Stress Adaptation Phenomena of Rhododendron Species in Alpine Tundra and Timberline of Changbai Mountain: Physiological Traits and Molecular Evolution
Abstract
1. Introduction
2. Results
2.1. Sampling Sites and Soil Chemical Properties
2.2. Adaptive Phenomena of Rhododendron Species in Harsh Alpine Environments
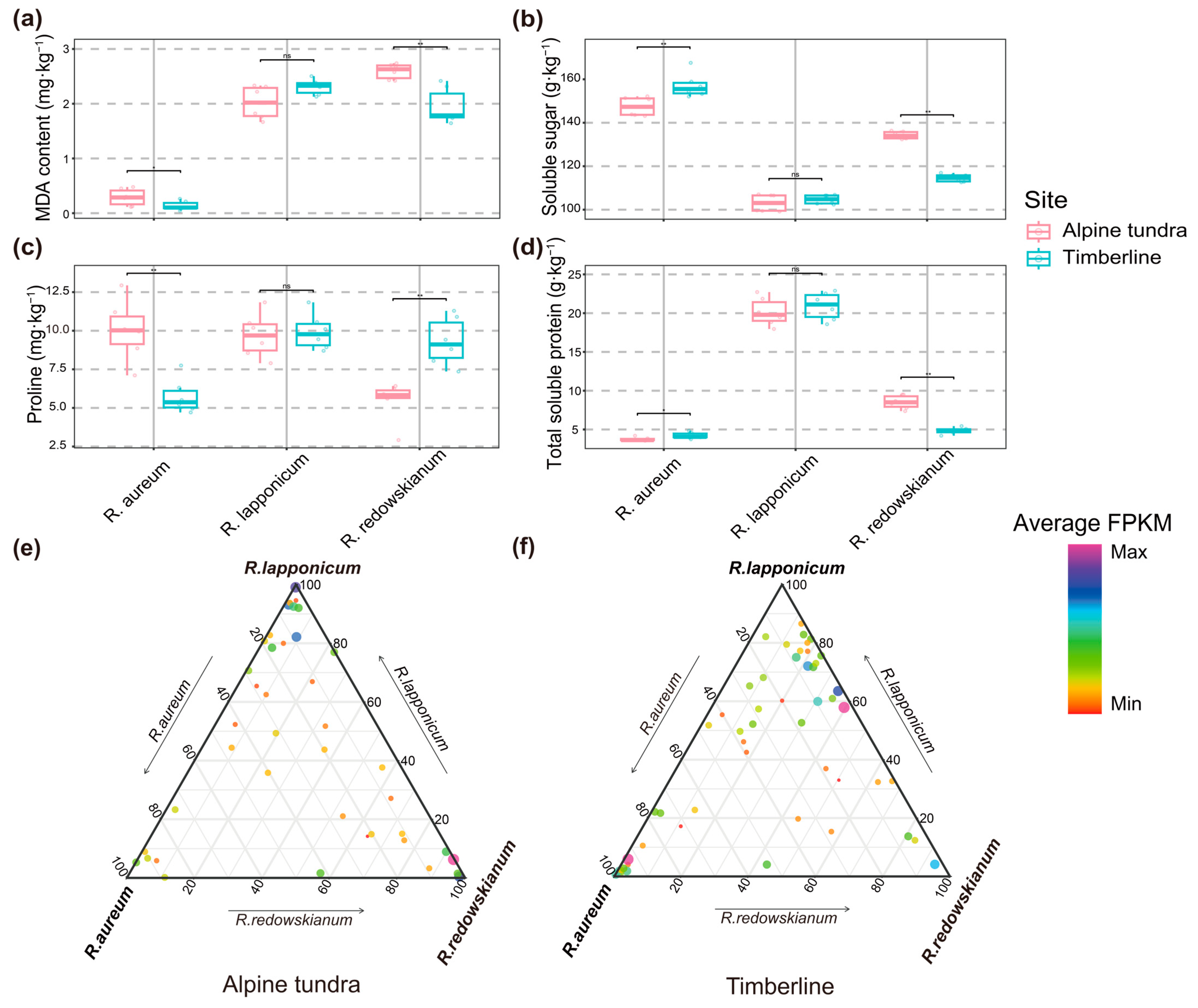
2.2.1. Leaf Physiological Adaptive Characteristics and Expression Levels of PSGs
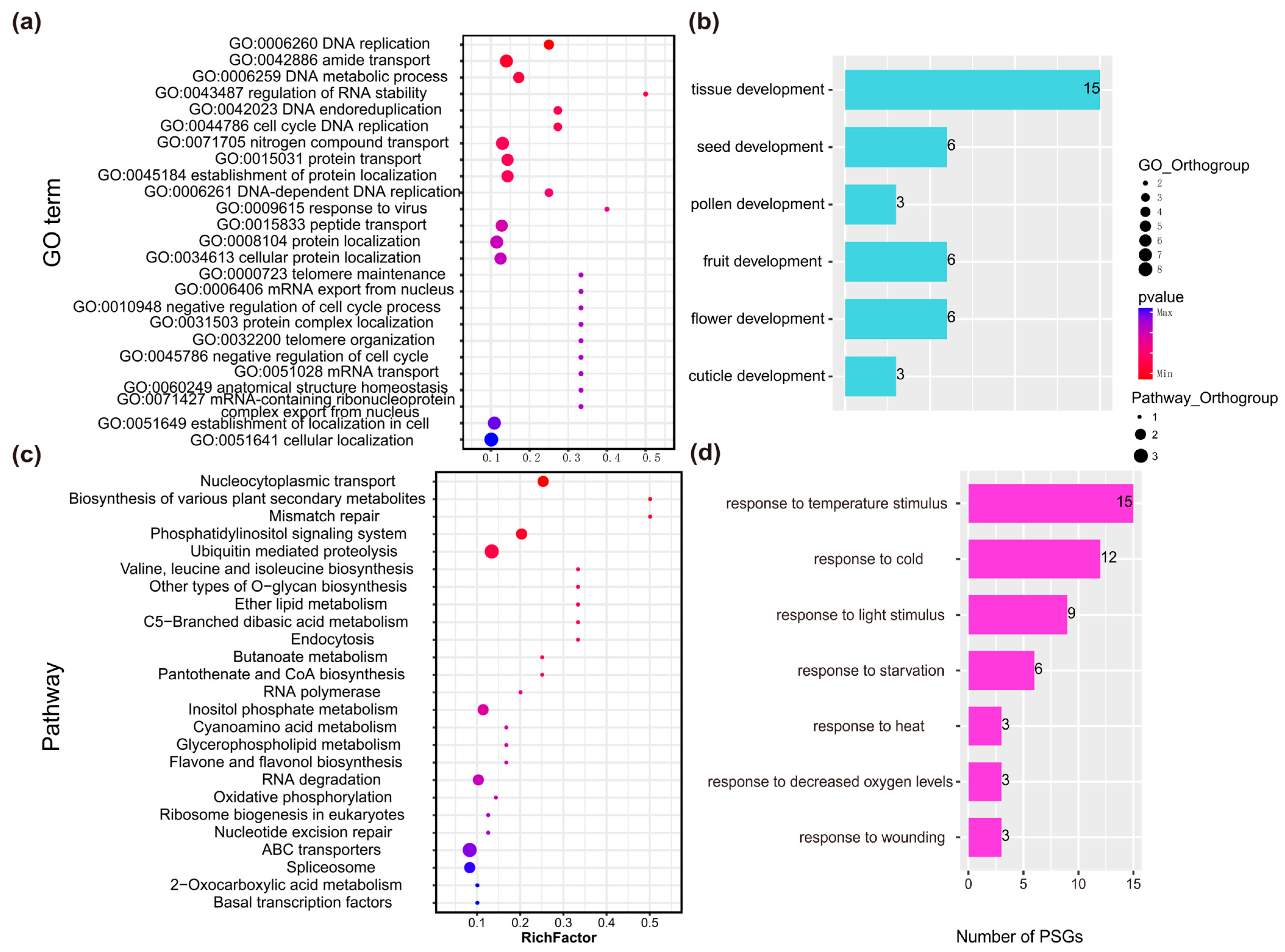
2.2.2. Functional Annotation of PSGs in Rhododendron Species
3. Discussion
3.1. Soil Nutrients and Physiological Adaptation of Rhododendron Species
3.2. The Molecular Mechanisms of Adaptation of Rhododendron Species
3.2.1. Metabolism and Basic Cellular Processes Are the Core Foundation of Adaptation
3.2.2. Tissue Plasticity and Reproductive Assurance Are Key Evolutionary Directions
3.2.3. Temperature Is One of the Dominant Selective Pressures
4. Materials & Methods
4.1. Experimental Materials and Sampling
4.2. Determination of Physicochemical and Physiological Indicators
4.2.1. Soil Physicochemical Properties
4.2.2. Leaf Area and Physiological Indicators
4.3. Identification and Annotation of Orthologs
4.3.1. Identification of Single-Copy Orthologs
4.3.2. Coding Sequence (CDS) Prediction
4.3.3. Multiple Sequence Alignment and Optimization
4.3.4. Selection Pressure Analysis
4.3.5. Functional Annotation
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PSGs | positively selected genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| SOC | soil total organic carbon |
| NH4+-N | ammonium nitrogen |
| NO3−-N | nitrate nitrogen |
| AP | available phosphorus |
| LA | leaf area |
| MDA | malondialdehyde |
| CDS | coding sequence |
| BSM | Branch Site Model |
References
- Xia, X.M.; Yang, M.Q.; Li, C.L.; Huang, S.X.; Jin, W.T.; Shen, T.T.; Wang, F.; Li, X.H.; Yoichi, W.; Zhang, L.H.; et al. Spatiotemporal Evolution of the Global Species Diversity of Rhododendron. Mol. Biol. Evol. 2022, 39, msab314. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.Q.; Fu, C.N.; Zhu, M.S.; Milne, R.; Yang, J.B.; Cai, J.; Qin, H.T.; Zheng, W.; Hollingsworth, P.M.; Li, D.Z.; et al. Resolution, conflict and rate shifts: Insights from a densely sampled plastome phylogeny for Rhododendron (Ericaceae). Ann. Bot. 2022, 130, 687–701. [Google Scholar] [CrossRef]
- Xia, X.M.; Du, H.L.; Hu, X.D.; Wu, J.J.; Yang, F.S.; Li, C.L.; Huang, S.X.; Wang, Q.; Liang, C.Z.; Wang, X.Q. Genomic insights into adaptive evolution of the species-rich cosmopolitan plant genus. Cell Rep. 2024, 43, 114745. [Google Scholar] [CrossRef]
- Ding, W.N.; Ree, R.H.; Spicer, R.A.; Xing, Y.W. Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 2020, 369, 578–581. [Google Scholar] [CrossRef]
- Liu, X.W.; Wang, Y.H.; Shen, S.K. Transcriptomic and metabolomic analyses reveal the altitude adaptability and evolution of different-colored flowers in alpine species. Tree Physiol. 2022, 42, 1100–1113. [Google Scholar] [CrossRef]
- Huang, X.; Li, C. An analysis on the ecology of alpine tundra landscape of Changbai Mountains. Acta Geogr. Sin. 1984, 39, 285–297. [Google Scholar] [CrossRef]
- Jin, Y.Y.; Xu, J.W.; He, H.S.; Li, M.H.; Tao, Y.; Zhang, Y.J.; Hu, R.; Gao, X.; Bai, Y.Y.; Wang, H.Y.; et al. The Changbai alpine shrub tundra will be replaced by herbaceous tundra under global climate change. Plants 2019, 8, 370. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.J.; Wu, Z.; Du, H.; Zong, S.; Ma, S. Potential distribution shifts of plant species under climate change in Changbai Mountains, China. Forests 2019, 10, 498. [Google Scholar] [CrossRef]
- Li, N.; Du, H.B.; Li, M.H.; Na, R.S.; Dong, R.K.; He, H.S.; Zong, S.W.; Huang, L.R.; Wu, Z.F. Deyeuxia angustifolia upward migration and nitrogen deposition change soil microbial community structure in an alpine tundra. Soil Biol. Biochem. 2023, 180, 109009. [Google Scholar] [CrossRef]
- Du, H.B.; Liu, J.; Li, M.H.; Buntgen, U.; Yang, Y.; Wang, L.; Wu, Z.F.; He, H.S. Warming-induced upward migration of the alpine treeline in the Changbai Mountains, northeast China. Glob. Chang. Biol. 2018, 24, 1256–1266. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Gong, F.S.; Xu, H.W.; Zhou, X.F. Molecular Mechanism of Exogenous ABA to Enhance UV-B Resistance in Rhododendron chrysanthum Pall. by Modulating Flavonoid Accumulation. Int. J. Mol. Sci. 2024, 25, 5248. [Google Scholar] [CrossRef]
- Cordero, R.R.; Seckmeyer, G.; Damiani, A.; Riechelmann, S.; Rayas, J.; Labbe, F.; Laroze, D. The world’s highest levels of surface UV. Photochem. Photobiol. Sci. 2014, 13, 70–81. [Google Scholar] [CrossRef]
- Ni, X.Y.; Yang, W.Q.; Tan, B.; Li, H.; He, J.; Xu, L.Y.; Wu, F.Z. Forest gaps slow the sequestration of soil organic matter: A humification experiment with six foliar litters in an alpine forest. Sci. Rep. 2016, 6, 19744. [Google Scholar] [CrossRef]
- Zhao, W.; Qi, X.J.; Lyu, J.W.; Yu, Z.X.; Chen, X. Characterization of microbial community structure in rhizosphere soils of Cowskin azalea (Georgi) on northern slope of Changbai Mountains, China. Chin. Geogr. Sci. 2016, 26, 78–89. [Google Scholar] [CrossRef]
- Zong, S.W.; Lembrechts, J.J.; Du, H.B.; He, H.S.; Wu, Z.F.; Li, M.H.; Rixen, C. Upward range shift of a dominant alpine shrub related to 50 years of snow cover change. Remote Sens. Environ. 2022, 268, 112773. [Google Scholar] [CrossRef]
- Hou, J.; Liu, M.; Yang, K.; Liu, B.; Liu, H.H.; Liu, J.Q. Genetic variation for adaptive evolution in response to changed environments in plants. J. Integr. Plant Biol. 2025, 67, 2265–2293. [Google Scholar] [CrossRef]
- Xu, W.Q.; Ren, C.Q.; Zhang, X.Y.; Comes, H.P.; Liu, X.H.; Li, Y.G.; Kettle, C.J.; Jalonen, R.; Gaisberger, H.; Ma, Y.Z.; et al. Genome sequences and population genomics reveal climatic adaptation and genomic divergence between two closely related sweetgum species. Plant J. 2024, 118, 1372–1387. [Google Scholar] [CrossRef]
- Chen, J.G.; Li, Y.B.; Yang, Y.; Sun, H. How cushion communities are maintained in alpine ecosystems: A review and case study on alpine cushion plant reproduction. Plant Divers. 2017, 39, 221–228. [Google Scholar] [CrossRef]
- Ruosch, M.; Spahni, R.; Joos, F.; Henne, P.D.; Van der Knaap, W.O.; Tinner, W. Past and future evolution of forests in Europe—Comparison of a dynamic vegetation model with palaeo data and observations. Glob. Chang. Biol. 2016, 22, 727–740. [Google Scholar] [CrossRef]
- Jin, Y.H.; Zhang, Y.J.; Xu, J.W.; Tao, Y.; He, H.S.; Guo, M.; Wang, A.L.; Liu, Y.X.; Niu, L.P. Comparative assessment of tundra vegetation changes between north and southwest slopes of Changbai Mountains, China, in response to global warming. Chin. Geogr. Sci. 2018, 28, 665–679. [Google Scholar] [CrossRef]
- Wang, C.; Ye, D.; Li, Y.; Hu, P.L.; Xu, R.; Wang, X.J. Genome-wide identification and bioinformatics analysis of the WRKY transcription factors and screening of candidate genes for anthocyanin biosynthesis in azalea (Rhododendron simsii). Front. Genet. 2023, 14, 1172321. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; You, J.; Li, J.; Zhao, W.; Xing, M.; Zhang, Y.; Ma, C.; Gong, Y.; Zhao, Y.; Chen, X. Divergent functional traits and gene expression profiles in native and encroaching plant species across an alpine elevational gradient. Front. Plant Sci. 2025, 16, 1656812. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Ni, Y.; Liang, W.; Wang, J.; Chu, H. Distinct soil bacterial communities along a small-scale elevational gradient in alpine tundra. Front. Microbiol. 2015, 6, 582. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Li, L.; Wang, X.; You, J.; Li, J.; Chen, X. Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 2020, 10, 12442. [Google Scholar] [CrossRef]
- Cansler, C.A.; McKenzie, D.; Halpern, C.B. Fire enhances the complexity of forest structure in alpine treeline ecotones. Ecosphere 2018, 9, e02091. [Google Scholar] [CrossRef]
- Shang, X.M.; Zhao, Z.Q.; Xiao, W.; Zeng, Y.K.; Li, M.D.; Jiang, X.; Dahro, B.; Chu, L.L.; Wang, M.; Li, C.L.; et al. The ctrCBL1/ctrCIPK6 complex of citrus phosphorylates ctrBBX32 to regulate ctrSTP1-mediated sugar accumulation and cold tolerance. Adv. Sci. 2025, 23, e08372. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhang, M.H.; Wu, C.Z.; Chen, C.; Meng, L.; Zhang, G.Q.; Zhuang, K.Y.; Shi, Q.H. SlWRKY51 regulates proline content to enhance chilling tolerance in tomato. Plant Cell Environ. 2024, 47, 5104–5114. [Google Scholar] [CrossRef]
- Hao, S.H.; Wang, Y.R.; Yan, Y.X.; Liu, Y.H.; Wang, J.Y.; Chen, S. A review on plant responses to salt stress and their mechanisms of salt resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Klaas, M.; Haiminen, N.; Grant, J.; Cormican, P.; Finnan, J.; Arojju, S.K.; Utro, F.; Vellani, T.; Parida, L.; Barth, S. Transcriptome characterization and differentially expressed genes under flooding and drought stress in the biomass grasses Phalaris arundinacea and Dactylis glomerata. Ann. Bot. 2019, 124, 717–730. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Z.Q.; Kong, X.J.; Khan, A.; Ullah, N.; Zhang, X. Plant coping with cold stress: Molecular and physiological adaptive mechanisms with future perspectives. Cells 2025, 14, 110. [Google Scholar] [CrossRef]
- Tang, D.; Yan, L.; Fisher, J.L.; Kang, H.B.; Dallongeville, P.; Wasaki, J.; Wang, X.; Lambers, H. Why are some invasive plant species so successful in nutrient-impoverished habitats in south-western Australia: A perspective based on their phosphorus-acquisition strategies. Funct. Ecol. 2025, 39, 635–652. [Google Scholar] [CrossRef]
- Tao, C.N.; Buswell, W.; Zhang, P.J.; Walker, H.; Johnson, I.; Field, K.; Schwarzenbacher, R.; Ton, J. A single amino acid transporter controls the uptake of priming-inducing beta-amino acids and the associated tradeoff between induced resistance and plant growth. Plant Cell 2022, 34, 4840–4856. [Google Scholar] [CrossRef] [PubMed]
- Koper, K.; de Oliveira, M.V.V.; Huss, S.; Hataya, S.; Soleymani, F.; Hawkins, C.; Rhee, S.Y.; Takasuka, T.E.; Nikoloski, Z.; Maeda, H.A. Mapping multi-substrate specificity of aminotransferases. Nat. Plants 2025, 11, 1863–1876. [Google Scholar] [CrossRef] [PubMed]
- Alhabsi, A.; Ling, Y.; Crespi, M.; Reddy, A.S.N.; Mahfouz, M. Alternative Splicing Dynamics in Plant Adaptive Responses to Stress. Annu. Rev. Plant Biol. 2025, 76, 687–717. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Allan, A.C.; Jones, J.J.; Geilfus, C.M. Why do plants blush when they are hungry? New Phytol. 2023, 239, 494–505. [Google Scholar] [CrossRef]
- Marathe, S.; Grotewold, E.; Otegui, M.S. Should I stay or should I go? Trafficking of plant extra-nuclear transcription factors. Plant Cell 2024, 36, 1524–1539. [Google Scholar] [CrossRef]
- Chen, Z.R.; Dong, Y.; Huang, X. Plant responses to UV-B radiation: Signaling, acclimation and stress tolerance. Stress Biol. 2022, 2, 51. [Google Scholar] [CrossRef]
- McDougall, K.L.; Khuroo, A.A.; Loope, L.L.; Parks, C.G.; Pauchard, A.; Reshi, Z.A.; Rushworth, I.; Kueffer, C. Plant invasions in mountains: Global lessons for better management. Mt. Res. Dev. 2011, 31, 380–387. [Google Scholar] [CrossRef]
- Carter, M.R.; Gregrorich, E.G. Soil Sampling and Methods of Analysis; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Xu, S.; Li, J.L.; Zhang, X.Q.; Wei, H.; Cui, L.J. Effects of heat acclimation pretreatment on changes of membrane lipid peroxidation, antioxidant metabolites, and ultrastructure of chloroplasts in two cool-season turfgrass species under heat stress. Environ. Exp. Bot. 2006, 56, 274–285. [Google Scholar] [CrossRef]
- Irigoyen, J.J.; Einerich, D.W.; Sánchez-Díaz, M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol. Plant 1992, 84, 55–60. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Blakesley, R.W.B.; Boezi, J.A. A new staining technique for proteins in polyacrylamide gels using coomassie brilliant blue G250. Anal. Biochem. 1977, 82, 580–582. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999, 99, 138–148. [Google Scholar]
- Altenhoff, A.M.; Dessimoz, C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009, 5, e1000262. [Google Scholar] [CrossRef]
- Gao, F.; Wang, N.; Li, H.Y.; Liu, J.S.; Fu, C.X.; Xiao, Z.H.; Wei, C.X.; Lu, X.D.; Feng, J.C.; Zhou, Y.J. Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 2016, 6, 34601. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Jin, D.C.; Niu, Z.W. Improvement of PAML algorithm and application. In Proceedings of the Information Computing and Applications, Tangshan, China, 15–18 October 2010; Part 2. pp. 360–366. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- N’Guessan, A.; Brito, I.L.; Serohijos, A.W.R.; Shapiro, B.J. Mobile gene sequence evolution within individual human gut microbiomes is better explained by gene-specific than host-specific selective pressures. Genome Biol. Evol. 2021, 13, evab142. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Samarfard, S.; Eskandarzade, N.; Afsharifar, A.; Eskandari, M.H.; Niazi, A.; Izadpanah, K.; Karbanowicz, T.P. Comparative phylogenetic analysis of SARS-CoV-2 spike protein-possibility effect on virus spillover. Brief. Bioinform. 2021, 22, bbab144. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Tao, S.Q.; Cao, B.; Tian, C.M.; Liang, Y.M. Comparative transcriptome analysis and identification of candidate effectors in two related rust species (Gymnosporangium yamadae and Gymnosporangium asiaticum). BMC Genom. 2017, 18, 651. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, X.T.; Zhang, T. Evaluation of a hybrid approach using UBLAST and BLASTX for metagenomic sequences annotation of specific functional genes. PLoS ONE 2014, 9, e110947. [Google Scholar] [CrossRef]
- Wang, L.G.; Lam, T.T.Y.; Xu, S.B.; Dai, Z.H.; Zhou, L.; Feng, T.Z.; Guo, P.F.; Dunn, C.W.; Jones, B.; Bradley, T.; et al. Treeio: An R package for phylogenetic tree input and output with richly annotated and associated data. Mol. Biol. Evol. 2020, 37, 599–603. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual comparisons by ranking methods. Biometr. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Lu, Y.P.; Liu, H.C.; Chen, W.; Yao, J.; Huang, Y.Q.; Zhang, Y.; He, X.Y. Conservation planning of the genus Rhododendron in Northeast China based on current and future suitable habitat distributions. Biodivers. Conserv. 2021, 30, 673–697. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; You, J.; Li, J.; Zhao, W.; Xing, M.; Zhang, Y.; Ma, C.; Gong, Y.; Zhao, Y.; Wubuli, A.; et al. Stress Adaptation Phenomena of Rhododendron Species in Alpine Tundra and Timberline of Changbai Mountain: Physiological Traits and Molecular Evolution. Plants 2025, 14, 3528. https://doi.org/10.3390/plants14223528
Yang Z, You J, Li J, Zhao W, Xing M, Zhang Y, Ma C, Gong Y, Zhao Y, Wubuli A, et al. Stress Adaptation Phenomena of Rhododendron Species in Alpine Tundra and Timberline of Changbai Mountain: Physiological Traits and Molecular Evolution. Plants. 2025; 14(22):3528. https://doi.org/10.3390/plants14223528
Chicago/Turabian StyleYang, Zhongzan, Jian You, Jiangnan Li, Wei Zhao, Ming Xing, Yujiao Zhang, Cui Ma, Yuqiao Gong, Yueming Zhao, Alimu Wubuli, and et al. 2025. "Stress Adaptation Phenomena of Rhododendron Species in Alpine Tundra and Timberline of Changbai Mountain: Physiological Traits and Molecular Evolution" Plants 14, no. 22: 3528. https://doi.org/10.3390/plants14223528
APA StyleYang, Z., You, J., Li, J., Zhao, W., Xing, M., Zhang, Y., Ma, C., Gong, Y., Zhao, Y., Wubuli, A., & Chen, X. (2025). Stress Adaptation Phenomena of Rhododendron Species in Alpine Tundra and Timberline of Changbai Mountain: Physiological Traits and Molecular Evolution. Plants, 14(22), 3528. https://doi.org/10.3390/plants14223528





