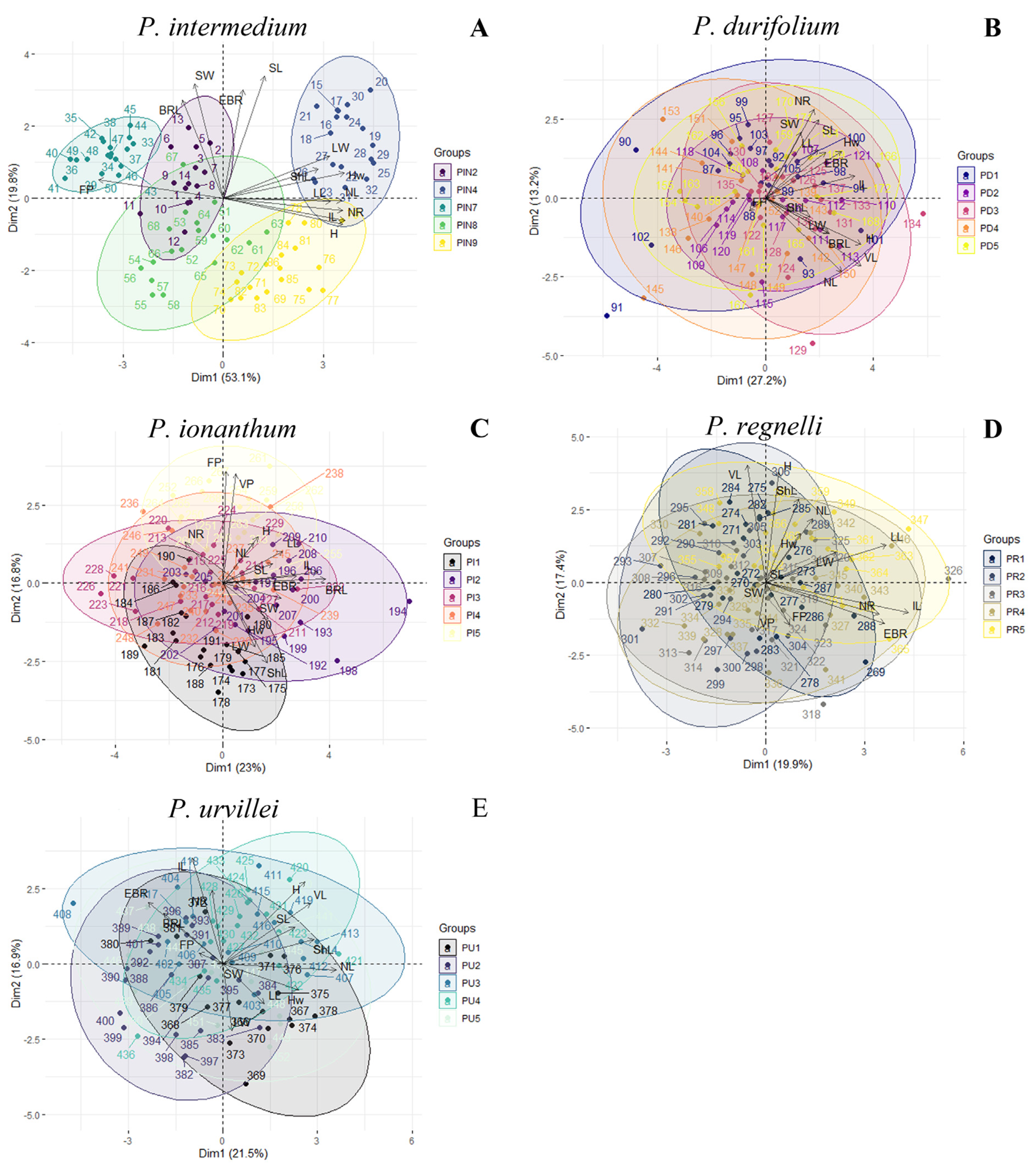

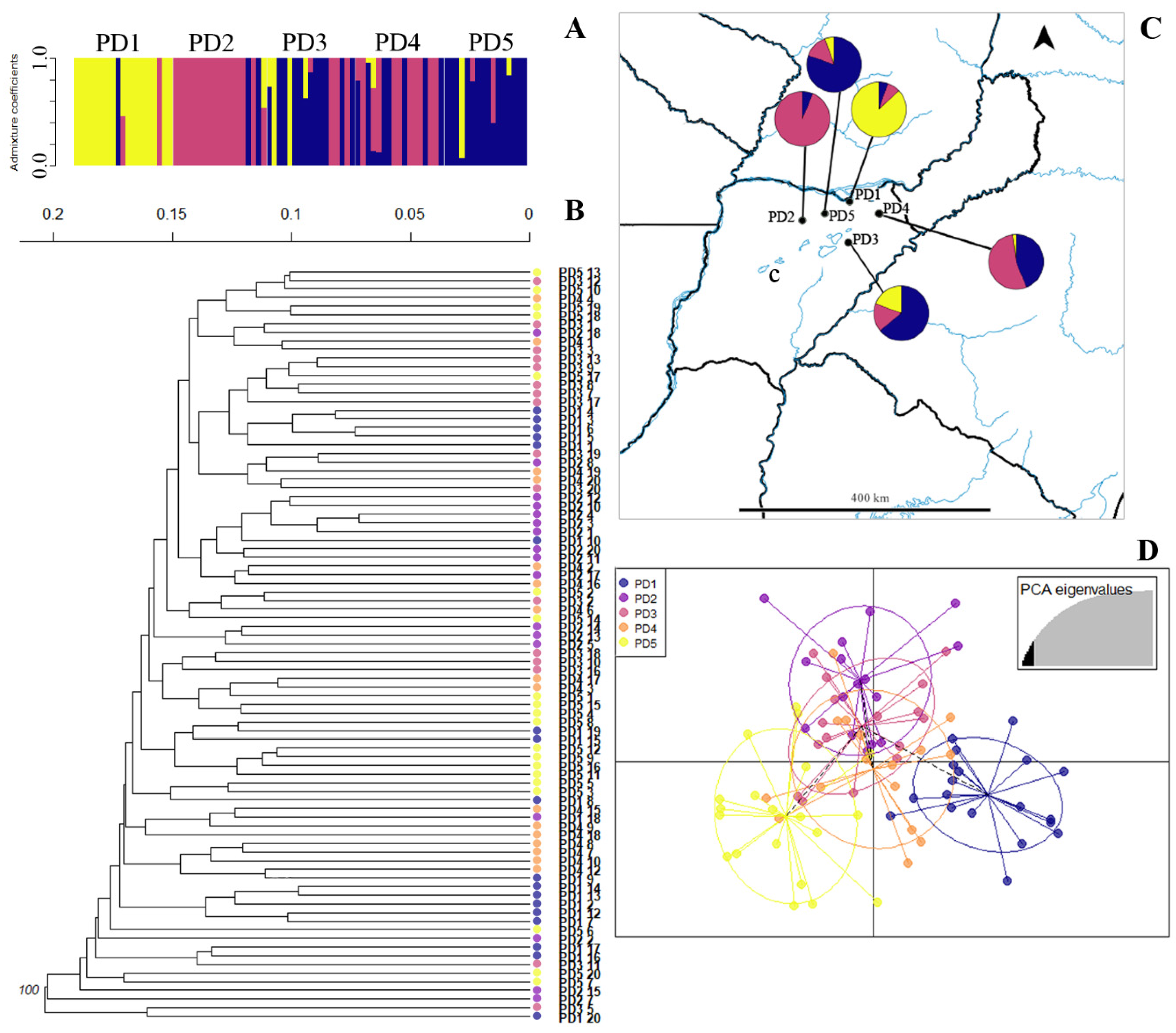
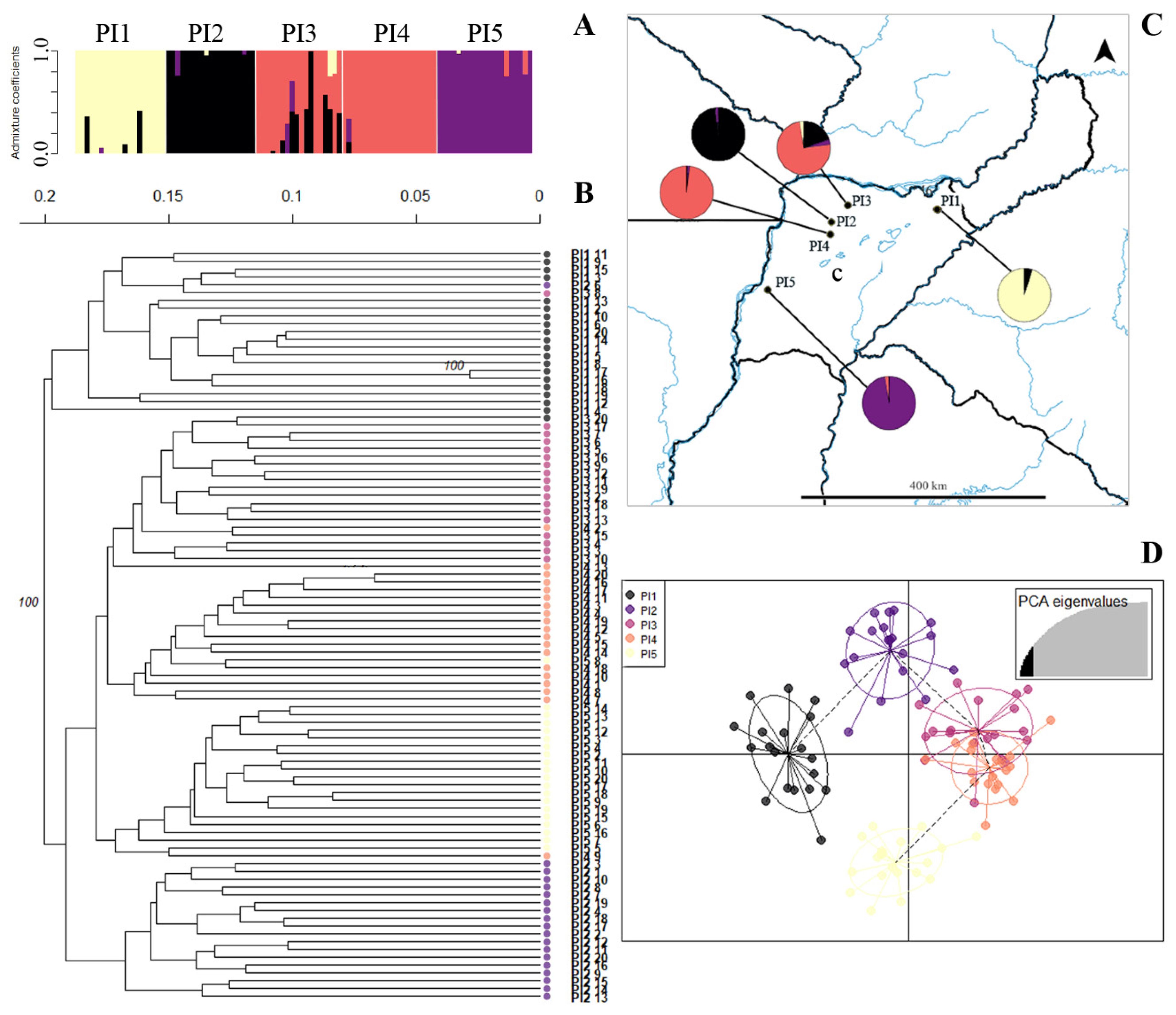

| Species | Pop | n | H | Hw | LL | LW | ShL | NL | IL | BRL | EBR | NR | VL | SL | SW | VP | FP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. intermedium | PIN2 | 14 | 187.9 ± 17.2 | 100.7 ± 9.2 | 34.2 ± 4.2 | 1.2 ± 0.1 | 26.4 ± 3.0 | 26.3 ± 3.2 | 25 ± 1.5 | 9.5 ± 1.0 | 110.6 ± 9.8 | 39.5 ± 2.5 | 16.5 ± 10.7 | 2.7 ± 0.2 | 1.6 ± 0.0 | 81.2 ± 9.1 | 114.8 ± 5.6 |
| PIN4 | 18 | 228.1 ± 17.2 | 131.1 ± 9.6 | 45.8 ± 5.5 | 1.9 ± 0.1 | 40.2 ± 5.6 | 36.6 ± 3.7 | 37.3 ± 2.7 | 9.0 ± 0.9 | 145.9 ± 36.4 | 77.6 ± 9.8 | 205.6 ± 11.8 | 3.0 ± 0.1 | 1.5 ± 0.0 | 62.0 ± 6.7 | 77.4 ± 3.5 | |
| PIN7 | 18 | 110.3 ± 21.1 | 71.4 ± 14.1 | 23.6 ± 2.9 | 1.2 ± 0.1 | 29.4 ± 2.4 | 11.0 ± 4.1 | 22.4 ± 2.4 | 10.9 ± 1.1 | 135.7 ± 10.9 | 33.4 ± 5.1 | 103.3 ± 18.9 | 2.7 ± 0.1 | 1.5 ± 0.0 | 112 ± 12.5 | 148.2 ± 8.7 | |
| PIN8 | 18 | 173.8 ± 31.3 | 85.6 ± 9.9 | 35.8 ± 7.5 | 1.4 ± 0.3 | 29.4 ± 5.5 | 23.9 ± 3.6 | 26.9 ± 3.5 | 8.2 ± 1.6 | 105.7 ± 21.5 | 45.9 ± 4.8 | 163.9 ± 29.3 | 2.5 ± 0.1 | 1.3 ± 0.1 | 92.1 ± 12.6 | 126.8 ± 16.2 | |
| PIN9 | 18 | 225.7 ± 25.6 | 106.8 ± 14.7 | 38.6 ± 10.2 | 1.4 ± 0.3 | 34.1 ± 3.8 | 28.3 ± 5.2 | 35.4 ± 4.8 | 8.1 ± 1.1 | 108.5 ± 14.8 | 71.2 ± 4.7 | 210.3 ± 19.1 | 2.4 ± 0.1 | 1.2 ± 0.1 | 66.1 ± 6.0 | 81.7 ± 4.2 | |
| P. durifolium | PD3 | 16 | 118.1 ± 17.4 | 57.9 ± 6.9 | 24.4 ± 6.4 | 0.9 ± 0.1 | 11.7 ± 2.9 | 23.2 ± 5.3 | 19.7 ± 2.4 | 8.6 ± 1.0 | 90.2 ± 11.9 | 19.4 ± 3.3 | 100 ± 14.6 | 3.0 ± 0.1 | 1.5 ± 0.1 | 62.4 ± 6.2 | 48.5 ± 6.5 |
| PD4 | 16 | 111.3 ± 21.3 | 53.6 ± 5.7 | 20.1 ± 3.0 | 0.8 ± 0.1 | 10.5 ± 2.3 | 18.9 ± 6.1 | 18.7 ± 2.3 | 7.7 ± 1.2 | 84.2 ± 14.6 | 17.2 ± 3.6 | 88.6 ± 19.1 | 3.0 ± 0.1 | 1.4 ± 0.1 | 57.4 ± 8.1 | 74.9 ± 6.7 | |
| PD5 | 19 | 114 ± 16.8 | 58.3 ± 6.3 | 25.6 ± 5.2 | 0.8 ± 0.1 | 10.7 ± 2.4 | 19.5 ± 4.6 | 18.7 ± 2.3 | 7.8 ± 1.1 | 104.5 ± 18.5 | 19.5 ± 3.0 | 89.9 ± 13.5 | 2.9 ± 0.1 | 1.4 ± 0.1 | 60.1 ± 4.2 | 74.4 ± 4.8 | |
| PD1 | 19 | 107.5 ± 19.6 | 58.7 ± 9.7 | 25.3 ± 4.8 | 0.8 ± 0.1 | 11.6 ± 2.9 | 16.5 ± 5.7 | 18.7 ± 2.0 | 7.4 ± 1.3 | 93.3 ± 18.9 | 17.7 ± 4.0 | 87.9 ± 16.6 | 2.9 ± 0.1 | 1.4 ± 0.1 | 68.3 ± 6.4 | 81.9 ± 5.6 | |
| PD2 | 16 | 118.7 ± 14.2 | 60.2 ± 12.0 | 25.7 ± 4.9 | 0.8 ± 0.1 | 9.8 ± 1.4 | 21.8 ± 4.4 | 18.9 ± 2.4 | 8.2 ± 1.4 | 98.6 ± 15.6 | 18.5 ± 3.1 | 98.6 ± 11.9 | 2.9 ± 0.1 | 1.4 ± 0.1 | 63.6 ± 5.2 | 76.9 ± 4.3 | |
| P. ionanthum | PI3 | 19 | 109.7 ± 21.2 | 67.7 ± 7.4 | 29.3 ± 5.0 | 0.7 ± 0.1 | 12.1 ± 2.0 | 25.8 ± 5.3 | 10.4 ± 1.7 | 9.3 ± 1.4 | 69.9 ± 12.3 | 2.1 ± 0.2 | 94.5 ± 21.1 | 4.2 ± 0.3 | 2 ± 0.0 | 62.1 ± 8.5 | 82.4 ± 6.2 |
| PI4 | 20 | 121.0 ± 18.8 | 69.1 ± 7.1 | 31.4 ± 5.2 | 0.8 ± 0.1 | 8.4 ± 1.5 | 25.8 ± 4.7 | 11.1 ± 1.3 | 9.5 ± 1.4 | 70.5 ± 14.4 | 2.3 ± 0.4 | 108.0 ± 16.8 | 4.4 ± 0.3 | 2.0 ± 0.0 | 56.6 ± 9.4 | 75.8 ± 7.4 | |
| PI5 | 19 | 126.3 ± 21.0 | 66.3 ± 9.7 | 31.0 ± 3.6 | 0.8 ± 0.1 | 9.0 ± 2.1 | 28.5 ± 5.3 | 11.8 ± 1.6 | 10.1 ± 1.4 | 73.3 ± 11.4 | 2.2 ± 0.3 | 116.1 ± 20.9 | 4.4 ± 0.3 | 2.0 ± 0.1 | 77.3 ± 8.8 | 92.8 ± 8.9 | |
| PI1 | 19 | 96.5 ± 6.7 | 67.8 ± 7.7 | 26.1 ± 3.0 | 0.8 ± 0.1 | 13.9 ± 2.4 | 23.0 ± 2.3 | 10.7 ± 1.3 | 9.7 ± 1.1 | 72.3 ± 11.2 | 2.0 ± 0.1 | 88.7 ± 3.4 | 4.2 ± 0.2 | 2.0 ± 0.0 | 51.6 ± 10.8 | 74.0 ± 9.3 | |
| PI2 | 19 | 102.1 ± 20.8 | 72.3 ± 7.6 | 32.3 ± 5.7 | 0.8 ± 0.1 | 15.3 ± 3.1 | 22.0 ± 6.4 | 11.9 ± 1.8 | 11.0 ± 1.8 | 82.1 ± 11.9 | 2.0 ± 0.1 | 89.3 ± 17.6 | 4.6 ± 0.2 | 2.2 ± 0.2 | 68.3 ± 8.2 | 83.3 ± 7.6 | |
| P. regnellii | PR2 | 18 | 183.3 ± 12.8 | 92.8 ± 7.6 | 34.5 ± 5.3 | 2.3 ± 0.2 | 14.9 ± 3.2 | 18.8 ± 2.6 | 12.4 ± 1.8 | 10.3 ± 1.2 | 144.9 ± 23.3 | 9.4 ± 1.3 | 164.1 ± 16.2 | 2.5 ± 0.0 | 1.5 ± 0.0 | 133.9 ± 16.0 | 165.0 ± 2.5 |
| PR1 | 20 | 185.6 ± 11.7 | 95.5 ± 2.8 | 33.8 ± 3.5 | 2.3 ± 0.2 | 18.0 ± 2.6 | 17.9 ± 2.00 | 14.1 ± 2.2 | 10.7 ± 1.1 | 179.8 ± 29.0 | 10.6 ± 2.2 | 163.9 ± 16.8 | 2.5 ± 0.0 | 1.5 ± 0.0 | 116.2 ± 8.8 | 161.2 ± 1.8 | |
| PR3 | 20 | 185 ± 10.9 | 99.0 ± 9.8 | 34.7 ± 5.4 | 2.3 ± 0.3 | 15.8 ± 1.9 | 19.2 ± 2.0 | 13.8 ± 2.3 | 11.0 ± 1.7 | 164.0 ± 33.2 | 10.3 ± 2.1 | 154.9 ± 14.6 | 2.5 ± 0.0 | 1.5 ± 0.0 | 142.3 ± 17.7 | 169.5 ± 5.7 | |
| PR4 | 20 | 184.5 ± 12.2 | 101.2 ± 7.2 | 35 ± 5.3 | 2.4 ± 0.3 | 14.3 ± 2.2 | 19.0 ± 3.0 | 14.4 ± 2.3 | 11.7 ± 1.6 | 163.4 ± 25.0 | 11.0 ± 2.1 | 157.4 ± 12.9 | 2.5 ± 0.0 | 1.5 ± 0.0 | 144.2 ± 12.7 | 167.6 ± 4.4 | |
| PR5 | 19 | 197.9 ± 8.5 | 112.1 ± 24.2 | 37.9 ± 5.5 | 2.2 ± 0.1 | 16.0 ± 1.7 | 20.3 ± 2.4 | 15.5 ± 2.4 | 12.2 ± 1.3 | 167.9 ± 24.2 | 11.8 ± 2.9 | 172.6 ± 11.9 | 2.5 ± 0.0 | 1.5 ± 0.0 | 131.4 ± 16.0 | 165.4 ± 4.9 | |
| P. urvillei | PU1 | 16 | 199.1 ± 8.4 | 135.3 ± 10.4 | 43.3 ± 5.1 | 1.6 ± 0.1 | 20.8 ± 2.1 | 20.4 ± 2.1 | 28.4 ± 4.9 | 11.4 ± 2.0 | 139.0 ± 24.0 | 14.4 ± 3.6 | 177.6 ± 18.3 | 2.9 ± 0.1 | 1.5 ± 0.0 | 43 ± 10.1 | 70.1 ± 3.6 |
| PU3 | 18 | 216.4 ± 11.2 | 118.1 ± 9.6 | 36 ± 2.7 | 1.2 ± 0.1 | 20.2 ± 2.4 | 19.4 ± 3.0 | 29.4 ± 3.3 | 10.5 ± 1.8 | 145.5 ± 23.1 | 14.8 ± 2.8 | 191.0 ± 13.6 | 2.9 ± 0.1 | 1.5 ± 0.0 | 52.8 ± 16 | 82.2 ± 14.8 | |
| PU4 | 17 | 213.1 ± 15.6 | 117 ± 9.6 | 34.4 ± 5.4 | 1.2 ± 0.1 | 22.7 ± 2.5 | 20.7 ± 2.1 | 31.4 ± 3.2 | 10.4 ± 1.1 | 133.7 ± 13.8 | 15.8 ± 1.6 | 183.0 ± 16.3 | 2.8 ± 0.1 | 1.5 ± 0.0 | 56.6 ± 23.2 | 85.6 ± 18 | |
| PU5 | 17 | 206.7 ± 12.2 | 124.9 ± 11.4 | 39.7 ± 5.6 | 1.4 ± 0.2 | 20.4 ± 2.4 | 19.5 ± 2.8 | 29.5 ± 4.0 | 11.1 ± 2.1 | 139.7 ± 20.6 | 14.1 ± 1.9 | 177.2 ± 17.7 | 2.8 ± 0.1 | 1.5 ± 0.0 | 72.1 ± 25 | 97.4 ± 26.4 | |
| PU2 | 20 | 196 ± 13.2 | 119 ± 12.5 | 35.8 ± 4.4 | 1.4 ± 0.2 | 19.6 ± 2.0 | 18.3 ± 2.5 | 28.5 ± 5.8 | 11.8 ± 2.2 | 144.6 ± 16.9 | 12.9 ± 2.4 | 171.2 ± 15.7 | 2.7 ± 0.2 | 1.5 ± 0.0 | 47.8 ± 17.3 | 79.6 ± 17.5 |
| Species | Pop | n | Molecular Diversity Indexes | Genotypic Diversity Indexes | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TB | CB | %PL | PB | G | D | E | H | |||
| P. intermedium | PIN2 | 14 | 81 | 3 | 0 | 1 | 1 | 0 | 1 | 0 |
| PIN4 | 18 | 75 | 3 | 0 | 3 | 1 | 0 | 1 | 0 | |
| PIN7 | 18 | 78 | 1 | 4.4 | 0 | 1 | 0 | 1 | 0.022 | |
| PIN8 | 18 | 78 | 1 | 2.2 | 0 | 1 | 0 | 1 | 0.008 | |
| PIN9 | 19 | 77 | 2 | 1.1 | 0 | 1 | 0 | 1 | 0.007 | |
| P. durifolium | PD1 | 19 | 123 | 0 | 77.4 | 0 | 19 | 1 | 1 | 1.28 |
| PD2 | 16 | 123 | 0 | 80.7 | 0 | 16 | 1 | 1 | 1.20 | |
| PD3 | 16 | 123 | 0 | 79.8 | 0 | 16 | 1 | 1 | 1.20 | |
| PD4 | 16 | 123 | 0 | 75.8 | 0 | 16 | 1 | 1 | 1.20 | |
| PD5 | 20 | 123 | 0 | 79.8 | 0 | 20 | 1 | 1 | 1.30 | |
| P. ionanthum | PI1 | 19 | 131 | 0 | 76.3 | 0 | 19 | 1 | 1 | 1.27 |
| PI2 | 19 | 132 | 2 | 76.3 | 0 | 19 | 1 | 1 | 1.27 | |
| PI3 | 19 | 134 | 1 | 81.3 | 0 | 19 | 1 | 1 | 1.27 | |
| PI4 | 19 | 134 | 1 | 80.6 | 1 | 19 | 1 | 1 | 1.27 | |
| PI5 | 20 | 131 | 0 | 74.8 | 0 | 20 | 1 | 1 | 1.30 | |
| P. regnellii | PR1 | 20 | 120 | 1 | 26.2 | 0 | 19 | 0.99 | 0.96 | 1.27 |
| PR2 | 18 | 120 | 0 | 25.4 | 0 | 18 | 1 | 1 | 1.26 | |
| PR3 | 20 | 120 | 0 | 21.3 | 0 | 13 | 0.91 | 0.59 | 1.01 | |
| PR4 | 20 | 116 | 0 | 14.8 | 0 | 8 | 0.77 | 0.47 | 0.72 | |
| PR5 | 19 | 115 | 1 | 15.6 | 0 | 9 | 0.81 | 0.48 | 0.79 | |
| P. urvillei | PU1 | 15 | 100 | 0 | 49.0 | 0 | 15 | 1 | 1 | 1.18 |
| PU2 | 20 | 101 | 0 | 53.9 | 0 | 20 | 1 | 1 | 1.30 | |
| PU3 | 19 | 100 | 0 | 42.2 | 0 | 19 | 1 | 1 | 1.28 | |
| PU4 | 18 | 100 | 0 | 46.1 | 0 | 18 | 1 | 1 | 1.26 | |
| PU5 | 18 | 102 | 0 | 55.9 | 0 | 18 | 1 | 1 | 1.26 | |
| Species | Source | df | SS | MS | Est. Var | RhoST | p |
|---|---|---|---|---|---|---|---|
| P. intermedium | Among Pops | 4 | 534.5 | 133.6 | 7.7 | 0.851 | 0.001 |
| Within Pops | 82 | 20.2 | 0.25 | 0.3 | |||
| P. durifolium | Among Pops | 4 | 188.6 | 47.2 | 1.7 | 0.084 | 0.001 |
| Within Pops | 82 | 1490.7 | 18.1 | 18.2 | |||
| P. ionanthum | Among Pops | 4 | 380.7 | 95.2 | 4.0 | 0.18 | 0.001 |
| Within Pops | 91 | 1666.8 | 18.3 | 18.3 | |||
| P. regnellii | Among Pops | 4 | 160.5 | 40.1 | 1.8 | 0.29 | 0.001 |
| Within Pops | 92 | 415.8 | 4.5 | 4.5 | |||
| P. urvillei | Among Pops | 4 | 200.4 | 50.1 | 2.3 | 0.20 | 0.001 |
| Within Pops | 85 | 793.8 | 9.3 | 9.3 |
| P. intermedium | |||||||||||
| Pop | PIN2 | PIN4 | PIN7 | PIN8 | PIN9 | ||||||
| PIN2 | - | *** | *** | *** | *** | ||||||
| PIN4 | 0.12 | - | *** | *** | *** | ||||||
| PIN7 | 0.07 | 0.13 | - | *** | *** | ||||||
| PIN8 | 0.07 | 0.13 | 0.04 | - | *** | ||||||
| PIN9 | 0.09 | 0.09 | 0.08 | 0.08 | - | ||||||
| P. durifolium | P. ionanthum | ||||||||||
| Pop | PD1 | PD2 | PD3 | PD4 | PD5 | Pop | PI1 | PI2 | PI3 | PI4 | PI5 |
| PD1 | - | ** | ** | ** | *** | PI1 | - | n.s. | n.s. | n.s. | *** |
| PD2 | 0.02 | - | n.s. | n.s. | *** | PI2 | 0.04 | - | n.s. | n.s. | *** |
| PD3 | 0.02 | 0.02 | - | n.s. | *** | PI3 | 0.04 | 0.03 | - | n.s. | *** |
| PD4 | 0.02 | 0.01 | 0.01 | - | *** | PI4 | 0.05 | 0.04 | 0.03 | - | *** |
| PD5 | 0.02 | 0.02 | 0.01 | 0.02 | - | PI5 | 0.05 | 0.05 | 0.04 | 0.04 | - |
| P. regnellii | P. urvillei | ||||||||||
| Pop | PR1 | PR2 | PR3 | PR4 | PR5 | Pop | PU1 | PU2 | PU3 | PU4 | PU5 |
| PR1 | - | n.s. | *** | *** | *** | PU1 | - | n.s. | n.s. | n.s. | n.s. |
| PR2 | 0.02 | - | n.s. | *** | *** | PU2 | 0.02 | - | n.s. | n.s. | n.s. |
| PR3 | 0.01 | 0.01 | - | *** | *** | PU3 | 0.02 | 0.03 | - | n.s. | n.s. |
| PR4 | 0.02 | 0.01 | 0.01 | - | *** | PU4 | 0.03 | 0.03 | 0.03 | - | n.s. |
| PR5 | 0.03 | 0.02 | 0.02 | 0.02 | - | PU5 | 0.02 | 0.03 | 0.03 | 0.03 | - |
| Species | Pop | N | rm ⁋ | Location and Habitat Type |
|---|---|---|---|---|
| P. durifolium | PD1 | 19 | S * ss | C, 27.627033° S, 56.747883° W. U. |
| PD2 | 18 | S ss | C, 27.931683° S, 57.5166° W. U. | |
| PD3 | 17 | S * ss | C, 28.290583° S, 56.770316° W. D. | |
| PD4 | 18 | S * ss | C, 27.825216° S, 56.2549° W. U. | |
| PD5 | 20 | S * ss | C, 27.560766° S, 57.153066° W. U. | |
| P. ionanthum | PI1 | 20 | S * ss | C, 27.821083° S, 56.27213° W. D. |
| PI2 | 20 | S * ss | C, 28.237416° S, 58.0542° W. U. | |
| PI3 | 16 | S ss | C, 27.753416° S, 57.76096° W. U. | |
| PI4 | 20 | S ss | C, 28.033733° S, 58.0362° W. U. | |
| PI5 | 20 | S ss | C, 29.1651° S, 59.096583° W. D. | |
| P. regnellii | PR1 | 20 | S sf | M, 26.54153° S, 54.7251° W. D. |
| PR2 | 19 | S sf | M, 27.052833° S, 55.25066° W. U. | |
| PR3 | 20 | S sf | M, 26.7271° S, 54.247416° W. D. | |
| PR4 | 20 | S sf | M, 27.374017° S, 54.42505° W. U. | |
| PR5 | 19 | S sf | M, 27.77713° S, 55.24996° W. U. | |
| P. urvillei | PU1 | 17 | S sf | M, 27.276883° S, 55.46225° W. D. |
| PU2 | 20 | S sf | C, 27.821083° S, 56.272133° W. D. | |
| PU3 | 20 | S sf | ER, 31.02038° S, 59.420366° W. D. | |
| PU4 | 20 | S sf | SF, 28.59555° S, 59.41675° W. D. | |
| PU5 | 18 | S sf | Ch, 26.40195° S, 59.37876° W. D. | |
| P. intermedium | PIN2 | 14 | A ap ps sf | C, 28.87166° S, 57.26405° W. D. |
| PIN4 | 18 | A ap ps sf | C, 27.615016° S, 58.75398° W. U. | |
| PIN7 | 18 | A ap ps sf | SF, 29.29074° S, 60.17697° W. U. | |
| PIN8 | 18 | A ap ps sf | SF, 28.15866° S, 60.74791° W. D. | |
| PIN9 | 19 | A ap ps sf | C, 27.38394° S, 57.78926° W. U. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reutemann, A.V.; Schedler, M.; Hojsgaard, D.H.; Brugnoli, E.A.; Zilli, A.L.; Acuña, C.A.; Honfi, A.I.; Martínez, E.J. The Role of Reproductive Modes in Shaping Genetic Diversity in Polyploids: A Comparative Study of Selfing, Outcrossing, and Apomictic Paspalum Species. Plants 2025, 14, 476. https://doi.org/10.3390/plants14030476
Reutemann AV, Schedler M, Hojsgaard DH, Brugnoli EA, Zilli AL, Acuña CA, Honfi AI, Martínez EJ. The Role of Reproductive Modes in Shaping Genetic Diversity in Polyploids: A Comparative Study of Selfing, Outcrossing, and Apomictic Paspalum Species. Plants. 2025; 14(3):476. https://doi.org/10.3390/plants14030476
Chicago/Turabian StyleReutemann, A. Verena, Mara Schedler, Diego H. Hojsgaard, Elsa A. Brugnoli, Alex L. Zilli, Carlos A. Acuña, Ana I. Honfi, and Eric J. Martínez. 2025. "The Role of Reproductive Modes in Shaping Genetic Diversity in Polyploids: A Comparative Study of Selfing, Outcrossing, and Apomictic Paspalum Species" Plants 14, no. 3: 476. https://doi.org/10.3390/plants14030476
APA StyleReutemann, A. V., Schedler, M., Hojsgaard, D. H., Brugnoli, E. A., Zilli, A. L., Acuña, C. A., Honfi, A. I., & Martínez, E. J. (2025). The Role of Reproductive Modes in Shaping Genetic Diversity in Polyploids: A Comparative Study of Selfing, Outcrossing, and Apomictic Paspalum Species. Plants, 14(3), 476. https://doi.org/10.3390/plants14030476










