Adaptive Traits and Molecular Mechanisms of Rhododendron Species in Changbai Mountains’ Alpine Tundra: A Phenotype–Transcriptome Study
Abstract
1. Introduction
2. Results
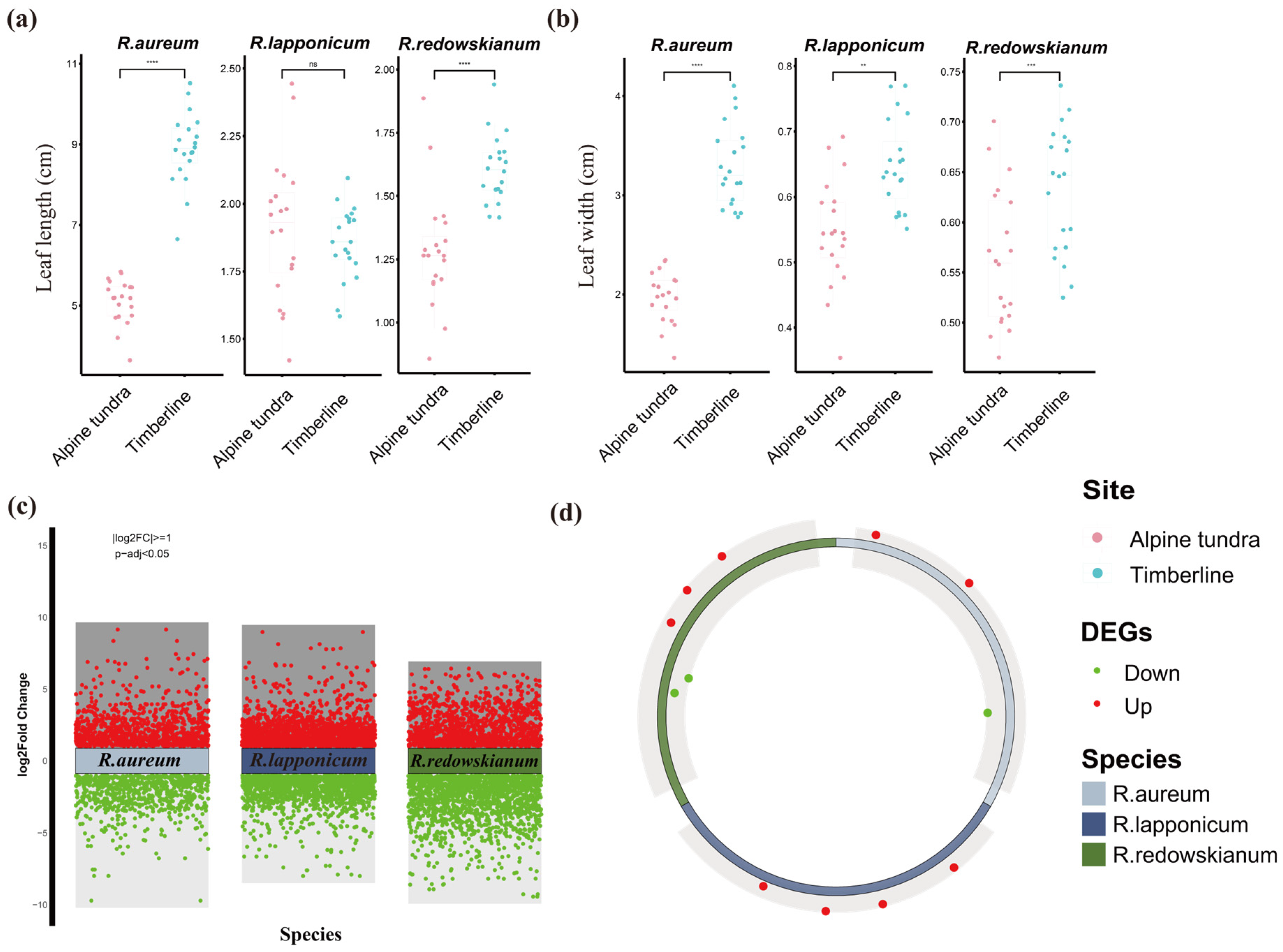
2.1. Leaf Phenotypic Responses of Rhododendron spp.
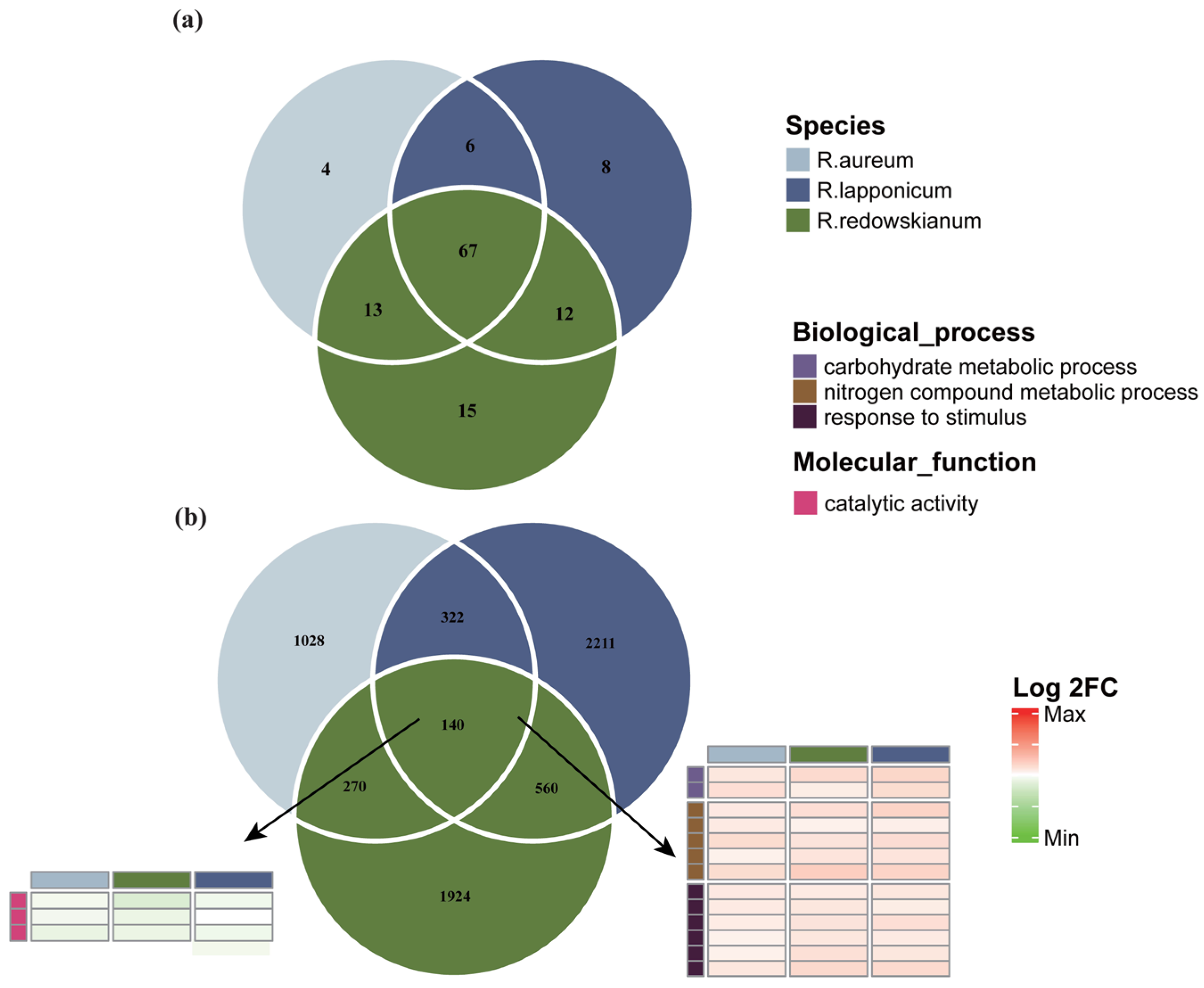
2.2. Characteristics of Differentially Expressed Genes (DEGs) in Rhododendron spp.
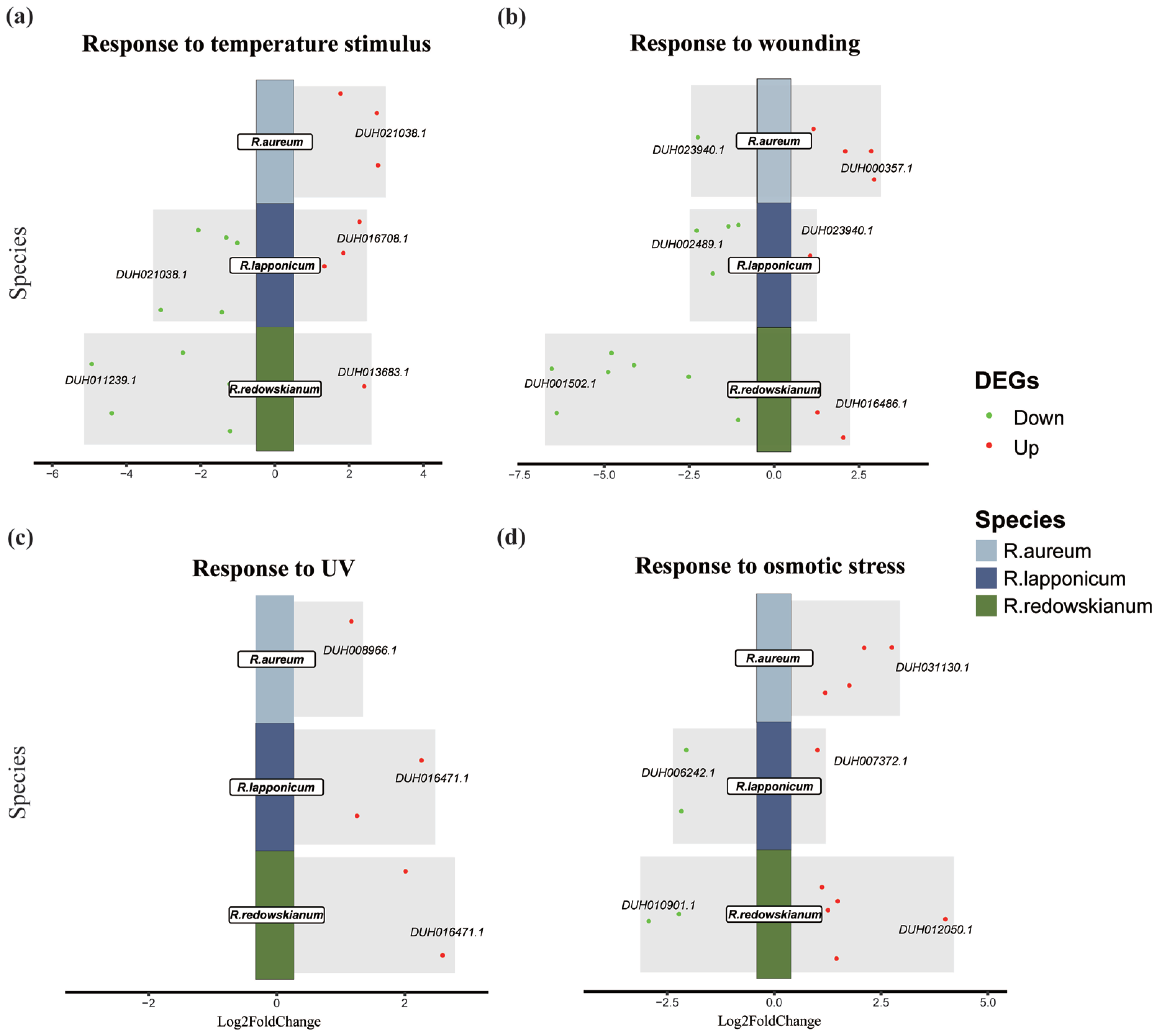
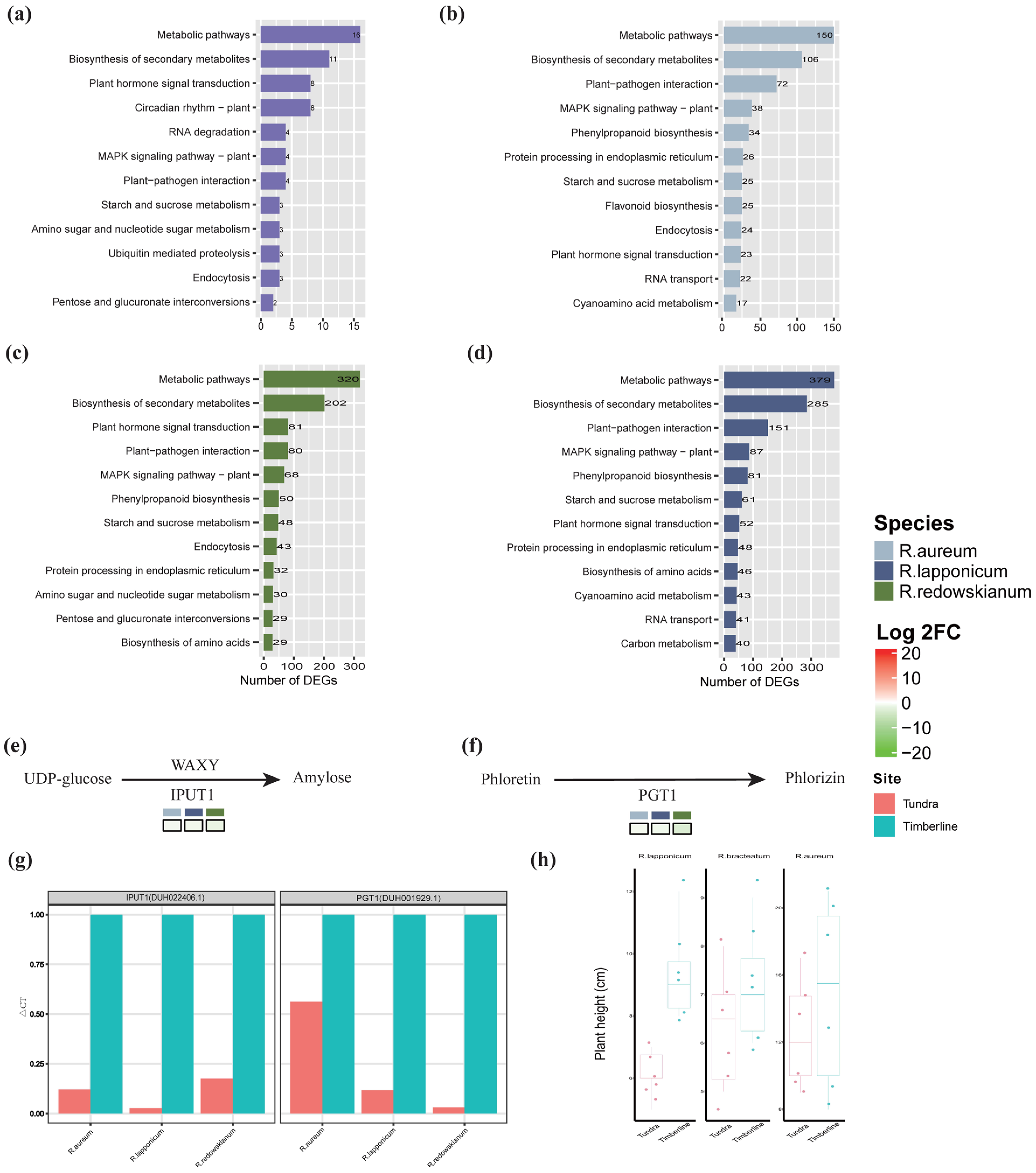
2.3. Molecular Biological Responses of Rhododendron spp. to Alpine Tundra
3. Discussion
3.1. Causes of Changes in Leaf Morphology
3.2. Adaptive Response of Rhododendron spp. in Alpine Tundra
3.3. Common Adaptive Mechanisms of Rhododendron spp.
4. Materials and Methods
4.1. Experimental Materials and RNA Extraction
4.2. Sequencing Library Construction and Sequencing
4.3. RNA-Seq Analysis and Data Processing
4.4. Data Collection and Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| RNA-seq | RNA sequencing |
| DEGs | differentially expressed genes |
| DELLA | DELLA domain-containing protein |
| EDS1 | Enhanced Disease Susceptibility 1 |
| UV | Ultraviolet |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-Activated Protein Kinase |
| IPUT1 | Inositol Phosphorylceramide Glucuronosyltransferase 1 |
| PGT1 | phlorizin synthase |
References
- Xia, X.M.; Yang, M.Q.; Li, C.L.; Huang, S.X.; Jin, W.T.; Shen, T.T.; Wang, F.; Li, X.H.; Yoichi, W.; Zhang, L.H.; et al. Spatiotemporal evolution of the global species diversity of Rhododendron. Mol. Biol. Evol. 2022, 39, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.L.; Li, Y.; Pei, F.; Gong, T.; Chen, T.J.; Chen, J.J.; Yang, J.L.; Li, Q.H.; Yu, S.S.; Zhu, P. Chromosome-scale genome assembly of provides insights into its evolution and terpenoid biosynthesis. BMC Plant Biol. 2022, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.L.; Ma, H.P.; Fan, P.C.; Gao, R.M.; Jia, Z.P. Antioxidant potential, total phenolic and total flavonoid contents of Rhododendron anthopogonoides and its protective effect on hypoxia-induced injury in PC12 cells. BMC Complement. Altern. Med. 2015, 15, 287. [Google Scholar] [CrossRef]
- Casati, M.; Kichey, T.; Closset, D.; Spicher, F.; Decocq, G. Is the invasive Rhododendron ponticum L. an emergent threat to mainland Atlantic forests? A population dynamics approach. For. Ecol. Manag. 2023, 549, 121463. [Google Scholar] [CrossRef]
- Zhou, Y.; Sha, M.Y.; Jin, H.Q.; Wang, L.F.; Zhang, J.; Xu, Z.F.; Tan, B.; Chen, L.H.; Wang, L.X.; Liu, S.N.; et al. The expansion of evergreen and deciduous shrubs changed the chemical characteristics and biological community of alpine meadows soil. Eur. J. Soil. Biol. 2023, 117, 103505. [Google Scholar] [CrossRef]
- Gyeltshen, N.; Gyeltshen, C.; Dema, S.; Ghimire, S.K.; Konig, N.; Hart, R. A swirling offering: Climate change impacts on incense and other useful alpine plants of Bhutan. Econ. Bot. 2025, 79, 231–246. [Google Scholar] [CrossRef]
- Mo, Z.Q.; Fu, C.N.; Zhu, M.S.; Milne, R.; Yang, J.B.; Cai, J.; Qin, H.T.; Zheng, W.; Hollingsworth, P.M.; Li, D.Z.; et al. Resolution, conflict and rate shifts: Insights from a densely sampled plastome phylogeny for Rhododendron (Ericaceae). Ann. Bot. 2022, 130, 687–701. [Google Scholar] [CrossRef]
- He, S.; Yuan, C.J.; Zhang, P.L.; Wang, H.D.; Luo, D.L.; Dai, X.Y. Study on the characteristics of genetic diversity of different populations of Guizhou endemic plant Rhododendron pudingense based on microsatellite markers. BMC Plant Biol. 2024, 24, 77. [Google Scholar] [CrossRef]
- Choudhary, S.; Thakur, S.; Majeed, A.; Bhardwaj, P. Adaptability of Rhododendrons in high altitude habitats. J. For. Res. 2021, 32, 449–460. [Google Scholar] [CrossRef]
- Cai, Y.F.; Wang, J.H.; Zhang, L.; Song, J.; Peng, L.C.; Zhang, S.B. Physiological and transcriptomic analysis highlight key metabolic pathways in relation to drought tolerance in Rhododendron delavayi. Physiol. Mol. Biol. Plants 2019, 25, 991–1008. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, W.G.; Hu, X.Y.; Shi, Y.H.; Liu, Y.; Zhong, Y.F.; Wang, P.; Deng, S.Y.; Niu, J.; Yu, X.D. Physiological and transcriptomic analysis revealed the involvement of crucial factors in heat stress response of Rhododendron hainanense. Gene 2018, 660, 109–119. [Google Scholar] [CrossRef]
- Zong, S.W.; Lembrechts, J.J.; Du, H.B.; He, H.S.; Wu, Z.F.; Li, M.H.; Rixen, C. Upward range shift of a dominant alpine shrub related to 50 years of snow cover change. Remote Sens. Environ. 2022, 268, 112773. [Google Scholar] [CrossRef]
- Li, K.J.; Liu, X.F.; Yang, L.; Shen, S.K. Alpine Rhododendron population contractions lead to spatial distribution mismatch with their pollinators under climate change. Sci. Total Environ. 2024, 926, 171832. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.C.; Zhu, X.F.; Song, W.Z.; Shi, X.P.; Huang, X.T. Predicting the potential distribution of 12 threatened medicinal plants on the Qinghai-Tibet Plateau, with a maximum entropy model. Ecol. Evol. 2024, 14, e11042. [Google Scholar] [CrossRef] [PubMed]
- Francon, L.; Roussel, E.; Lopez-Saez, J.; Saulnier, M.; Stoffel, M.; Corona, C. Alpine shrubs have benefited more than trees from 20th century warming at a treeline ecotone site in the French Pyrenees. Agric. For. Meteorol. 2023, 329, 109284. [Google Scholar] [CrossRef]
- Yu, F.Y.; Wang, T.J.; Groen, T.A.; Skidmore, A.K.; Yang, X.F.; Ma, K.P.; Wu, Z.F. Climate and land use changes will degrade the distribution of Rhododendrons in China. Sci. Total Environ. 2019, 659, 515–528. [Google Scholar] [CrossRef]
- Zhang, J.H.; Li, K.J.; Liu, X.F.; Yang, L.; Shen, S.K. Interspecific variance of suitable habitat changes for four alpine Rhododendron species under climate change: Implications for their reintroductions. Forests 2021, 12, 1520. [Google Scholar] [CrossRef]
- Huang, X.; Li, C. An analysis on the ecology of alpine tundra landscape of Changbai Mountains. Acta Geogr. Sin. 1984, 39, 285–297. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, Y.; Xu, J.; Tao, Y.; He, H.; Guo, M.; Wang, A.; Liu, Y.; Niu, L. Comparative assessment of tundra vegetation changes between north and southwest slopes of Changbai Mountains, China, in response to global warming. Chin. Geogr. Sci. 2018, 28, 665–679. [Google Scholar] [CrossRef]
- Jin, Y.Y.; Xu, J.W.; He, H.S.; Li, M.H.; Tao, Y.; Zhang, Y.J.; Hu, R.; Gao, X.; Bai, Y.Y.; Wang, H.Y.; et al. The Changbai alpine shrub tundra will be replaced by herbaceous tundra under global climate change. Plants 2019, 8, 370. [Google Scholar] [CrossRef]
- Lu, Y.P.; Liu, H.C.; Chen, W.; Yao, J.; Huang, Y.Q.; Zhang, Y.; He, X.Y. Conservation planning of the genus Rhododendron in Northeast China based on current and future suitable habitat distributions. Biodivers. Conserv. 2021, 30, 673–697. [Google Scholar] [CrossRef]
- Tikhonova, N.A.; Polezhaeva, M.A. Morphological and genetic differentiation of populations of Rhododendron aureum Georgi. (Ericaceae) in the mountains of Southern Siberia and on the Kamchatka Peninsula. Contemp. Probl. Ecol. 2023, 16, 575–582. [Google Scholar] [CrossRef]
- Zhao, W.; Qi, X.J.; Lyu, J.W.; Yu, Z.X.; Chen, X. Characterization of microbial community structure in rhizosphere soils of Cowskin Azalea (Rhododendron aureum Georgi) on northern slope of Changbai Mountains, China. Chin. Geogr. Sci. 2016, 26, 78–89. [Google Scholar] [CrossRef]
- Simin, T.; Tang, J.; Holst, T.; Rinnan, R. Volatile organic compound emission in tundra shrubs—Dependence on species characteristics and the near-surface environment. Environ. Exp. Bot. 2021, 184, 104387. [Google Scholar] [CrossRef] [PubMed]
- Chernonosov, A.A.; Karpova, E.A.; Karakulov, A.V. Metabolomic profiling of three Rhododendron species from Eastern Siberia by liquid chromatography with high-resolution mass spectrometry. S. Afr. J. Bot. 2023, 157, 622–634. [Google Scholar] [CrossRef]
- Flagmeier, M.; Long, D.G.; Genney, D.R.; Hollingsworth, P.M.; Ross, L.C.; Woodin, S.J. Fifty years of vegetation change in oceanic-montane liverwort-rich heath in Scotland. Plant Ecol. Divers. 2014, 7, 457–470. [Google Scholar] [CrossRef]
- Gasperini, D.; Greenland, A.; Hedden, P.; Dreos, R.; Harwood, W.; Griffiths, S. Genetic and physiological analysis of Rht8 in bread wheat: An alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J. Exp. Bot. 2012, 63, 6057. [Google Scholar] [CrossRef]
- Kalberer, S.R.; Leyva-Estrada, N.; Krebs, S.L.; Arora, R. Frost dehardening and rehardening of floral buds of deciduous azaleas are influenced by genotypic biogeography. Environ. Exp. Bot. 2007, 59, 264–275. [Google Scholar] [CrossRef]
- Yu, W.; Gong, F.S.; Xu, H.W.; Zhou, X.F. Molecular mechanism of exogenous ABA to enhance UV-B resistance in Rhododendron chrysanthum Pall. by modulating flavonoid accumulation. Int. J. Mol. Sci. 2024, 25, 5248. [Google Scholar] [CrossRef]
- Malfasi, F.; Cannone, N. Climate warming persistence triggered tree ingression after shrub encroachment in a high alpine tundra. Ecosystems 2020, 23, 1657–1675. [Google Scholar] [CrossRef]
- Ma, Y.Z.; Mao, X.X.; Wang, J.; Zhang, L.; Jiang, Y.Z.; Geng, Y.Y.; Ma, T.; Cai, L.M.; Huang, S.Q.; Hollingsworth, P.; et al. Pervasive hybridization during evolutionary radiation of Rhododendron subgenus Hymenanthes in mountains of southwest China. Natl. Sci. Rev. 2022, 9, nwac276. [Google Scholar] [CrossRef] [PubMed]
- Zhongzan, Y.; Jian, Y.; Jiangnan, L.; Wei, Z.; Ming, X.; Yujiao, Z.; Ma, C.; Yuqiao, G.; Yueming, Z.; Xia, C. Divergent functional traits and gene expression profiles in native and encroaching plant species across an alpine elevational gradient. Front. Plant Sci. 2025, 16, 1656812. [Google Scholar] [CrossRef]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, V.; Quevedo, M.; Nardeli, S.M.; Meena, S.K.; Monte, E.; Kindgren, P. Convergent antisense transcription primes hosting genes for stress responsiveness in plants. Mol. Plant 2025, 18, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, M.; Yang, K.; Liu, B.; Liu, H.H.; Liu, J.Q. Genetic variation for adaptive evolution in response to changed environments in plants. J. Integr. Plant Biol. 2025, 67, 2265–2293. [Google Scholar] [CrossRef]
- Ge, X.L.; Zhang, L.; Du, J.J.; Wen, S.S.; Qu, G.Z.; Hu, J.J. Transcriptome analysis of Populus euphratica under salt treatment and PeERF1 gene enhances salt tolerance in transgenic Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2022, 23, 3727. [Google Scholar] [CrossRef]
- Wang, P.C.; Zhao, Y.; Li, Z.P.; Hsu, C.C.; Liu, X.; Fu, L.W.; Hou, Y.J.; Du, Y.; Xie, S.J.; Zhang, C.G.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Yan, Y.Z.; Shi, M.M.; Liu, Y.Q. Effect of an atmospheric pressure plasma jet on the structure and physicochemical properties of waxy and normal maize starch. Polymers 2019, 11, 8. [Google Scholar] [CrossRef]
- Ortega-Albero, N.; González-Orenga, S.; Vicente, O.; Rodríguez-Burruezo, A.; Fita, A. Responses to salt stress of the interspecific hybrid Solanum insanum × Solanum melongena and its parental species. Plants 2023, 12, 295. [Google Scholar] [CrossRef]
- Li, Q.; He, X.; Huang, X.J.; Zhang, L. Effects of altitude and leaf age on leaf shape in an alpine shrub: The relevance for the leaf area estimation model. Pol. J. Ecol. 2022, 70, 33–43. [Google Scholar] [CrossRef]
- Zhao, B.Q.; Liu, Q.Y.; Wang, B.S.; Yuan, F. Roles of phytohormones and their signaling pathways in leaf development and stress responses. J. Agric. Food Chem. 2021, 69, 3566–3584. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Guan, C.M.; Jiao, Y.L. Molecular mechanisms of leaf morphogenesis. Mol. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Leigh, A.; Sevanto, S.; Close, J.D.; Nicotra, A.B. The influence of leaf size and shape on leaf thermal dynamics: Does theory hold up under natural conditions? Plant Cell Environ. 2017, 40, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Demmings, E.M.; Williams, B.R.; Lee, C.R.; Barba, P.; Yang, S.S.; Hwang, C.F.; Reisch, B.I.; Chitwood, D.H.; Londo, J.P. Quantitative trait locus analysis of leaf morphology indicates conserved shape loci in grapevine. Front. Plant Sci. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Li, Y.G.; Yang, Y.H.; Hu, Y.L.; Liu, H.L.; He, M.; Yang, Z.Y.; Kong, F.J.; Liu, X.; Hou, X.L. DELLA and EDS1 form a feedback regulatory module to fine-tune plant growth-defense tradeoff in Arabidopsis. Mol. Plant 2019, 12, 1485–1498. [Google Scholar] [CrossRef]
- Guo, Q.; Li, X.; Niu, L.; Jameson, P.E.; Zhou, W. Transcription-associated metabolomic adjustments in maize occur during combined drought and cold stress. Plant Physiol. 2021, 186, 677–695. [Google Scholar] [CrossRef]
- Eshel, G.; Duppen, N.; Wang, G.N.; Oh, D.H.; Kazachkova, Y.; Herzyk, P.; Amtmann, A.; Gordon, M.; Chalifa-Caspi, V.; Oscar, M.A.; et al. Positive selection and heat-response transcriptomes reveal adaptive features of the Brassicaceae desert model, Anastatica hierochuntica. New Phytol. 2022, 236, 1006–1026. [Google Scholar] [CrossRef]
- Jin, Y.H.; Xu, J.W.; He, H.S.; Tao, Y.; Wang, H.Y.; Zhang, Y.J.; Hu, R.; Gao, X.; Bai, Y.Y.; Zhao, C.; et al. Effects of catastrophic wind disturbance on formation of forest patch mosaic structure on the western and southern slopes of Changbai Mountain. For. Ecol. Manag. 2021, 481, 118746. [Google Scholar] [CrossRef]
- Hu, C.C.; Tian, C.G.; Chen, C.J.; Song, W.; Yue, X.; Liu, X.Y. Increased carbon cost for nitrogen assimilation in plants under a warming climate. Nat. Geosci. 2025, 18, 1133–1137. [Google Scholar] [CrossRef]
- Xiong, R.Y.; Wang, H.X.; Mao, F.Q.; Tao, L.; Tan, X.M.; Pan, X.H.; Zeng, Y.J.; Zeng, Y.H. The mechanism of carbon and nitrogen metabolism under low temperature and low light stress after heading in late indica rice. Plant Physiol. Biochem. 2025, 218, 109316. [Google Scholar] [CrossRef]
- Ramos-Madrigal, J.; Fritz, G.J.; Schroeder, B.; Smith, B.; Sánchez-Barreiro, F.; Caroe, C.; Runge, A.K.W.; Boer, S.; Mcgrath, K.; Vieira, F.G.; et al. The genomic origin of early maize in eastern North America. Cell 2025, 188, 33–43.e16. [Google Scholar] [CrossRef]
- Liu, X.F.; Han, S.H.; Makowski, D.; Wang, X.H.; Fu, Z.; Ciais, P. Response of rice quality to climate warming: A meta-analysis. Field Crops Res. 2025, 331, 109995. [Google Scholar] [CrossRef]
- Zhou, K.; Hu, L.Y.; Li, Y.T.S.; Chen, X.F.; Zhang, Z.J.; Liu, B.B.; Li, P.M.; Gong, X.Q.; Ma, F.W. MdUGT88F1-mediated phloridzin biosynthesis regulates apple development and Valsa canker resistance. Plant Physiol. 2019, 180, 2290–2305. [Google Scholar] [CrossRef] [PubMed]
- Tartaglio, V.; Rennie, E.A.; Cahoon, R.; Wang, G.; Baidoo, E.; Mortimer, J.C.; Cahoon, E.B.; Scheller, H.V. Glycosylation of inositol phosphorylceramide sphingolipids is required for normal growth and reproduction in Arabidopsis. Plant J. 2017, 89, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Harguindeguy, N.; Díaz, S.; Garnier, E.; Lavorel, S.; Poorter, H.; Jaureguiberry, P.; Bret-Harte, M.S.; Cornwell, W.K.; Craine, J.M.; Gurvich, D.E.; et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013, 61, 167–234. [Google Scholar] [CrossRef]
- Li, R.Q.; Li, Y.R.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wang, X.Y.; Gao, Y.; Wu, X.P.; Wen, X.H.; Li, D.Q.; Zhou, H.; Li, Z.; Liu, B.; Wei, J.F.; Chen, F.; et al. High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol. J. 2021, 19, 2544–2560. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D.; Warnes, G. qvalue: Q-Value Estimation for False Discovery Rate Control, Version 2.26.0; R Package. 2021. Available online: http://github.com/jdstorey/qvalue (accessed on 3 February 2022).
- Ginestet, C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A—Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; You, J.; Li, J.; Zhao, W.; Xing, M.; Gong, Y.; Chen, X. Adaptive Traits and Molecular Mechanisms of Rhododendron Species in Changbai Mountains’ Alpine Tundra: A Phenotype–Transcriptome Study. Plants 2025, 14, 3602. https://doi.org/10.3390/plants14233602
Yang Z, You J, Li J, Zhao W, Xing M, Gong Y, Chen X. Adaptive Traits and Molecular Mechanisms of Rhododendron Species in Changbai Mountains’ Alpine Tundra: A Phenotype–Transcriptome Study. Plants. 2025; 14(23):3602. https://doi.org/10.3390/plants14233602
Chicago/Turabian StyleYang, Zhongzan, Jian You, Jiangnan Li, Wei Zhao, Ming Xing, Yuqiao Gong, and Xia Chen. 2025. "Adaptive Traits and Molecular Mechanisms of Rhododendron Species in Changbai Mountains’ Alpine Tundra: A Phenotype–Transcriptome Study" Plants 14, no. 23: 3602. https://doi.org/10.3390/plants14233602
APA StyleYang, Z., You, J., Li, J., Zhao, W., Xing, M., Gong, Y., & Chen, X. (2025). Adaptive Traits and Molecular Mechanisms of Rhododendron Species in Changbai Mountains’ Alpine Tundra: A Phenotype–Transcriptome Study. Plants, 14(23), 3602. https://doi.org/10.3390/plants14233602



