Induction of Autopolyploidy and Preliminary Investigation of the Dwarfing Mechanism in Hedychium coccineum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Colchicine Induction
2.3. Polyploid Identification of H. coccineum
2.4. Growth, Developmental Traits, and Morphological Measurements of Polyploid Plants
2.5. Cytological Observation of Polyploid Plants
2.6. RNA Extraction and Transcriptome Sequencing
2.7. Cloning, Sequence Alignment, and Phylogenetic Analysis of HcPCNA1
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.9. Arabidopsis thaliana Genetic Transformation
2.10. Virus-Induced Gene Silencing (VIGS) of HcPCNA1
2.11. Data Processing
3. Results
3.1. Effects of Colchicine Treatment Conditions on Polyploid Induction Efficiency and Tissue Growth in H. coccineum
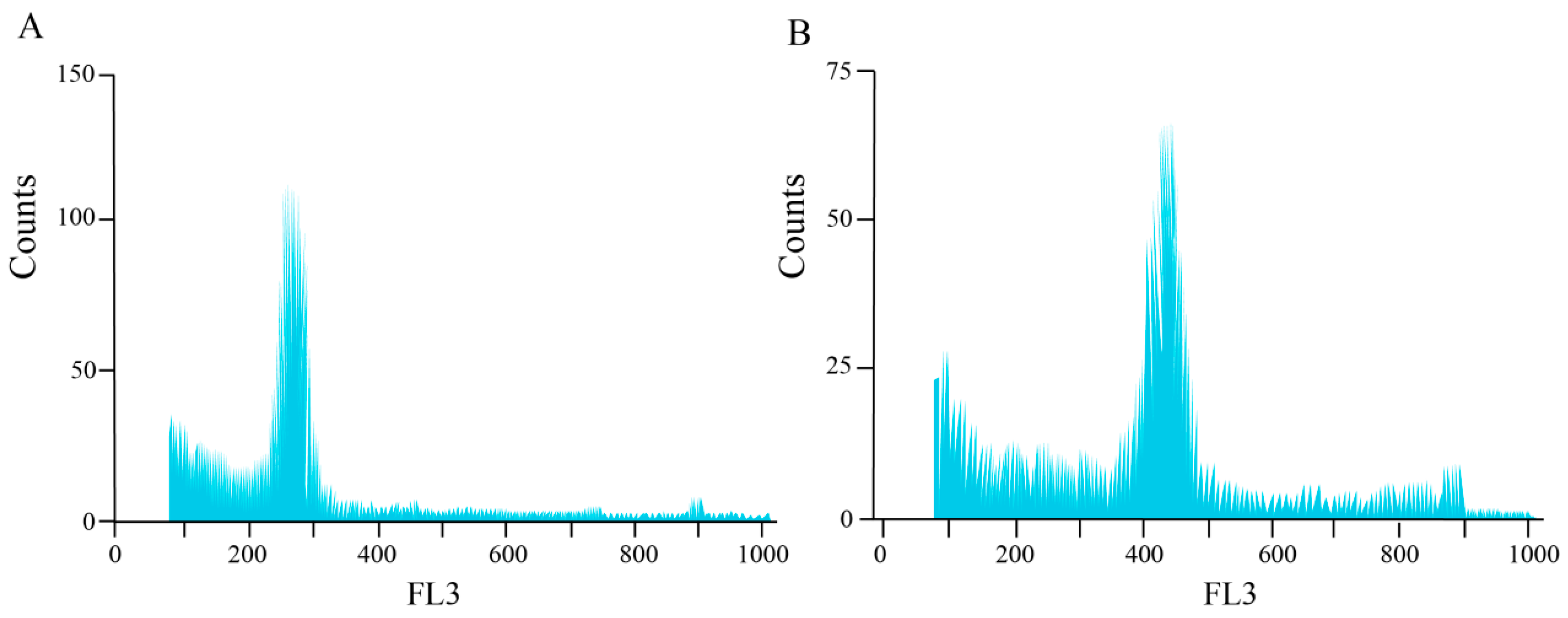
3.2. Chromosome Number and Ploidy Analysis of Mutagenized Materials of H. coccineum
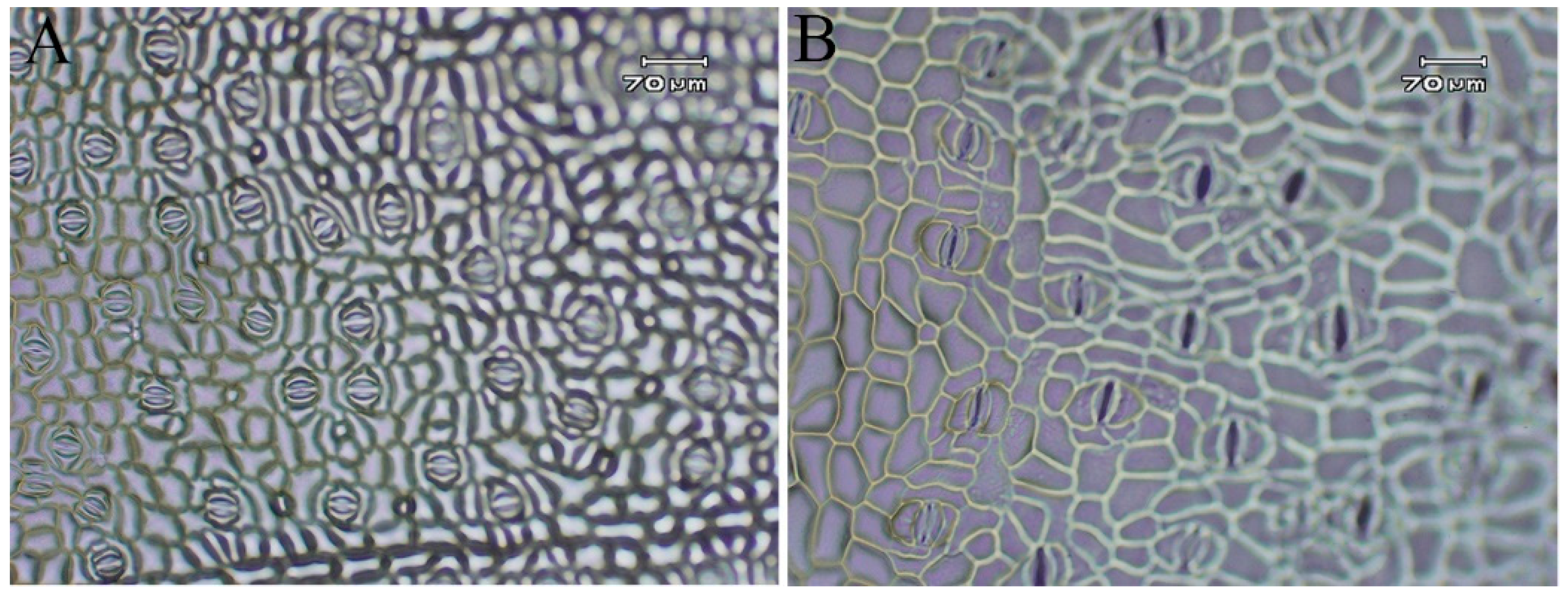
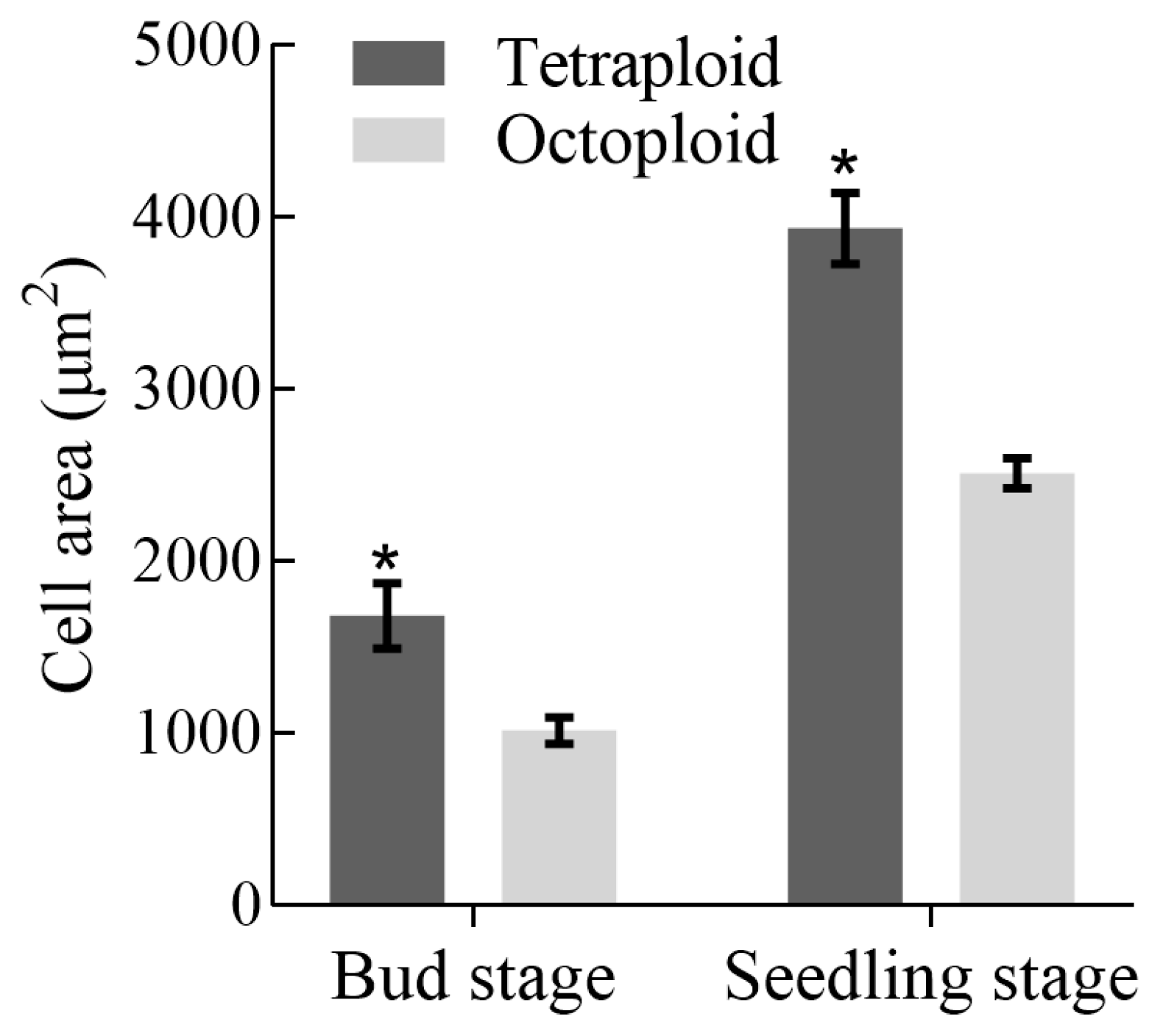
3.3. Analysis of Growth and Developmental Characteristics and Morphological Traits of Tetraploid and Octoploid H. coccineum
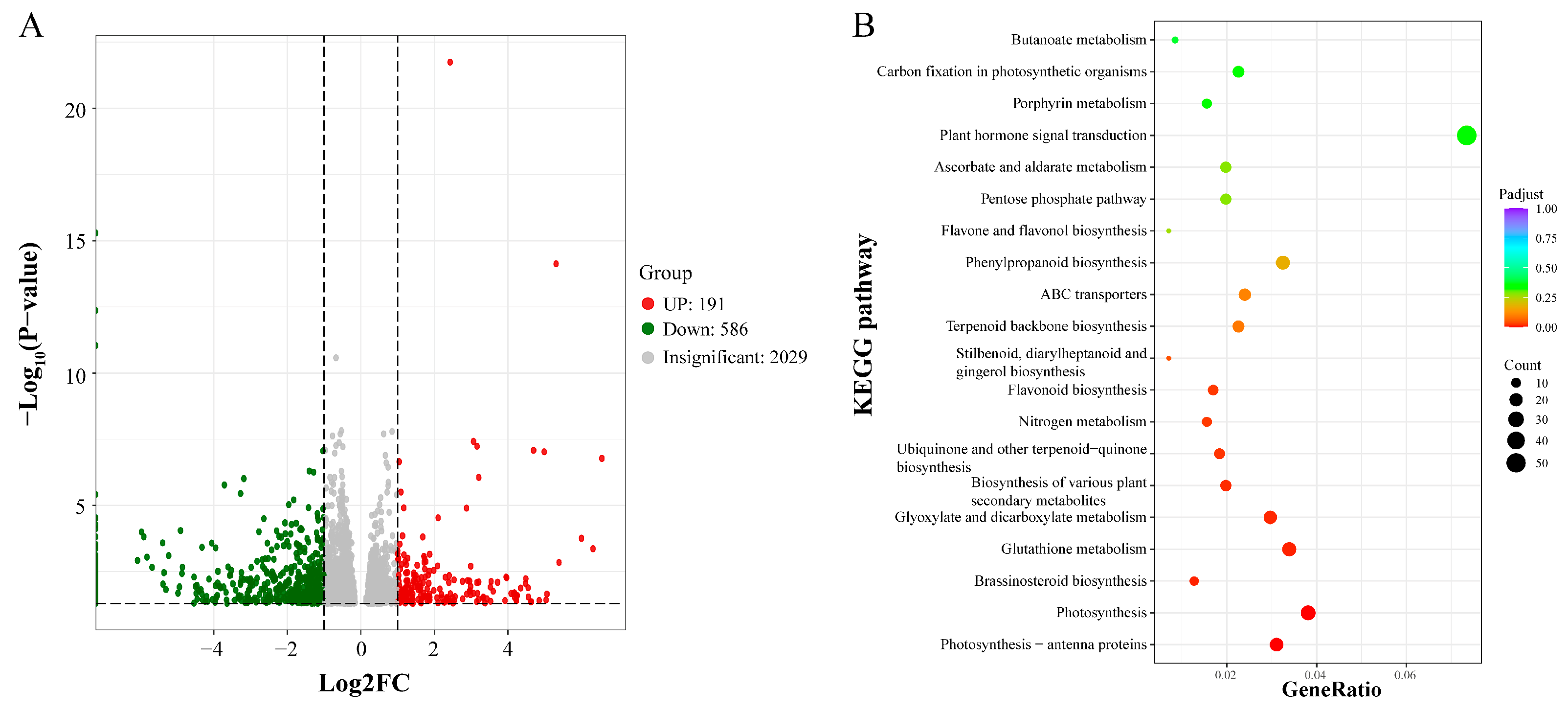
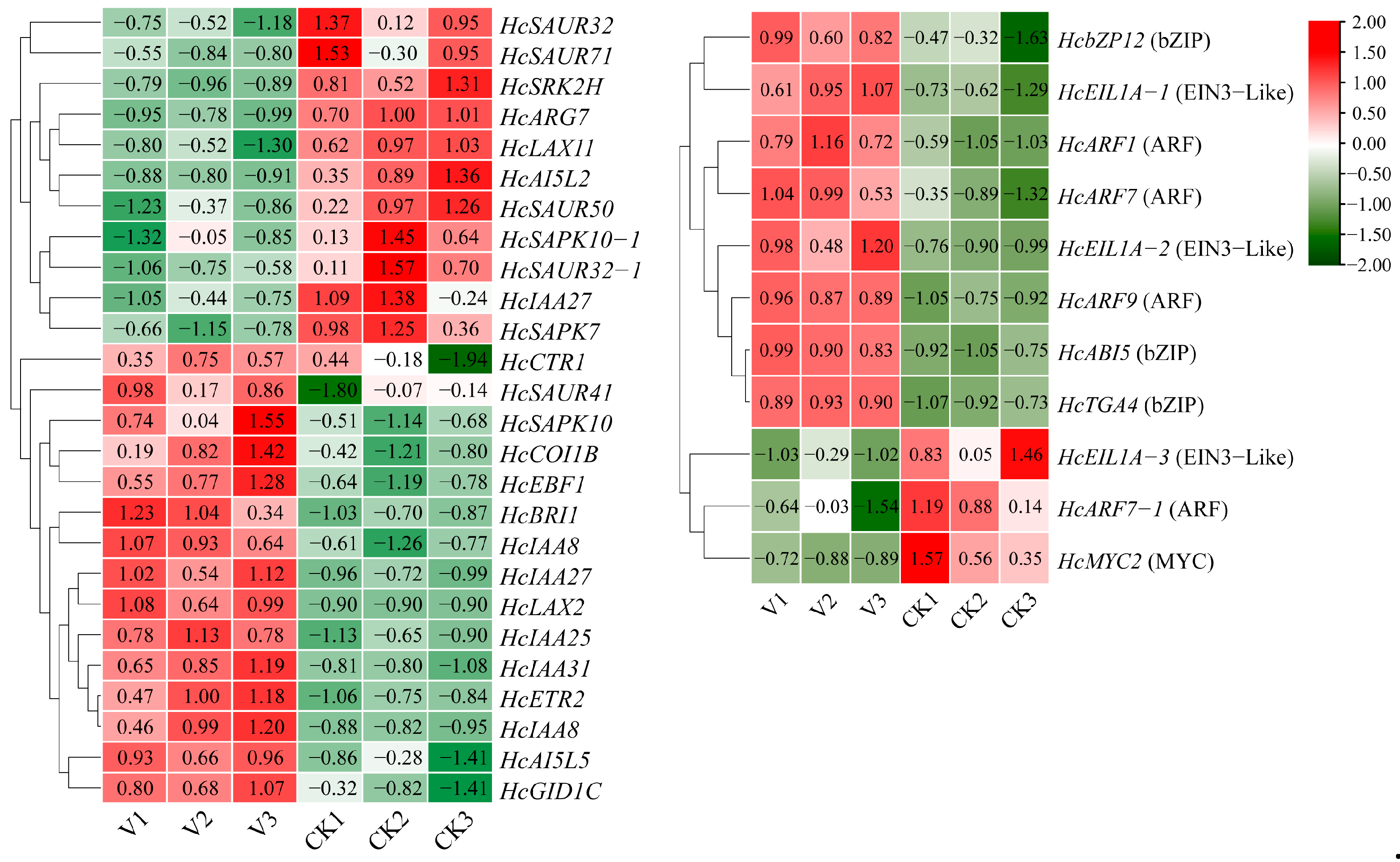
3.4. Transcriptomic Analysis of Tetraploid and Octoploid H. coccineum Plants
3.5. Functional Validation and Analysis of the Key Dwarfing Regulatory Gene HcPCNA1 in H. coccineum
3.5.1. Gene Structure and Phylogenetic Analysis of HcPCNA1
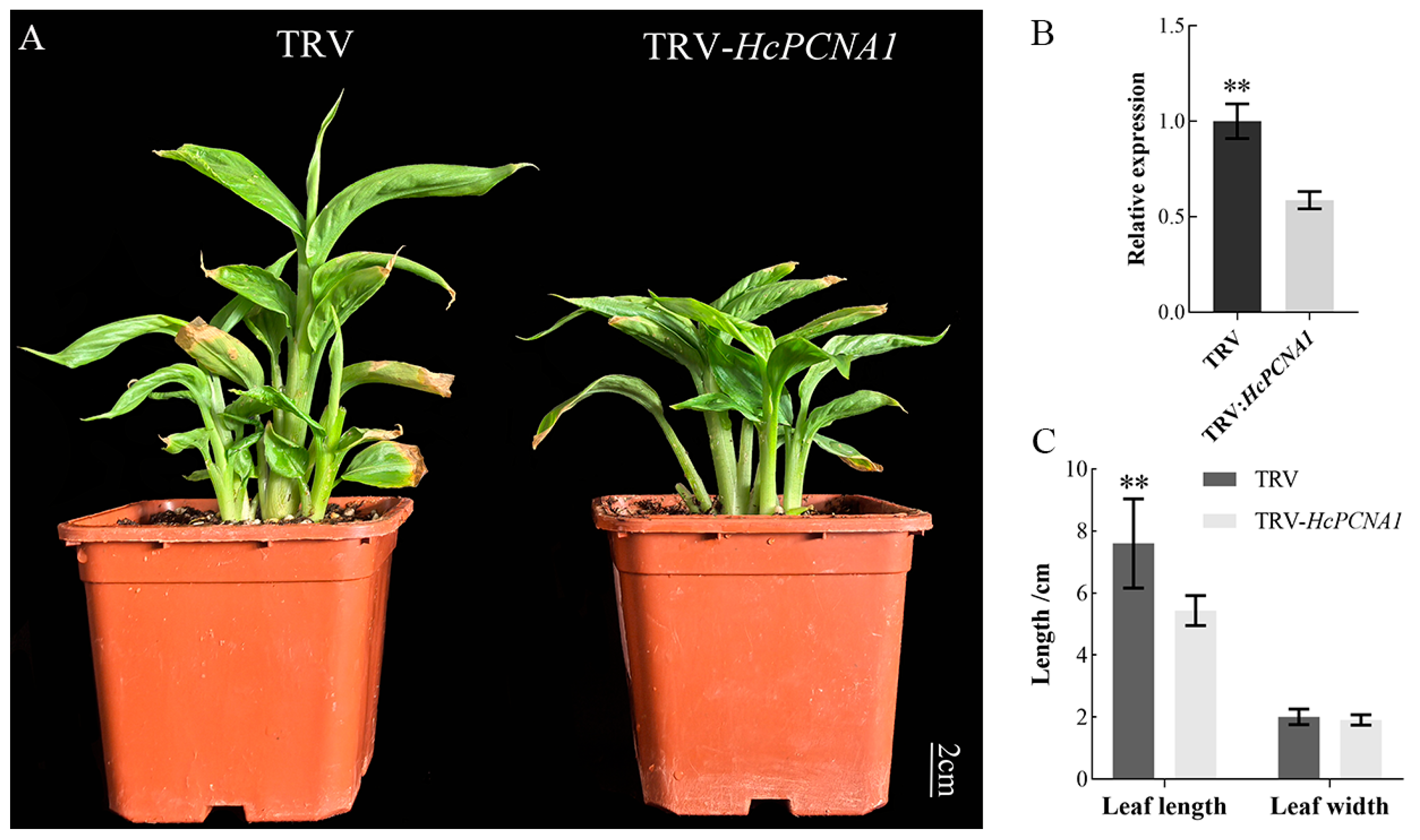
3.5.2. Overexpression and Virus-Induced Silencing of HcPCNA1 Significantly Affect Plant Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ran, Y.Q.; Xie, D.; Long, D.; Jin, L.; Chen, G.; Yang, B.; Tang, D.X.; Tian, M.Y. Phytochemical characterization, antioxidant, enzyme inhibitory and anti-inflammatory activities of Hedychium coccineum Buch.-Ham. ex Sm. rhizome. Biochem. Syst. Ecol. 2025, 122, 105022. [Google Scholar] [CrossRef]
- Tu, H.Y.; Zhang, A.L.; Xiao, W.; Lin, Y.R.; Shi, J.H.; Wu, Y.W.; Wu, S.T.; Zhong, C.H.; Mo, S.X. Induction and identification of tetraploid Hedychium coronarium through thin cell layer culture. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 135, 395–406. [Google Scholar] [CrossRef]
- Liao, L.Y.; Ge, L.Q.; He, X.H.; Li, T.; Huang, B.; Zhao, H.X.; Li, C.L.; Han, Q.L. Prevention and control of ginger blast by two fumigants and their effects on a soil bacterial community and the metabolic components of ginger. Agriculture 2024, 14, 1439. [Google Scholar] [CrossRef]
- Deng, B.; Wang, X.; Long, X.; Fang, R.; Zhou, S.G.; Zhang, J.; Peng, X.L.; An, Z.Y.; Huang, W.X.; Tang, W.Z.; et al. Plant hormone metabolome and transcriptome analysis of dwarf and wild-type banana. J. Plant Growth Regul. 2021, 41, 2386–2405. [Google Scholar] [CrossRef]
- Antonio, S.M.; Robert, S. The pillars of land plants: New insights into stem development. Curr. Opin. Plant Biol. 2018, 45, 11–17. [Google Scholar] [CrossRef]
- Zenil-Ferguson, R.; Burleigh, J.G.; Freyman, W.A.; Igić, B.; Mayrose, I.; Goldberg, E.E. Interaction among ploidy, breeding system and lineage diversification. New Phytol. 2019, 224, 1252–1265. [Google Scholar] [CrossRef]
- Huy, N.P.; Luan, V.Q.; Tung, H.T.; Hien, V.T.; Ngan, H.T.M.; Duy, P.N.; Nhut, D.T. In vitro polyploid induction of Paphiopedilum villosum using colchicine. Sci. Hortic. 2019, 252, 283–290. [Google Scholar] [CrossRef]
- Iannicelli, J.; Guariniello, J.; Tossi, V.E.; Regalado, J.J.; Di Ciaccio, L.; Van Baren, C.M.; Álvarez, S.P.; Escandón, A.S. The “polyploid effect” in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- Li, J.X.; Hoshino, Y. Exploring the impact of ploidy levels on fruit attributes of haskap (Lonicera caerulea L. subsp. edulis (Turcz. ex Herder) Hultén). Euphytica 2025, 221, 74. [Google Scholar]
- Du, K.; Liao, T.; Ren, Y.Y.; Geng, X.N.; Kang, X.Y. Molecular mechanism of vegetative growth advantage in allotriploid Populus. Int. J. Mol. Sci. 2020, 21, 441. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.Q.; Wang, F.; Zhang, Z.H. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Liu, R.; Gao, C.Y.; Jin, J.Z.; Wang, Y.H.; Jia, X.Q.; Ma, H.; Zhang, Y.X.; Zhang, H.X.; Qi, B.X.; Xu, J.F. Induction and identification of tetraploids of pear plants (Pyrus bretschneideri and Pyrus betulaefolia). Sci. Hortic. 2022, 304, 111322. [Google Scholar] [CrossRef]
- Dong, R.Q.; Pei, Y.; Hong, L.Y.; Nie, J.L.; Chen, Y.H.; Yan, H.L.; Liu, Y.J. Tetraploid induction identification and transcriptome preliminary analysis of black currant (Ribes rubrum L.). Plant Cell Tissue Organ Cult. (PCTOC) 2023, 155, 861–872. [Google Scholar] [CrossRef]
- Wei, Y.H.; Zhang, Y.F.; Li, J.X.; Gong, F.F.; Hua, W.H.; Liu, H.W.; Jin, W.C.; Tang, K.Y.; Sun, J.W.; Yan, H.J.; et al. Unravelling the role of phytosulfokine (PSK) in regulating rose petal growth through cell proliferation. Plant J. 2025, 123, e70424. [Google Scholar] [CrossRef]
- Ma, Z.Q.; Buckley, T.N.; Sack, L. The determination of leaf size on the basis of developmental traits. New Phytol. 2025, 246, 461–480. [Google Scholar] [CrossRef]
- Hepworth, J.; Lenhard, M. Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 2014, 17, 36–42. [Google Scholar] [CrossRef]
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef]
- Wang, T.; Liu, L.; Wang, X.J.; Liang, L.X.; Yue, J.J.; Li, L.B. Comparative analyses of anatomical structure, phytohormone levels, and gene expression profiles reveal potential dwarfing mechanisms in shengyin bamboo (Phyllostachys edulis f. tubaeformis). Int. J. Mol. Sci. 2018, 19, 1697. [Google Scholar] [CrossRef] [PubMed]
- Cotias-de-Oliveira, A.L.P.; GuedesM, L.S.; Barrre, E.C. Chromosome numbers for Anthurium and Philodendron spp. (Araceae) occurring in Bahia. Braz. Genet. Mol. Biol. 1999, 22, 237–242. [Google Scholar] [CrossRef]
- Chen, B.Y.; Wang, C.H.; Tian, Y.K.; Chu, Q.G.; Hu, C.H. Anatomical characteristics of young stems and mature leaves of dwarf pear. Sci. Hortic. 2015, 186, 172–179. [Google Scholar] [CrossRef]
- Feng, L.; Liang, X.H.; Zhou, Y.; Zhang, Y.; Liu, J.R.; Cai, M.; Wang, J.; Cheng, T.R.; Zhang, Q.X.; Pan, H.T. Functional Analysis of Aux/IAAs and SAURs on Shoot Growth of Lagerstroemia indica through Virus-Induced Gene Silencing (VIGS). Forests 2020, 11, 1288. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Mukherjee, I. Chromosome studies of some species of Hedychium. Shokubutsugaku Zasshi 1970, 83, 237–241. [Google Scholar] [CrossRef]
- Yu, F.; Kress, W.; Gao, J.Y. Morphology, distribution, and chromosome counts of two varieties of Hedychium villosum (Zingiberaceae). J. Syst. Evol. 2010, 48, 344–349. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, R.Y.; Zhou, L.; Wu, X.Y.; Xu, N.; Ma, X.H.; Zhang, Y. Genome-wide identification analysis of the auxin response factors family in Nicotiana tabacum and the function of NtARF10 in leaf size regulation. J. Plant Biol. 2021, 64, 281–297. [Google Scholar] [CrossRef]
- He, Y.; Li, L.Y.; Shi, W.B.; Tan, J.H.; Luo, X.X.; Zheng, S.Y.; Chen, W.T.; Li, J.; Zhuang, C.X.; Jiang, D.G. Florigen repression complexes involving rice CENTRORADIALIS2 regulate grain size. Plant Physiol. 2022, 190, 1260–1274. [Google Scholar] [CrossRef]
- Salih, H.; He, S.P.; Li, H.G.; Peng, Z.; Du, X.M. Investigation of the EIL/EIN3 transcription factor gene family members and their expression levels in the early stage of cotton Fiber development. Plants 2020, 9, 128. [Google Scholar] [CrossRef]
- Singh, B.; Yun, S.Y.; Gil, Y.; Park, M.H. The role of colchicine in plant breeding. Int. J. Mol. Sci. 2025, 26, 6743. [Google Scholar] [CrossRef]
- Mohammadi, V.; Talebi, S.; Ahmadnasab, M.; Mollahassanzadeh, H. The effect of induced polyploidy on phytochemistry, cellular organelles and the expression of genes involved in thymol and carvacrol biosynthetic pathway in thyme (Thymus vulgaris). Front. Plant Sci. 2023, 14, 1228844. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Thakur, L.; Thakur, M.; Bhardwaj, V.; Gupta, M. In vitro polyploidy induction in ginger (Zingiber officinale Rosc.) cv. Himgiri: Morpho-physiological, biochemical, and molecular evaluation of regenerants. Vitr. Cell. Dev. Biol.-Plant 2024, 60, 693–710. [Google Scholar]
- Lin, J.N.; Zhang, B.N.; Zou, J.T.; Luo, Z.; Yang, H.; Zhou, P.; Chen, X.Y.; Zhou, W. Induction of tetraploids in Paper Mulberry (Broussonetia papyrifera (L.) L’Hér. ex Vent.) by colchicine. BMC Plant Biol. 2023, 23, 574. [Google Scholar] [CrossRef] [PubMed]
- Mallón, R.; Rodríguez-Oubiña, J.; González, M.L. In vitro propagation of the endangered plant Centaurea ultreiae: Assessment of genetic stability by cytological studies, flow cytometry and RAPD analysis. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 101, 31–39. [Google Scholar] [CrossRef]
- Taratima, W.; Rohmah, K.N.; Plaikhuntod, K.; Maneerattanarungroj, P.; Trunjaruen, A. Optimal protocol for in vitro polyploid induction of Cymbidium aloifolium (L.) Sw. BMC Plant Biol. 2023, 23, 295. [Google Scholar] [CrossRef]
- Bian, G.L.; Yi, Y.; Song, Z.Q.; Huang, Y.M.; Mao, Q.X.; Qin, J.; Shang, X.L. Colchicine-induced polyploidization influences the morphological, physiological, and biochemical characteristics of Cyclocarya paliurus. Plants 2025, 14, 2778. [Google Scholar] [CrossRef]
- Obando-González, R.I.; Martínez-Hernández, L.E.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; Ruiz-Medrano, R.; Ramírez-Pool, J.A.; Xoconostle-Cázares, B. Plant growth enhancement in colchicine-treated tomato seeds without polyploidy induction. Plant Mol. Biol. 2025, 115, 3. [Google Scholar] [CrossRef] [PubMed]
- Yakushiji, H.; Yamasaki, A.; Kobayashi, S.; Kaneyoshi, J.; Azuma, A.; Sugiura, H.; Sato, A. Characteristics of a dwarf octoploid mutation arising from a nonaploid persimmon cultivar. Hortic. J. 2016, 85, 209–216. [Google Scholar] [CrossRef]
- Wen, Y.B.; Liu, H.J.; Meng, H.W.; Qiao, L.J.; Zhang, G.; Cheng, Z.Q.; Cheng, Z.H. In vitro induction and phenotypic variations of autotetraploid garlic (Allium sativum L.) with dwarfism. Front. Plant Sci. 2022, 13, 917910. [Google Scholar] [CrossRef]
- Chen, F.B.; Gao, J.; Li, W.B.; Liu, Y.; Fang, P.; Peng, Z.H. Colchicine-induced tetraploidy influences morphological and cytological characteristics and enhances accumulation of anthocyanins in a red-fleshed radish (Raphanus sativus L.). Hortic. Environ. Biotechnol. 2021, 62, 937–948. [Google Scholar] [CrossRef]
- Chen, S.Y.; Zhang, Y.; Zhang, T.; Zhan, D.J.; Pang, Z.W.; Zhao, J.; Zhang, J.F. Comparative transcriptomic, anatomical and phytohormone analyses provide new insights into hormone-mediated tetraploid dwarfing in hybrid sweetgum (Liquidambar styraciflua × L. formosana). Front. Plant Sci. 2022, 13, 924044. [Google Scholar] [CrossRef]
- Wang, L.H.; Luo, Z.; Wang, L.L.; Deng, W.P.; Wei, H.R.; Liu, P.; Liu, M.J. Morphological, cytological and nutritional changes of autotetraploid compared to its diploid counterpart in Chinese jujube (Ziziphus jujuba Mill.). Sci. Hortic. 2019, 249, 263–270. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.B.; Qi, S.Z.; Dong, M.L.; Wang, Z.W.; Li, Y.X.; Chen, S.Y.; Li, B.L.; Zhang, J.F. Ploidy and hybridity effects on leaf size, cell size and related genes expression in triploids, diploids and their parents in Populus. Planta 2019, 249, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.L.; Van de Peer, Y. Polyploidy: A biological force from cells to ecosystems. Trends Cell Biol. 2020, 30, 688–694. [Google Scholar] [CrossRef]
- Sedov, E.N.; Sedysheva, G.A.; Serova, Z.M.; Gorbacheva, N.G. Autogamy of polyploid apple varieties and forms. Russ. Agric. Sci. 2014, 40, 253–256. [Google Scholar] [CrossRef]
- Chester, M.; Gallagher, J.P.; Symonds, V.V.; Cruz da Silva, A.V.; Mavrodiev, E.V.; Leitch, A.R.; Soltis, P.S.; Soltis, D.E. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 2012, 109, 1176–1181. [Google Scholar] [CrossRef]
- Dieckman, L. Something’s gotta give: How PCNA alters its structure in response to mutations and the implications on cellular processes. Prog. Biophys. Mol. Biol. 2021, 163, 46–59. [Google Scholar] [CrossRef]
- Mondol, T.; Stodola, J.L.; Galletto, R.; Burgers, P.M. PCNA accelerates the nucleotide incorporation rate by DNA polymerase δ. Nucleic Acids Res. 2019, 47, 1977–1986. [Google Scholar] [CrossRef]
- Lv, J.H.; Feng, Y.; Zhai, L.M.; Jiang, L.Z.; Wu, Y.; Huang, Y.M.; Yu, R.Q.; Wu, T.; Zhang, X.Z.; Wang, Y.; et al. MdARF3 switches the lateral root elongation to regulate dwarfing in apple plants. Hortic. Res. 2024, 11, uhae051. [Google Scholar] [CrossRef]
- Kong, X.C.; Wang, F.; Wang, Z.Y.; Gao, X.H.; Geng, S.F.; Deng, Z.Y.; Zhang, S.; Fu, M.X.; Cui, D.D.; Liu, S.S.; et al. Grain yield improvement by genome editing of TaARF12 that decoupled peduncle and rachis development trajectories via differential regulation of gibberellin signalling in wheat. Plant Biotechnol. J. 2023, 21, 1990–2001. [Google Scholar] [CrossRef]
- Li, C.P.; Wen, H.; Wu, Y.Y.; Li, Y.T.; Feng, X.Q.; Li, X.W.; Xu, S.Y.; Jiang, D.; Zhang, B.C.; Li, M.; et al. OsRF2b interacting with OsbZIP61 modulates nitrogen use efficiency and grain yield via heterodimers in rice. Plant Biotechnol. J. 2025, 23, 3300–3312. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.P.; Zhang, L.; Zhu, L.F.; Zhang, X.L.; He, X. The HD-Zip transcription factor GhHB12 represses plant height by regulating the auxin signaling in cotton. J. Integr. Agric. 2023, 22, 2015–2024. [Google Scholar] [CrossRef]
- Zhao, Z.C.; Lu, Q.M.; Gao, Z.P.; Kong, X.L.; Zhang, X.B.; Chen, L.; Hu, Y.G. Exogenous GA3 significantly improved the grain filling process and yield traits of Rht15 dwarf lines in durum wheat. Eur. J. Agron. 2025, 162, 127430. [Google Scholar] [CrossRef]
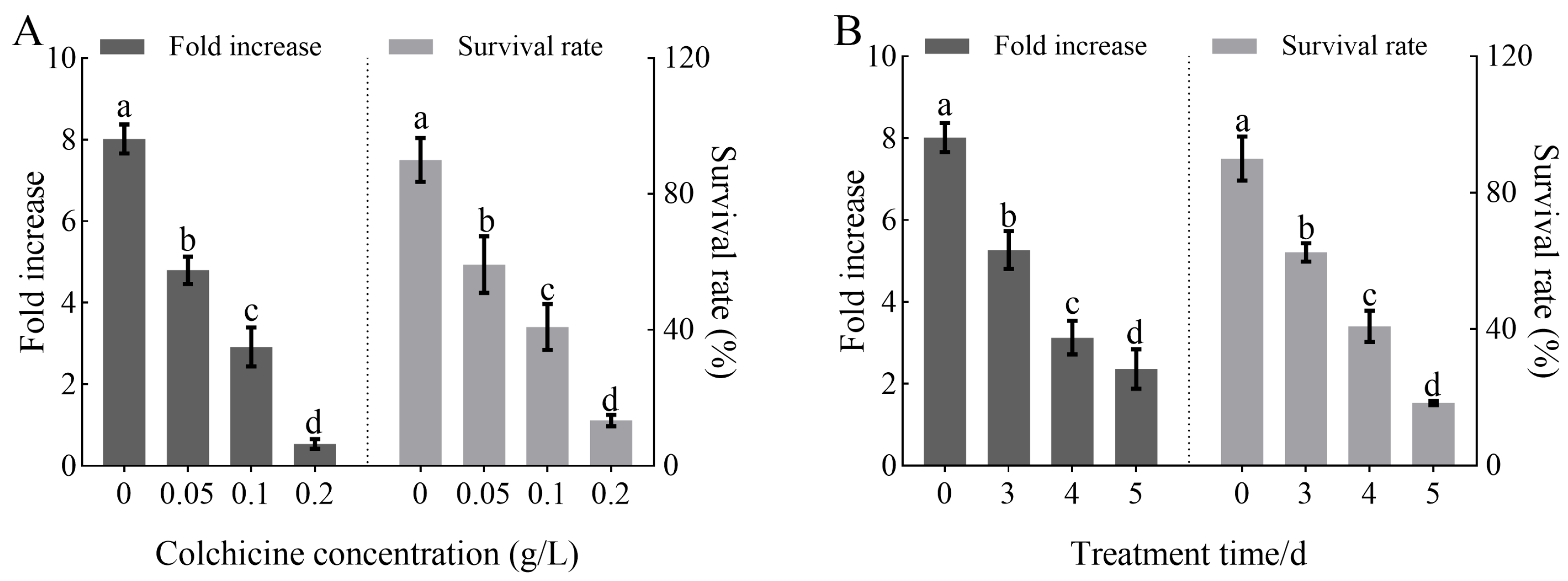
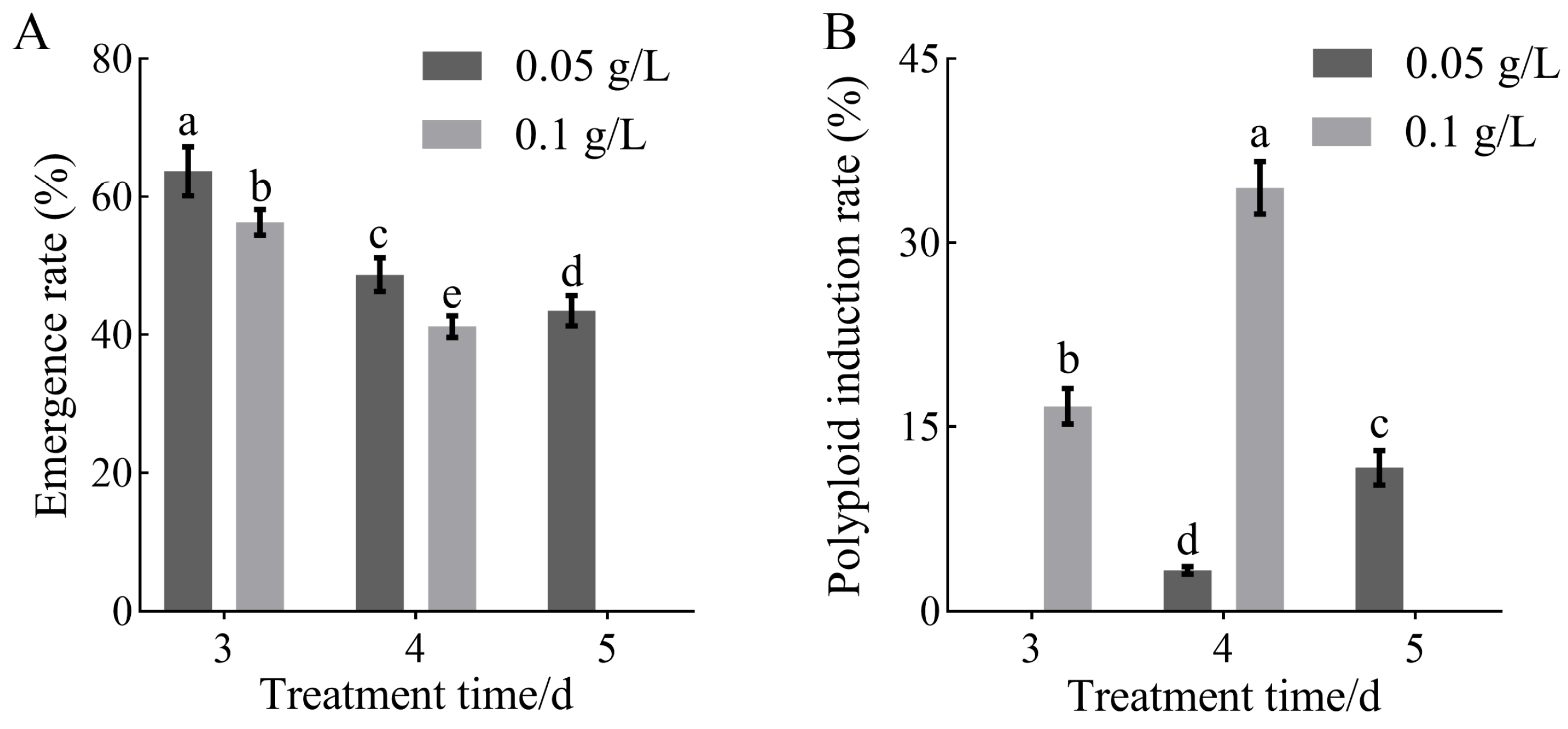








| Treatment Concentration (g/L) | Treatment Time (d) | Fold Increase | Survival Rate (%) |
|---|---|---|---|
| 0 | 0 | 8.02 ± 0.28 a | 90.00 ± 3.26 a |
| 0.05 | 3 | 5.85 ± 0.25 b | 75.00 ± 3.38 b |
| 4 | 4.76 ± 0.46 c | 65.00 ± 3.32 c | |
| 5 | 3.83 ± 0.32 d | 37.50 ± 2.24 e | |
| 0.1 | 3 | 3.49 ± 0.24 e | 60.00 ± 3.26 c |
| 4 | 2.33 ± 0.23 f | 45.00 ± 2.21 d | |
| 5 | 1.24 ± 0.27 g | 17.50 ± 1.18 g | |
| 0.2 | 3 | 0.96 ± 0.14 h | 27.50 ± 2.21 f |
| 4 | 0.53 ± 0.10 h | 12.50 ± 1.12 g | |
| 5 | 0.00 ± 0.00 h | 0.00 ± 0.00 h |
| Traits | Tetraploid | Octoploid |
|---|---|---|
| Vegetative growth period (months) | 4~5 | 4~5 |
| Bud break period (months) | 2~3 | 2~3 |
| Dormancy period (months) | December to February of the following year | Almost no dormancy |
| Flowering period (months) | June to October | Mid-May to early June |
| Flowering period of an inflorescence (day) | 8~10 | 2~3 |
| Flowering period of a single flower (day) | 2~3 | 1.5~2.5 |
| Leaf emergence rate (days per leaf) | 3.68 | 4.90 |
| Plant height (cm) | 183.09 ± 39.24 a | 72.98 ± 6.30 b |
| Plant spread (m2) | 1.95 ± 0.67 a | 0.30 ± 0.11 b |
| Number of tillers(branches) | 29.89 ± 11.31 a | 20.60 ± 7.40 a |
| Number of internodes (number) | 15.78 ± 2.99 a | 10.5 ± 0.58 b |
| Pseudostem diameter (cm) | 16.25 ± 2.42 a | 16.65 ± 0.78 a |
| Average internode length (cm) | 11.99 ± 3.60 a | 6.95 ± 0.38 b |
| Leaf length (cm) | 42.58 ± 3.34 a | 20.38 ± 1.11 b |
| Leaf width (cm) | 5.01 ± 0.42 a | 3.63 ± 0.29 b |
| Leaf angle (°) | 55.36 ± 4.03 b | 71.67 ± 5.17 a |
| Stomata Density (no./mm2) | 75.43 ± 2.23 a | 36.84 ± 1.72 b |
| Stomata Length (μm) | 32.20 ± 1.86 a | 52.37 ± 4.71 b |
| Stomata Width (μm) | 30.52 ± 2.88 a | 78.30 ± 8.01 b |
| Inflorescence length (cm) | 25.38 ± 5.31 a | 8.15 ± 0.26 b |
| Inflorescence width (cm) | 14.13 ± 1.84 a | 4.95 ± 0.13 b |
| Number of bracteoles (number) | 42.75 ± 2.22 a | 19.00 ± 1.73 b |
| Flower length (cm) | 5.43 ± 0.10 a | 4.58 ± 0.17 b |
| Flower width (cm) | 3.75 ± 0.17 a | 2.93 ± 0.19 b |
| Bract length (cm) | 3.65 ± 0.31 a | 2.85 ± 0.19 b |
| Bract width (cm) | 1.38 ± 0.25 a | 1.29 ± 0.09 a |
| Bracteole length (cm) | 1.98 ± 0.21 a | 1.76 ± 0.05 a |
| Bracteole width (cm) | 1.14 ± 0.15 a | 1.16 ± 0.11 a |
| Calyx length (cm) | 2.89 ± 0.14 a | 2.65 ± 0.25 a |
| Calyx width (cm) | 0.60 ± 0.07 b | 0.80 ± 0.04 a |
| Corolla tube length (cm) | 3.03 ± 0.10 a | 1.65 ± 0.12 b |
| Corolla tube width (cm) | 0.19 ± 0.02 b | 0.33 ± 0.03 a |
| Lobe length (cm) | 3.15 ± 0.27 a | 2.93 ± 0.05 a |
| Lobe width (cm) | 0.23 ± 0.03 a | 0.32 ± 0.05 a |
| lip petal length (cm) | 2.48 ± 0.19 a | 2.84 ± 0.17 a |
| lip petal width (cm) | 1.85 ± 0.18 a | 1.76 ± 0.09 a |
| Lateral petal length (cm) | 2.60 ± 0.16 a | 2.88 ± 0.10 a |
| Lateral petal width (cm) | 0.43 ± 0.06 a | 0.29 ± 0.03 b |
| Pistil length (cm) | 8.00 ± 0.08 a | 5.68 ± 0.17 b |
| Pistil width (cm) | 0.19 ± 0.02 b | 0.33 ± 0.03 a |
| Stamen length (cm) | 5.16 ± 0.11 a | 3.91 ± 0.09 b |
| Stamen width (cm) | 0.21 ± 0.02 a | 0.28 ± 0.03 a |
| Anther length (cm) | 0.73 ± 0.09 b | 0.91 ± 0.03 a |
| Anther width (cm) | 0.21 ± 0.02 a | 0.28 ± 0.03 a |
| Filament length (cm) | 4.45 ± 0.07 a | 2.90 ± 0.08 b |
| Filament width (cm) | 0.14 ± 0.02 b | 0.21 ± 0.02 a |
| Sample | Clean Reads | Clean Bases | GC Content | Q30 | Total Mapping Reads |
|---|---|---|---|---|---|
| T1 | 39,578,990 | 7,994,174,796 | 49.42% | 94.46% | 68.99% |
| T2 | 38,755,407 | 7,827,821,973 | 49.31% | 94.61% | 68.89% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Jin, F.; Liang, X.; Qiu, J.; Wang, Q.; Yu, Y.; Yu, R.; Fan, Y. Induction of Autopolyploidy and Preliminary Investigation of the Dwarfing Mechanism in Hedychium coccineum. Plants 2025, 14, 3573. https://doi.org/10.3390/plants14233573
Wang F, Jin F, Liang X, Qiu J, Wang Q, Yu Y, Yu R, Fan Y. Induction of Autopolyploidy and Preliminary Investigation of the Dwarfing Mechanism in Hedychium coccineum. Plants. 2025; 14(23):3573. https://doi.org/10.3390/plants14233573
Chicago/Turabian StyleWang, Fang, Feixuan Jin, Xuanguo Liang, Jiangming Qiu, Qing Wang, Yunyi Yu, Rangcai Yu, and Yanping Fan. 2025. "Induction of Autopolyploidy and Preliminary Investigation of the Dwarfing Mechanism in Hedychium coccineum" Plants 14, no. 23: 3573. https://doi.org/10.3390/plants14233573
APA StyleWang, F., Jin, F., Liang, X., Qiu, J., Wang, Q., Yu, Y., Yu, R., & Fan, Y. (2025). Induction of Autopolyploidy and Preliminary Investigation of the Dwarfing Mechanism in Hedychium coccineum. Plants, 14(23), 3573. https://doi.org/10.3390/plants14233573





