Genome-Wide Identification of the AGC Kinase Family in Tetraploid Potato (Solanum tuberosum L.) Cultivar ‘Qingshu No. 9’ and Functional Analysis of StD6PK in Response to Late Blight (Phytophthora infestans)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Processing
2.2. Identification of AGC Gene Family in Tetraploid Potato ‘Qingshu No. 9’
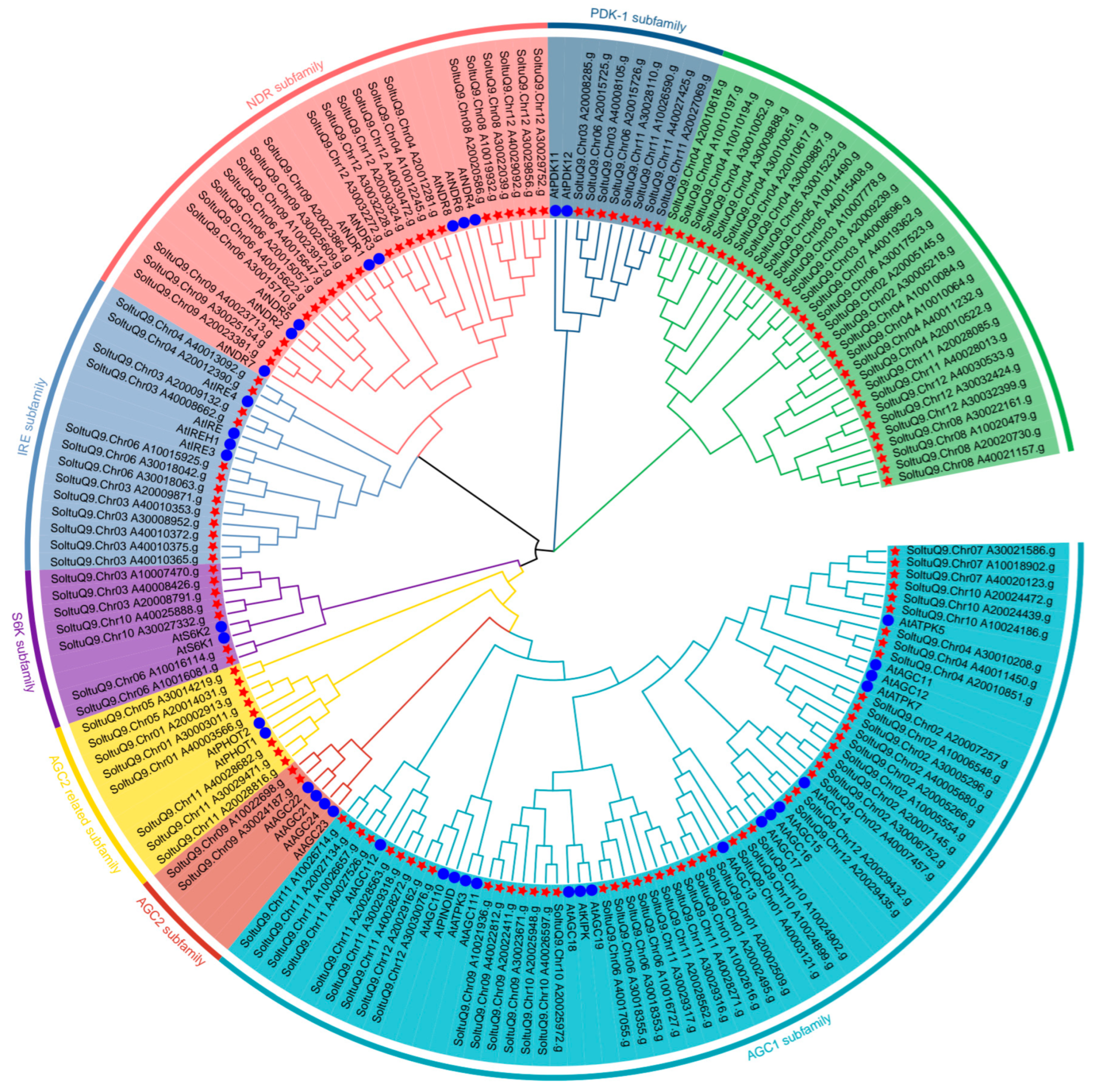
2.3. Phylogenetic Analysis, Protein Conserved Motif Analysis, and Physico-Chemical Characterization of Potato AGC Protein
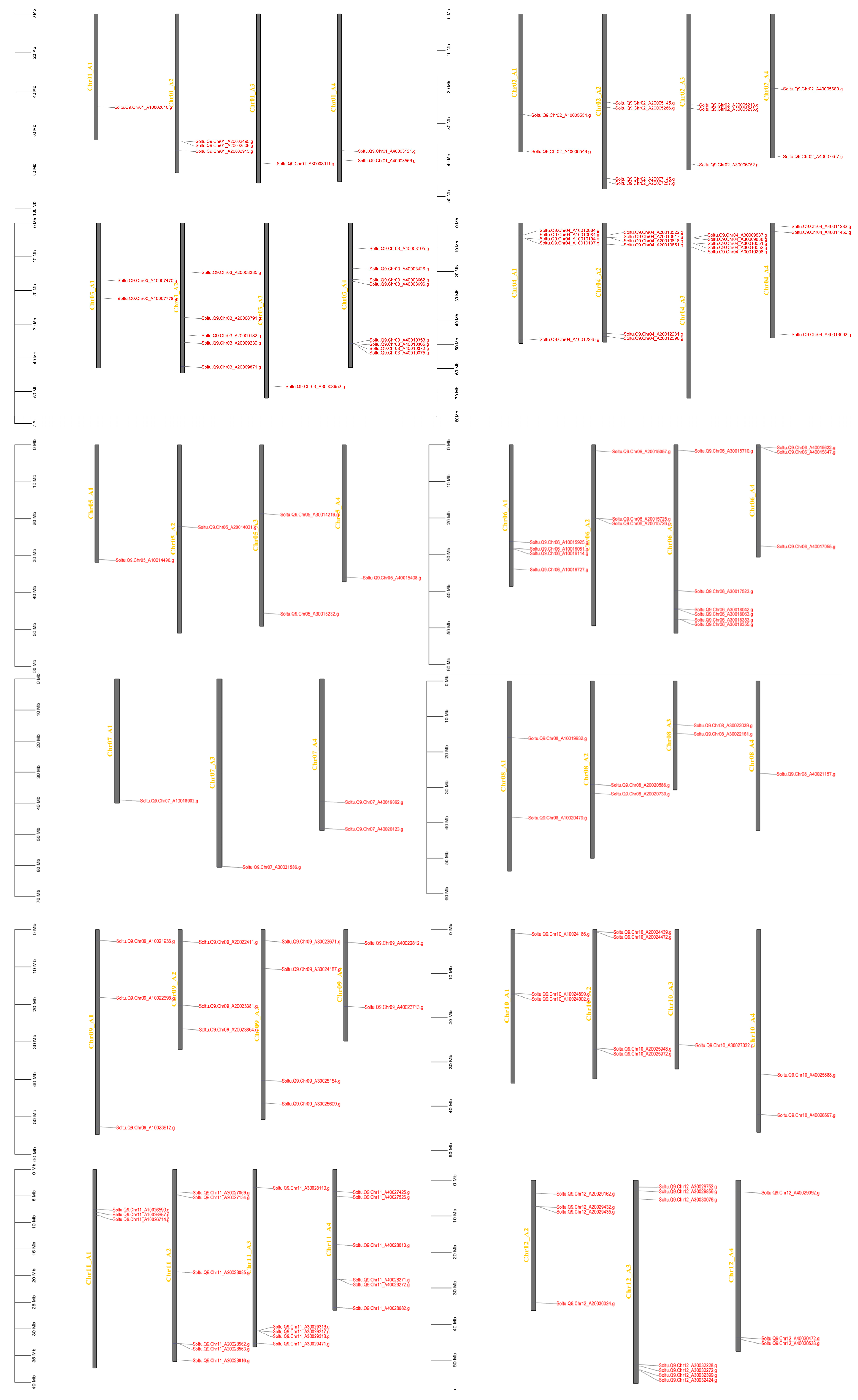
2.4. Collinearity Analysis in AGC
2.5. Analysis of Cis-Acting Elements
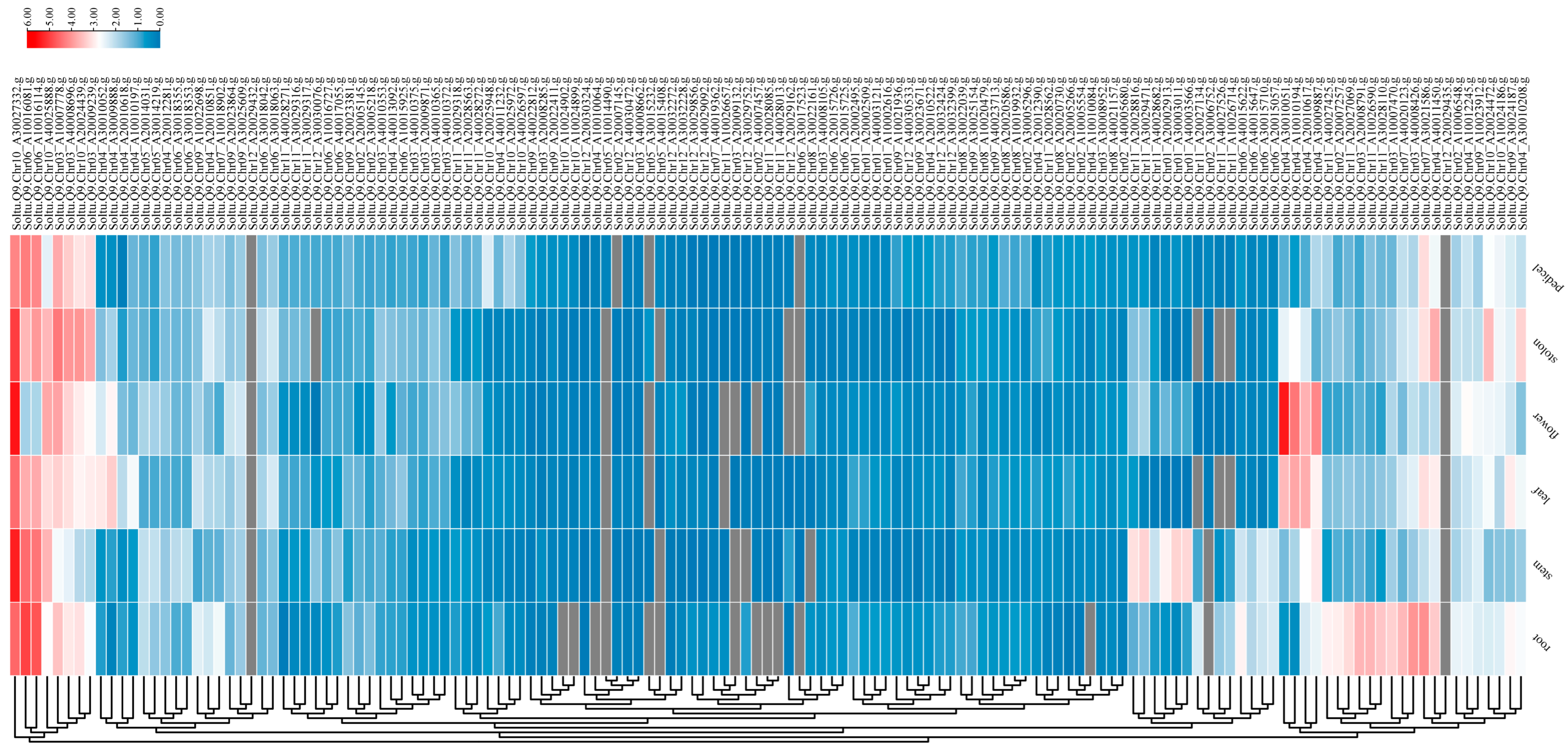
2.6. Tissue Expression Profile Analysis
2.7. Expression Analysis of AGC Kinase Family Under Late-Blight Stress in Potato
2.8. qRT-PCR Analysis of StAGCs Under Late-Blight Stress in Potato
2.9. Subcellular Localization of StD6PKConstruction and Localization Observation of StD6PK Subcellular Carriers
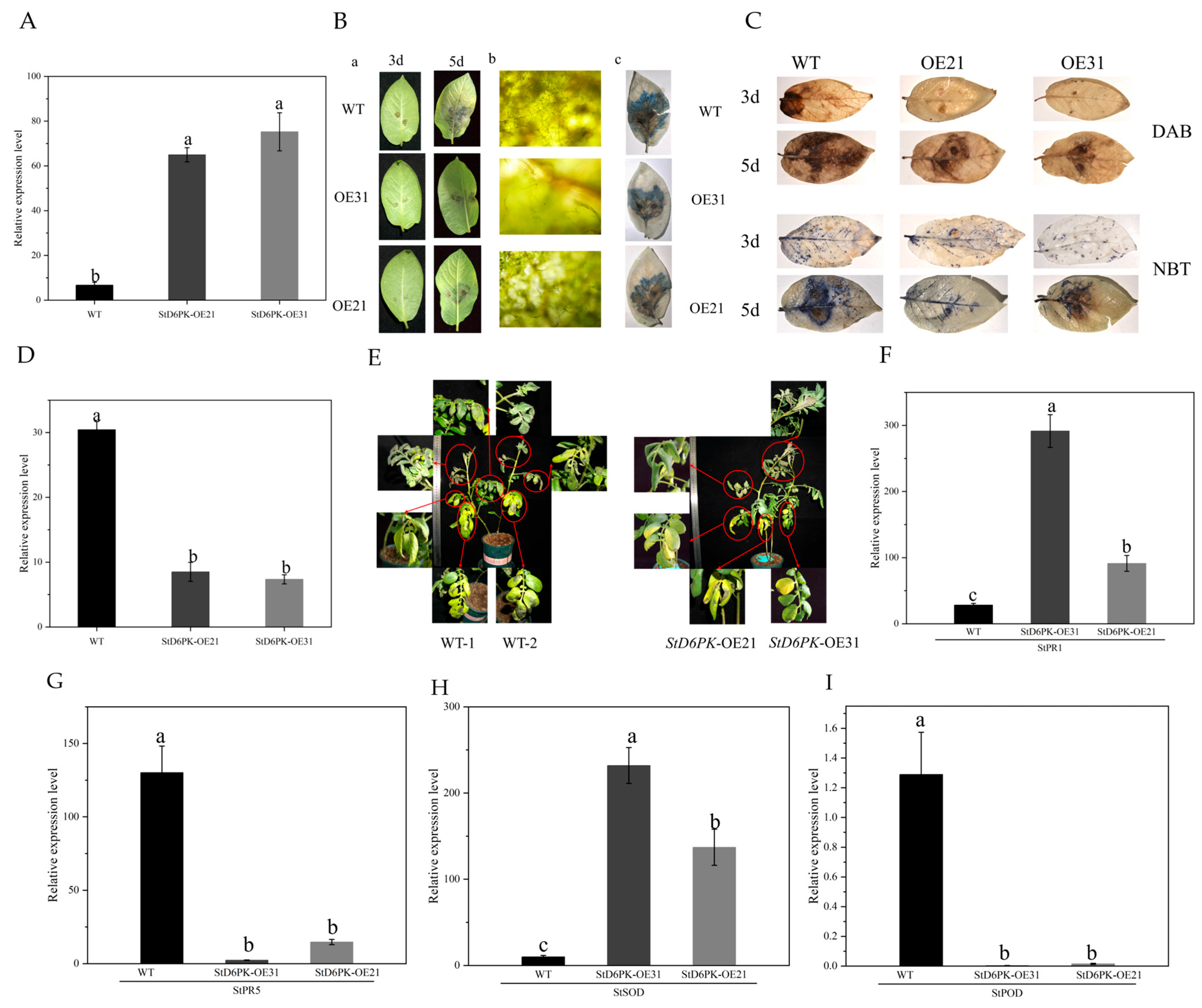
2.10. Generation of Transgenic StD6PK Potato Plants
2.11. Inoculation of Phytophthora Infestans Zoospore Fluid, Trypan Blue, and DAB and NBT Staining
3. Results
3.1. Identification of AGC Gene Family Members in Potato ‘Qingshu No. 9’ and Analysis of Their Physico-Chemical Properties
3.2. Analysis of AGC Gene Family Structure and Conserved Motifs in ‘Qingshu No. 9’
3.3. Chromosome Distribution and Collinearity Analysis of AGC Family Genes in ‘Qingshu No. 9’
3.4. Analysis of Cis-Acting Elements of AGC Gene Family in ‘Qingshu No. 9’
3.5. Expression Patterns of AGC Gene Family Members in Different Tissues of ‘Qingshu No. 9’
3.6. Expression Analysis of StAGCs Genes Under Potato Late-Blight Stress
3.7. Subcellular Localization of StD6PK
3.8. Overexpression of StD6PK Enhances Resistance to Late Blight in Potato
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bilbrough, T.; Piemontese, E.; Seitz, O. Dissecting the role of protein phosphorylation: A chemical biology toolbox. Chem. Soc. Rev. 2022, 51, 5691–5730. [Google Scholar] [CrossRef]
- Pearce, L.R.; Komander, D.; Alessi, D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010, 11, 9–22. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, X.; Zhou, M.; Yang, J.; Ke, S.; Li, Y. Genome-Wide identification of the AGC protein kinase gene family related to Photosynthesis in Rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 12557. [Google Scholar] [CrossRef]
- Garcia, A.V.; Al-Yousif, M.; Hirt, H. Role of AGC kinases in plant growth and stress responses. Cell. Mol. Life Sci. 2012, 69, 3259–3267. [Google Scholar] [CrossRef]
- Rademacher, E.H.; Offringa, R. Evolutionary adapt ations of plant AGC kinases: From light signaling to cell polarity regulation. Front. Plant Sci. 2012, 3, 250. [Google Scholar] [CrossRef]
- Zegzouti, H.; Anthony, R.G.; Jahchan, N.; Bögre, L.; Christensen, S.K. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 6404–6409. [Google Scholar] [CrossRef] [PubMed]
- Bogre, L.; Okresz, L.; Henriques, R.; Anthony, R.G. Growth signaling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003, 8, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Sunghan, K.; Delauney, A.J.; Verma, S. Arabidopsis target of rapamycin Interacts with raptor, Which Regulates the Activity of S6 Kinase in Response to Osmotic Stress Signals. Plant Cell 2006, 18, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Franziska, T.; Frederic, Z.; Kozma, S.C.; Thomas, G.; Ferenc, N. Phytohormones Participate in an S6 Kinase Signal Transduction Pathway in Arabidopsis. Plant Physiol. 2004, 134, 1527–1535. [Google Scholar] [CrossRef]
- Marc, B.; Alba, G.R.; Laia, C.; Bernat, M.; Ferga, H.; Sanata, T.; Adrián, P.; Monte-Bello, C.; Caldana, C.; Henriques, R. 40S Ribosomal protein S6 kinase integrates daylength perception and growth regulation in Arabidopsis thaliana. Plant Physiol. 2024, 195, 3039–3052. [Google Scholar] [CrossRef]
- Wei, Y.; Tan, X.; Tian, T.; Luo, X.; Ren, M. Ribosomal S6 kinases 2 mediates potato resistance to late blight, through WRKY59 transcription factor. Int. J. Biol. Macromol. 2024, 277, 134581. [Google Scholar] [CrossRef]
- Pislariu, C.I.; Dickstein, R. An IRE-Like AGC Kinase Gene, MtIRE, Has Unique Expression in the Invasion Zone of Developing Root Nodules in Medicago truncatula. Plant Physiol. 2007, 144, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Shimura, Y.; Okada, K. The IRE gene encodes a protein kinase homologue and modulates root hair growth in Arabidopsis. Plant J. 2002, 30, 289–299. [Google Scholar] [CrossRef]
- Coppinger, P.; Repetti, P.P.; Day, B.; Dahlbeck, D.; Mehlert, A.; Staskawicz, B.J. Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 2004, 40, 225–237. [Google Scholar] [CrossRef]
- Day, B.; Dahlbeck, D.; Staskawicz, B.J. NDR1 Interaction with RIN4 Mediates the Differential Activation of Multiple Disease Resistance Pathways in Arabidopsis. Plant Cell 2006, 18, 2782–2791. [Google Scholar] [CrossRef]
- Cheng, Y.; Qin, G.; Dai, X.; Zhao, Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 21017–21022. [Google Scholar] [CrossRef]
- Christensen, S.K.; Dagenais, N.; Chory, J.; Weigel, D. Regulation of auxin response by the protein kinase PINOID. Cell 2000, 18, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Glanc, M.; Van Gelderen, K.; Hoermayer, L.; Tan, S.; Naramoto, S.; Zhang, X.; Domjan, D.; Včelařová, L.; Hauschild, R.; Johnson, A.; et al. AGC kinases and MAB4/MEL proteins maintain PIN polarity by limiting lateral diffusion in plant cells. Curr. Biol. 2021, 31, 1918–1930.e5. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, Y.-L.; Shi, X.; Li, H.; Yuan, X.; Li, S.; Zhang, Y. A positive feedback circuit for ROP-mediated polar growth. Mol. Plant 2021, 14, 395–410. [Google Scholar] [CrossRef]
- Sukumar, P.; Edwards, K.S.; Rahman, A.; DeLong, A.; Muday, G.K. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in arabidopsis. Plant Physiol. 2009, 150, 722–735. [Google Scholar] [CrossRef]
- Zhang, Y.; He, J.; McCormick, S. Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes. Plant J. 2009, 58, 474–484. [Google Scholar] [CrossRef]
- Han, J.; Liu, C.-X.; Liu, J.; Wang, C.-R.; Wang, S.-C.; Miao, G. AGC kinases OXI1 and AGC2-2 regulate camalexin secretion and disease resistance by phosphorylating transporter PDR6. Plant Physiol. 2024, 195, 1835–1850. [Google Scholar] [CrossRef]
- Christie, J.M.; Blackwood, L.; Petersen, J.; Sullivan, S. Plant flavoprotein photoreceptors. Plant Cell Physiol. 2015, 56, 401–413. [Google Scholar] [CrossRef]
- Eckstein, A.; Grzyb, J.; Hermanowicz, P.; Zgłobicki, P.; Łabuz, J.; Strzałka, W.; Dziga, D.; Banaś, A.K. Arabidopsis phototropins participate in the regulation of dark-induced leaf senescence. Int. J. Mol. Sci. 2021, 22, 1836. [Google Scholar] [CrossRef]
- Inoue, S.; Kaiserli, E.; Zhao, X.; Waksman, T.; Takemiya, A.; Okumura, M.; Takahashi, H.; Seki, M.; Shinozaki, K.; Endo, Y.; et al. CIPK23 regulates blue light-dependent stomatal opening in Arabidopsis thaliana. Plant J. 2020, 104, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, Y.-P.; Zhu, J.-D.; Wang, Y.-X.; Yang, M.-Y.; Yan, H.-R.; Lv, Q.-Y.; Cheng, K.; Zhao, X.; Zhang, X. Phototropin 1 mediates high-intensity blue light-induced chloroplast accumulation response in a root phototropism 2-dependent manner in arabidopsis phot2 mutant plants. Front. Plant Sci. 2021, 12, 704618. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, J.; Sheng, Y.; Tian, Y.; Zhang, Y.; Zhou, C.; Zhao, X.; Zhang, X. Phototropin2-mediated hypocotyl phototropism is negatively regulated by JAC1 and RPT2 in Arabidopsis. Plant Physiol. Biochem. 2021, 164, 289–298. [Google Scholar] [CrossRef]
- Galen, C.; Rabenold, J.J.; Liscum, E. Functional ecology of a blue light photoreceptor: Effects of phototropin-1 on root growth enhance drought tolerance in Arabidopsis thaliana. New Phytol. 2007, 173, 91–99. [Google Scholar] [CrossRef]
- Shrestha, B.; Darapuneni, M.; Stringam, B.L.; Lombard, K.; Djaman, K. Irrigation Water and Nitrogen Fertilizer Management in Potato (Solanum tuberosum L.): A Review. Agronomy 2023, 13, 2566. [Google Scholar] [CrossRef]
- Ortiz, V.; Phelan, S.; Mullins, E. A temporal assessment of nematode community structure and diversity in the rhizo-sphere of cisgenic Phytophthora infestans-resistant potatoes. BMC Ecol. 2016, 16, 55. [Google Scholar] [CrossRef]
- Xie, C.H. Status and development of potato industry. J. Huazhong Agric. Univ. Soc. Sci. Ed. 2012, 97, 1–4. [Google Scholar]
- Gedlu, D.; Hailu, N.; Kefelegn, H. Integrated management of potato late blight (Phytophthora infestans (Mont) de Bary) through resistant varieties and fungicides in North Shewa, Ethiopia. J. Plant Pathol. 2023, 105, 95–106. [Google Scholar] [CrossRef]
- Bekele, B.; Abate, E.; Asefa, A.; Dickinson, M. Incidence of potato viruses and bacterial wilt disease in the west Amhara sub-region of Ethiopia. J. Plant Pathol. 2011, 93, 149–157. [Google Scholar]
- Rodriguez, D.; Uribe, P.; Benavides, C.A. Response of commercial potato genotypes Solanum tuberosum L. to Phytophthora infestans (Mont.) de Bary late blight attack. Rev. Cienc. Agric. 2023, 40, e1200. [Google Scholar] [CrossRef]
- Wang, W.; Long, Y. A review of biocontrol agents in controlling late blight of potatoes and tomatoes caused by Phytophthora infestans and the underlying mechanisms. Pest Manag. Sci. 2023, 79, 4715–4725. [Google Scholar] [CrossRef] [PubMed]
- Fry, W.E.; Goodwin, S.B. Resurgence of the irish potato famine fungus. BioScience 1997, 47, 363–371. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Groves, C.T.; Parra, G.R. PCR amplification of the Irish potato famine pathogen from historic specimens. Nature 2001, 411, 695–697. [Google Scholar] [CrossRef] [PubMed]
- Bourke, P.M.A. Emergence of potato blight. Nature 1964, 203, 805–808. [Google Scholar] [CrossRef]
- Savary, S.; Bregaglio, S.; Willocquet, L.; Gustafson, D.; Mason D’Croz, D.; Sparks, A.; Castilla, N.; Djurle, A.; Allinne, C.; Sharma, M.; et al. Crop health and its global impacts on the components of food security. Food Secur. 2017, 9, 311–327. [Google Scholar] [CrossRef]
- Liao, W.; Zhuo, G.; Ni, M.; Wang, J.; Zhang, Y.; Qu, J.; Liu, J.; Yin, Z. The introduction, evaluation and utilization of full powder processed potato new variety “Qingshu No. 9”. Tibet J. Agric. Sci. 2012, 34, 36–41. [Google Scholar]
- Tian, P.; Zheng, J.; Wang, B.; Jiao, W.; Sang, J.; Ma, Z.; Han, P.; Zhang, H.; Song, Y.; Meng, Y.; et al. Genome-wide DNA methylation landscape and its association with the transcriptome reprogramming in potato in response to Phytophthora infestans infection. Hortic. Res. 2025, uhaf297. [Google Scholar] [CrossRef]
- Wang, F.; Xia, Z.; Zou, M.; Zhao, L.; Jiang, S.; Zhou, Y.; Zhang, C.; Ma, Y.; Bao, Y.; Sun, H.; et al. The autotetraploid potato genome provides insights into highly heterozygous species. Plant Biotechnol. J. 2022, 20, 1996–2005. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- He, M.; Zhou, Y.; Ye, G.; Zheng, J.; Meng, Y.; Wang, J.; Shan, W. Serial Transcriptome Analysis Reveals Genes Associated with Late Blight Resistance in Potato Cultivar Qingshu 9. Agronomy 2021, 11, 1919. [Google Scholar] [CrossRef]
- Nathalie, N.; Jean, F.; Lucien, H.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar]
- Lv, C.; Bao, Y.; Xu, M.; Deng, K.; Zhao, L.; Zhao, Y.; Zhou, Y.; Feng, Y.; Wang, F. Genome-Wide Identification of the StPYL Gene Family and Analysis of the Functional Role of StPYL9a-like in Salt Tolerance in Potato (Solanum tuberosum L.). Plants 2025, 14, 2731. [Google Scholar] [CrossRef]
- Bouaziz, D.; Ayadi, M.; Bidani, A.; Rouis, S.; Nouri-Ellouz, O.; Jellouli, R.; Drira, N.; Gargouri-Bouzid, R. A stable cytosolic expression of VH antibody fragment directed against PVY NIa protein in transgenic potato plant confers partial protection against the virus. Plant Sci. 2009, 176, 489–496. [Google Scholar] [CrossRef]
- Bézier, A.; Lambert, B.; Baillieul, F. Study of defense-related gene expression in grapevine leaves and berries infected with Botrytis cinerea. Eur. J. Plant Pathol. 2002, 108, 111–120. [Google Scholar] [CrossRef]
- Thomas, C.; Meyer, D.; Wolff, M.; Himber, C.; Alioua, M.; Steinmetz, A. Molecular characterization and spatial expression of the sunflower ABP1 gene. Plant Mol. Biol. 2002, 52, 1025–1036. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Z.; Kang, L.; Li, M.; Zhang, J.; Feng, Y.; Yin, J.; Gong, X.; Zhao, J. Small G Protein StRab5b positively regulates potato resistance to Phytophthora infestans. Front. Plant Sci. 2023, 13, 1065627. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zou, M.; Zhao, L.; Xia, Z.; Wang, J. Genome-Wide Association Mapping of Late Blight Tolerance Trait in Potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 714575. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.; Tiwari, J.K.; Kumari, C.; Zinta, R.; Sharma, S.; Thakur, A.K.; Buckseth, T.; Dalamu, D.; Singh, R.K.; Kumar, V. Screening of wild species and transcriptome profiling to identify differentially regulated genes in response to late blight resistance in potato. Front. Plant Sci. 2023, 14, 1212135. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB gene family in potato (Solanum tuberosum L.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes 2018, 9, 54. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Li, C.; Li, G.; Wang, P.; Peng, Z.; Cheng, L.; Li, H.; Zhang, Z.; Li, Y.; Huang, W.; et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 2022, 15, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Tang, C.-N.; Abdullah, W.M.A.N.W.; Wee, C.-Y.; Yusof, Z.N.B.; Yap, W.-S.; Cheng, W.-H.; Baharum, N.A.; Ong-Abdullah, J.; Loh, J.-Y.; Lai, K.-S. Promoter cis-element analyses reveal the function of αVPE in drought stress response of Arabidopsis. Biology 2023, 12, 430. [Google Scholar] [CrossRef] [PubMed]
- Sazegari, S.; Niazi, A.; Ahmadi, F.S. A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 2015, 11, 101–106. [Google Scholar] [CrossRef]
- Zheng, K.; Pang, L.; Xue, X.; Gao, P.; Zhao, H.; Wang, Y.; Han, S. Genome-wide comprehensive survey of the subtilisin-like proteases gene family associated with rice caryopsis development. Front. Plant Sci. 2022, 13, 943184. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, P.; Ding, Y.; Wang, Z.; Ma, Z.; Gagnon, E.; Jia, Y.; Cheng, L.; Bao, Z.; Liu, Z.; et al. Ancient hybridization underlies tuberization and radiation of the potato lineage. Cell 2025, 188, 5249–5265.e15. [Google Scholar] [CrossRef]
- Zhang, S.; Broome, M.; Lawton, M.; Hunter, T.; Lamb, C. atpk1, a novel ribosomal protein kinase gene from Arabidopsis. II. Functional and biochemical analysis of the encoded protein. J. Biol. Chem. 1994, 269, 17593–17599. [Google Scholar] [CrossRef]
- Huang, Y.-X.; Yin, Y.-G.; Sanuki, A.; Fukuda, N.; Ezura, H.; Matsukura, C. Phosphoenolpyruvate carboxykinase (PEPCK) deficiency affects the germination, growth and fruit sugar content in tomato (Solanum lycopersicum L.). Plant Physiol. Biochem. 2015, 96, 417–425. [Google Scholar] [CrossRef]
- Kagawa, T.; Kimura, M.; Wada, M. Blue Light-Induced Phototropism of Inflorescence Stems and Petioles Is Mediated by Phototropin Family Members phot1 and phot2. Plant Cell Physiol. 2009, 50, 1774–1785. [Google Scholar] [CrossRef]
- Anthony, R.G.; Henriques, R.; Helfer, A.; Mészáros, T.; Rios, G.; Testerink, C.; Munnik, T.; Deák, M.; Koncz, C.; Bögre, L. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004, 23, 572–581. [Google Scholar] [CrossRef]
- Chen, M.; Yang, Z.; Peng, Y.; Sun, L.; Liu, X.; Sun, L.; Tan, S. Gravacin as an inhibitor of the auxin transport-activating protein kinase D6PK in Arabidopsis. Front. Plant Sci. 2025, 16, 1563571. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, X.; Yu, Q.; Peng, L.; Chen, L.; Liu, J.; Wang, J.; Li, X.; Yang, Y. Cytosolic ABA Receptor Kinases phosphorylate the D6 PROTEIN KINASE leading to its stabilization which promotes Arabidopsis growth. Plant Cell Environ. 2024, 47, 3030–3045. [Google Scholar] [CrossRef]
- Arshad, W.; Steinbrecher, T.; Wilhelmsson, P.K.; Fernandez-Pozo, N.; Pérez, M.; Mérai, Z.; Rensing, S.A.; Chandler, J.O.; Leubner-Metzger, G. Aethionema arabicum dimorphic seed trait resetting during transition to seedlings. Front. Plant Sci. 2024, 15, 1358312. [Google Scholar] [CrossRef]
- Li, S.; Tahir, M.M.; Wu, T.; Xie, L.; Zhang, X.; Mao, J.; Ayyoub, A.; Xing, L.; Zhang, D.; Shao, Y. Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG During Adventitious Root Formation in Apple. Int. J. Mol. Sci. 2022, 23, 976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, L.; Yin, H.; Xu, Z.; Zhao, Y.; Gao, M.; Wu, H.; Chen, Y.; Wang, Y. D6 protein kinase in root xylem benefiting resistance to Fusarium reveals infection and defense mechanisms in tung trees. Hortic. Res. 2021, 8, 240. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Lin, F.-W.; Cheng, K.-T.; Chang, C.-H.; Hung, S.-C.; Efferth, T.; Chen, Y.-R. XCP1 cleaves Pathogenesis-related protein 1 into CAPE9 for systemic immunity in Arabidopsis. Nat. Commun. 2023, 14, 4697. [Google Scholar] [CrossRef]
- Feng, L.; Wei, S.; Li, Y. Thaumatin-like Proteins in Legumes: Functions and Potential Applications—A Review. Plants 2024, 13, 1124. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, T.; Wang, D.; Gou, R.; Jiang, Y.; Zeng, Z.; Zhang, G.; Zhang, X.; Wei, Z. Salicylic acid influences the resistance of colored calla lily to soft rot (Pectobacterium carotovorum) via the TGA2-LNC88940-miR528-SOD module. Plant J. 2025, 123, e70392. [Google Scholar]
- Kong, L.; Ma, X.; Zhang, C.; Kim, S.-I.; Li, B.; Xie, Y.; Yeo, I.-C.; Thapa, H.; Chen, S.; Devarenne, T.P.; et al. Dual phosphorylation of DGK5-mediated PA burst regulates ROS in plant immunity. Cell 2024, 187, 609–623.e21. [Google Scholar] [CrossRef]
- Barbosa, I.C.; Zourelidou, M.; Willige, B.C.; Weller, B.; Schwechheimer, C. D6 PROTEIN KINASE Activates Auxin Transport-Dependent Growth and PIN-FORMED Phosphorylation at the Plasma Membrane. Dev. Cell 2014, 29, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Bassukas, A.E.L.; Xiao, Y.; Barbosa, I.C.R.; Mergner, J.; Grill, P.; Michalke, B.; Kuster, B.; Schwechheimer, C. D6PK plasma membrane polarity requires a repeated CXX(X)P motif and PDK1-dependent phosphorylation. Nat. Plants 2024, 10, 300–314. [Google Scholar] [CrossRef]
- Kulich, I.; Schmid, J.; Teplova, A.; Qi, L.; Friml, J. Rapid translocation of NGR proteins driving polarization of PIN-activating D6 protein kinase during root gravitropism. eLife 2024, 12, RP91523. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. Intervention of Phytohormone Pathways by Pathogen Effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Lv, C.; Zhao, Y.; Bao, Y.; Wang, F. Genome-Wide Identification of the AGC Kinase Family in Tetraploid Potato (Solanum tuberosum L.) Cultivar ‘Qingshu No. 9’ and Functional Analysis of StD6PK in Response to Late Blight (Phytophthora infestans). Plants 2025, 14, 3818. https://doi.org/10.3390/plants14243818
Zhou Y, Lv C, Zhao Y, Bao Y, Wang F. Genome-Wide Identification of the AGC Kinase Family in Tetraploid Potato (Solanum tuberosum L.) Cultivar ‘Qingshu No. 9’ and Functional Analysis of StD6PK in Response to Late Blight (Phytophthora infestans). Plants. 2025; 14(24):3818. https://doi.org/10.3390/plants14243818
Chicago/Turabian StyleZhou, Yifan, Chunna Lv, Yihan Zhao, Yuting Bao, and Fang Wang. 2025. "Genome-Wide Identification of the AGC Kinase Family in Tetraploid Potato (Solanum tuberosum L.) Cultivar ‘Qingshu No. 9’ and Functional Analysis of StD6PK in Response to Late Blight (Phytophthora infestans)" Plants 14, no. 24: 3818. https://doi.org/10.3390/plants14243818
APA StyleZhou, Y., Lv, C., Zhao, Y., Bao, Y., & Wang, F. (2025). Genome-Wide Identification of the AGC Kinase Family in Tetraploid Potato (Solanum tuberosum L.) Cultivar ‘Qingshu No. 9’ and Functional Analysis of StD6PK in Response to Late Blight (Phytophthora infestans). Plants, 14(24), 3818. https://doi.org/10.3390/plants14243818




