Elevated Soybean Seed Oil Phenotype Associated with a Single Nucleotide Polymorphism in GmNFR1α
Abstract
1. Introduction
2. Results
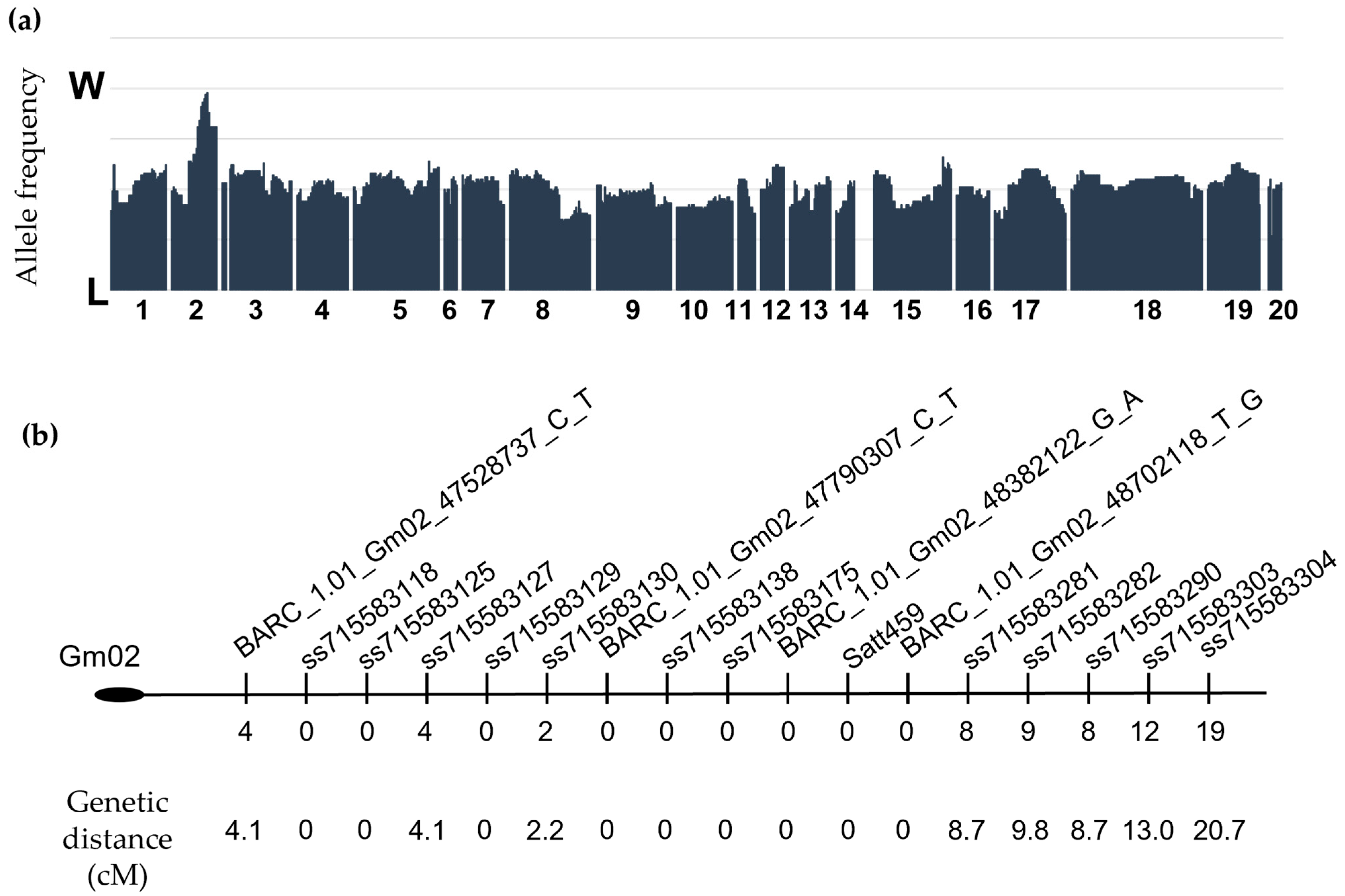
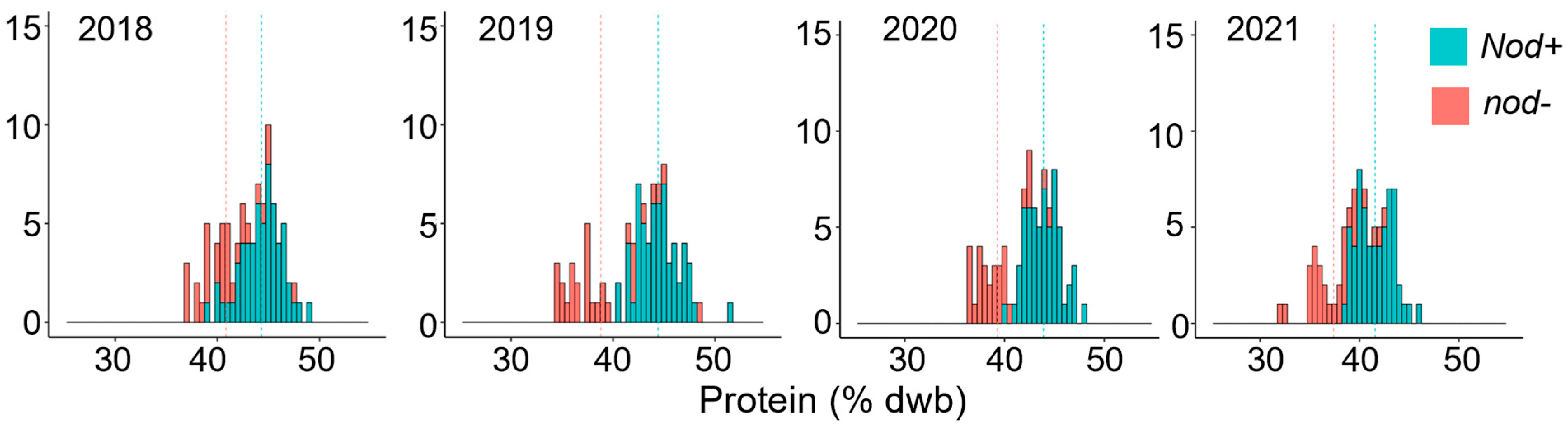
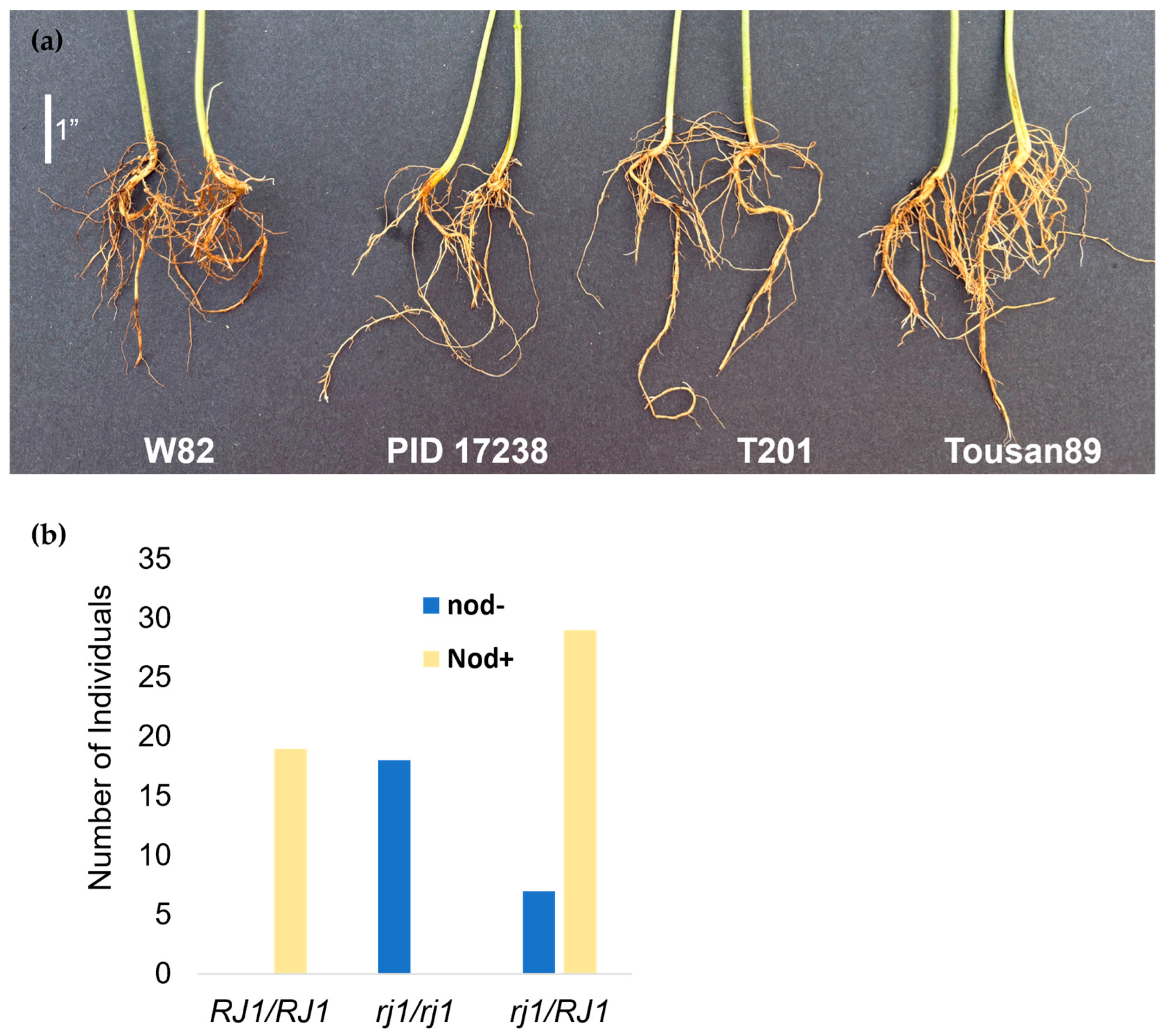
2.1. A Locus Conferring Low Protein and High Oil Content Maps to Chromosome 2
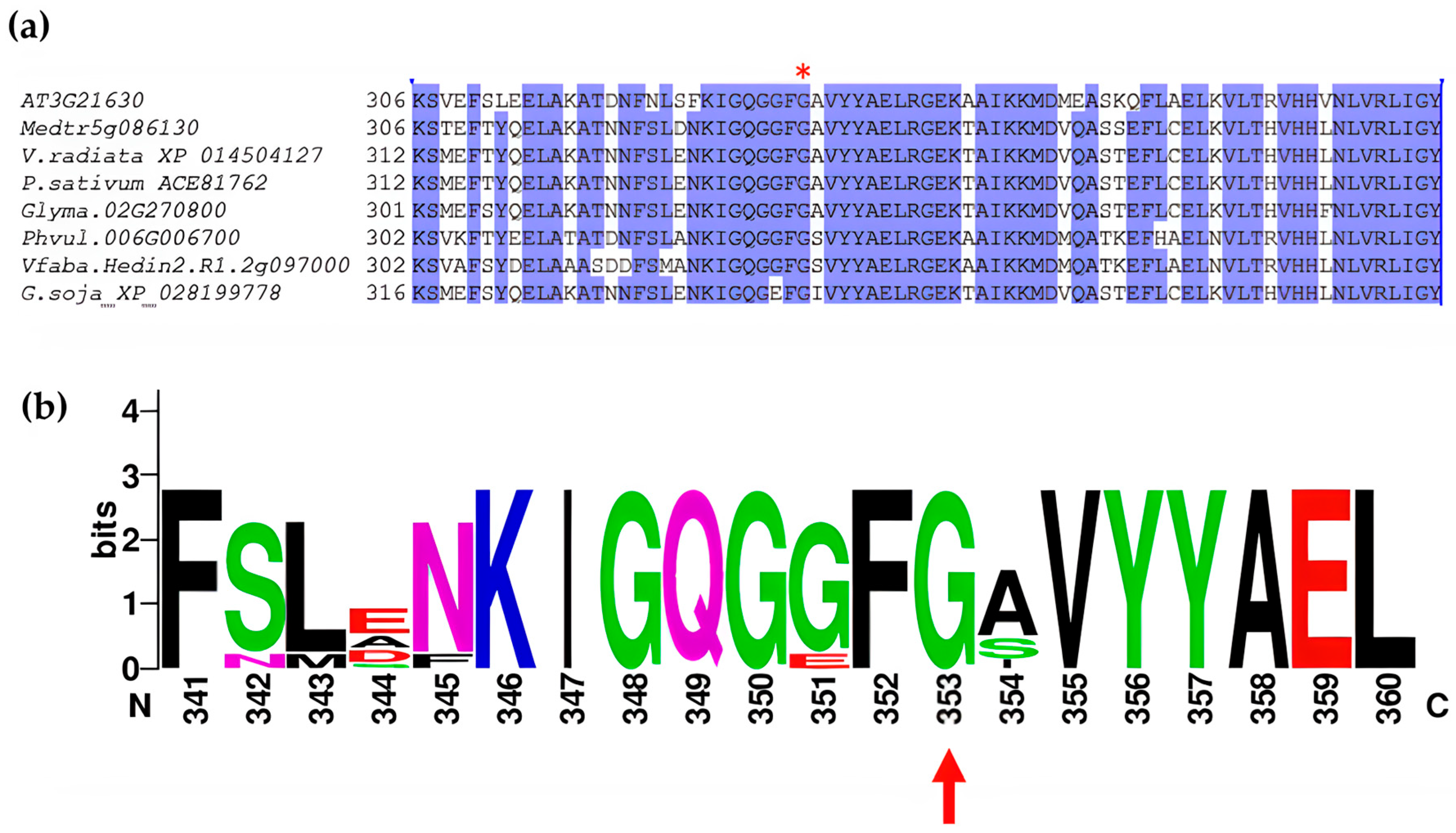
2.2. Mutation in NFR1alpha
3. Discussion
4. Methods and Materials
4.1. Plant Materials and Growth Conditions
4.2. Genetic Mapping, Sequencing, and Genotyping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safdar, B.; Zhou, H.; Li, H.; Cao, J.; Zhang, T.; Ying, Z.; Liu, X. Prospects for Plant-Based Meat: Current Standing, Consumer Perceptions, and Shifting Trends. Foods 2022, 11, 3770. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, T.; Li, H.; Chen, T.; Liu, X. Control of Beany Flavor from Soybean Protein Raw Material in Plant-Based Meat Analog Processing. Foods 2023, 12, 923. [Google Scholar] [CrossRef]
- Liu, A.; Cheng, S.-S.; Yung, W.-S.; Li, M.-W.; Lam, H.-M. Genetic Regulations of the Oil and Protein Contents in Soybean Seeds and Strategies for Improvement. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2022; Volume 102, pp. 259–293. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, L.; Li, S.; Wang, W.; Ding, Y.; Swarm, S.A.; Li, L.; Wang, X.; Tang, X.; Zhang, Z.; et al. Elevation of Soybean Seed Oil Content Through Selection for Seed Coat Shininess. Nat. Plants 2018, 4, 30–35. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q.; et al. Simultaneous Changes in Seed Size, Oil Content and Protein Content Driven by Selection of SWEET Homologues During Soybean Domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Xing, X.; Popp, M.; Chen, P.; Manjarrez-Sandoval, P.; Gbur, E. Evaluation of High-Oil and High-Protein Soybean Using Component Pricing. J. Crop Improv. 2018, 32, 264–280. [Google Scholar] [CrossRef]
- Guo, B.; Sun, L.; Jiang, S.; Ren, H.; Sun, R.; Wei, Z.; Hong, H.; Luan, X.; Wang, J.; Wang, X.; et al. Soybean Genetic Resources Contributing to Sustainable Protein Production. Theor. Appl. Genet. 2022, 135, 4095–4121. [Google Scholar] [CrossRef] [PubMed]
- Sebolt, A.M.; Shoemaker, R.C.; Diers, B.W. Analysis of a Quantitative Trait Locus Allele from Wild Soybean That Increases Seed Protein Concentration in Soybean. Crop Sci. 2000, 40, 1438–1444. [Google Scholar] [CrossRef]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular Mapping and Genomics of Soybean Seed Protein: A Review and Perspective for the Future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- Fliege, C.E.; Ward, R.A.; Vogel, P.; Nguyen, H.; Quach, T.; Guo, M.; Viana, J.P.G.; Dos Santos, L.B.; Specht, J.E.; Clemente, T.E.; et al. Fine Mapping and Cloning of the Major Seed Protein Quantitative Trait Loci on Soybean Chromosome 20. Plant J. 2022, 110, 114–128. [Google Scholar] [CrossRef]
- Gupta, S.K.; Manjaya, J.G. Advances in Improvement of Soybean Seed Composition Traits Using Genetic, Genomic and Biotechnological Approaches. Euphytica 2022, 218, 99. [Google Scholar] [CrossRef]
- Lyu, X.; Sun, C.; Lin, T.; Wang, X.; Li, S.; Zhao, S.; Gong, Z.; Wei, Z.; Yan, C.; Ma, C. Systemic Regulation of Soybean Nodulation and Nitrogen Fixation by Nitrogen via Isoflavones. Front. Plant Sci. 2022, 13, 968496. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhu, H. Genetic and Molecular Mechanisms Underlying Symbiotic Specificity in Legume-Rhizobium Interactions. Front. Plant Sci. 2018, 9, 313. [Google Scholar] [CrossRef]
- Indrasumunar, A.; Searle, I.; Lin, M.; Kereszt, A.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Nodulation Factor Receptor Kinase 1α Controls Nodule Organ Number in Soybean (Glycine max L. Merr). Plant J. 2011, 65, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K. Soybean Protein and Oil Variants Identified Through a Forward Genetic Screen for Seed Composition. Plants 2022, 11, 2966. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K. Soybean Oil-Quality Variants Identified by Large-Scale Mutagenesis. Int. J. Agron. 2012, 2012, 569817. [Google Scholar] [CrossRef]
- Nelson, R.L.; Johnson, E.O.C. Registration of the High-Yielding Soybean Germplasm Line LG04-6000. J. Plant Regist. 2012, 6, 212–215. [Google Scholar] [CrossRef]
- Song, Q.J.; Marek, L.F.; Shoemaker, R.C.; Lark, K.G.; Concibido, V.C.; Delannay, X.; Specht, J.E.; Cregan, P.B. A New Integrated Genetic Linkage Map of the Soybean. Theor. Appl. Genet. 2004, 109, 122–128. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting Deleterious Amino Acid Substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Yao, K.; Wang, Y.; Li, X.; Ji, H. Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response. Int. J. Mol. Sci. 2023, 24, 13621. [Google Scholar] [CrossRef]
- Lee, W.K.; Jeong, N.; Indrasumunar, A.; Gresshoff, P.M.; Jeong, S.-C. Glycine Max Non-Nodulation Locus Rj1: A Recombinogenic Region Encompassing a SNP in a Lysine Motif Receptor-like Kinase (GmNFR1α). Theor. Appl. Genet. 2011, 122, 875–884. [Google Scholar] [CrossRef]
- Weber, C.R. Nodulating and Nonnodulating Soybean Isolines: I. Agronomic and Chemical Attributes1. Agron. J. 1966, 58, 43–46. [Google Scholar] [CrossRef]
- Clevinger, E.M.; Biyashev, R.; Haak, D.; Song, Q.; Pilot, G.; Saghai Maroof, M.A. Identification of Quantitative Trait Loci Controlling Soybean Seed Protein and Oil Content. PLoS ONE 2023, 18, e0286329. [Google Scholar] [CrossRef]
- Araya, S.; Elia, P.; Quigley, C.V.; Song, Q. Genetic Variation and Genetic Complexity of Nodule Occupancy in Soybean Inoculated with USDA110 and USDA123 Rhizobium Strains. BMC Genomics 2023, 24, 520. [Google Scholar] [CrossRef]
- Yang, S.; Tang, F.; Gao, M.; Krishnan, H.B.; Zhu, H. R Gene-Controlled Host Specificity in the Legume–Rhizobia Symbiosis. Proc. Natl. Acad. Sci. USA 2010, 107, 18735–18740. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Rogers, A.; Nelson, R.; Long, S.P. Testing the “Source–Sink” Hypothesis of down-Regulation of Photosynthesis in Elevated [CO2] in the Field with Single Gene Substitutions in Glycine Max. Agric. For. Meteorol. 2004, 122, 85–94. [Google Scholar] [CrossRef]
- Chan, Y.O.; Dietz, N.; Zeng, S.; Wang, J.; Flint-Garcia, S.; Salazar-Vidal, M.N.; Škrabišová, M.; Bilyeu, K.; Joshi, T. The Allele Catalog Tool: A Web-Based Interactive Tool for Allele Discovery and Analysis. BMC Genomics 2023, 24, 107. [Google Scholar] [CrossRef]
- Hayashi, M.; Saeki, Y.; Haga, M.; Harada, K.; Kouchi, H.; Umehara, Y. Rj (Rj) Genes Involved in Nitrogen-Fixing Root Nodule Formation in Soybean. Breed. Sci. 2012, 61, 544–553. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Xiang, S.; Wang, W.; Shu, Y.; Li, Z.; Wang, S.; Chen, L.; Yang, X.; Zhao, T. Transcriptomic and Photosynthetic Responses to Grafting of the Nod1 Gene in Nodulated and Non-Nodulated Soybeans. G3 GenesGenomesGenetics 2021, 11, jkab209. [Google Scholar] [CrossRef] [PubMed]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Neff, M.M.; Neff, J.D.; Chory, J.; Pepper, A.E. dCAPS, a Simple Technique for the Genetic Analysis of Single Nucleotide Polymorphisms: Experimental Applications in Arabidopsis Thaliana Genetics. Plant J. 1998, 14, 387–392. [Google Scholar] [CrossRef]
- Richards, E.; Reichardt, M.; Rogers, S. Preparation of Genomic DNA from Plant Tissue. Curr. Protoc. Mol. Biol. 1994, 27, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kendig, K.I.; Baheti, S.; Bockol, M.A.; Drucker, T.M.; Hart, S.N.; Heldenbrand, J.R.; Hernaez, M.; Hudson, M.E.; Kalmbach, M.T.; Klee, E.W.; et al. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Front. Genet. 2019, 10, 736. [Google Scholar] [CrossRef] [PubMed]
| Williams-82 | Williams-82 × 17238 | |||
|---|---|---|---|---|
| Year | Protein | Oil | Protein | Oil |
| 2020 | 41.61 ± 2.1 | 18.92 ± 0.9 | 32.91 ± 1.1 ** | 20.54 ± 0.5 ** |
| 2021 | 41.97 ± 2.4 | 21.03 ± 1 | 35.5 ± 2.9 * | 22.21 ± 0.4 |
| 2022 | 43.1 ± 1.6 | 20.3 ± 0.8 | 31.2 ± 2.0 ** | 23.1 ± 0.84 ** |
| 2023 | 38.8 ± 1.4 | 21.6 ± 0.7 | 30.5 ± 2.0 ** | 25.0 ± 0.9 ** |
| 2024 | 38.09 ± 1.3 | 23.9 ± 0.5 | 29.6 ± 0.9 ** | 24.8 ± 0.7 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patibandla, S.V.; Carrero-Colón, M.; Song, Q.; Qin, Q.; Clevinger, E.; Zhu, H.; Maroof, M.A.S.; Hudson, K. Elevated Soybean Seed Oil Phenotype Associated with a Single Nucleotide Polymorphism in GmNFR1α. Plants 2025, 14, 3676. https://doi.org/10.3390/plants14233676
Patibandla SV, Carrero-Colón M, Song Q, Qin Q, Clevinger E, Zhu H, Maroof MAS, Hudson K. Elevated Soybean Seed Oil Phenotype Associated with a Single Nucleotide Polymorphism in GmNFR1α. Plants. 2025; 14(23):3676. https://doi.org/10.3390/plants14233676
Chicago/Turabian StylePatibandla, Sri Veda, Militza Carrero-Colón, Qijian Song, Quilin Qin, Elizabeth Clevinger, Hongyan Zhu, M. A. Saghai Maroof, and Karen Hudson. 2025. "Elevated Soybean Seed Oil Phenotype Associated with a Single Nucleotide Polymorphism in GmNFR1α" Plants 14, no. 23: 3676. https://doi.org/10.3390/plants14233676
APA StylePatibandla, S. V., Carrero-Colón, M., Song, Q., Qin, Q., Clevinger, E., Zhu, H., Maroof, M. A. S., & Hudson, K. (2025). Elevated Soybean Seed Oil Phenotype Associated with a Single Nucleotide Polymorphism in GmNFR1α. Plants, 14(23), 3676. https://doi.org/10.3390/plants14233676





