Genome-Wide Identification and Expression Analysis of Mitochondrial Dicarboxylate Carriers (DICs) in Medicago Under Aluminum Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of the Dicarboxylate Carriers in Medicago
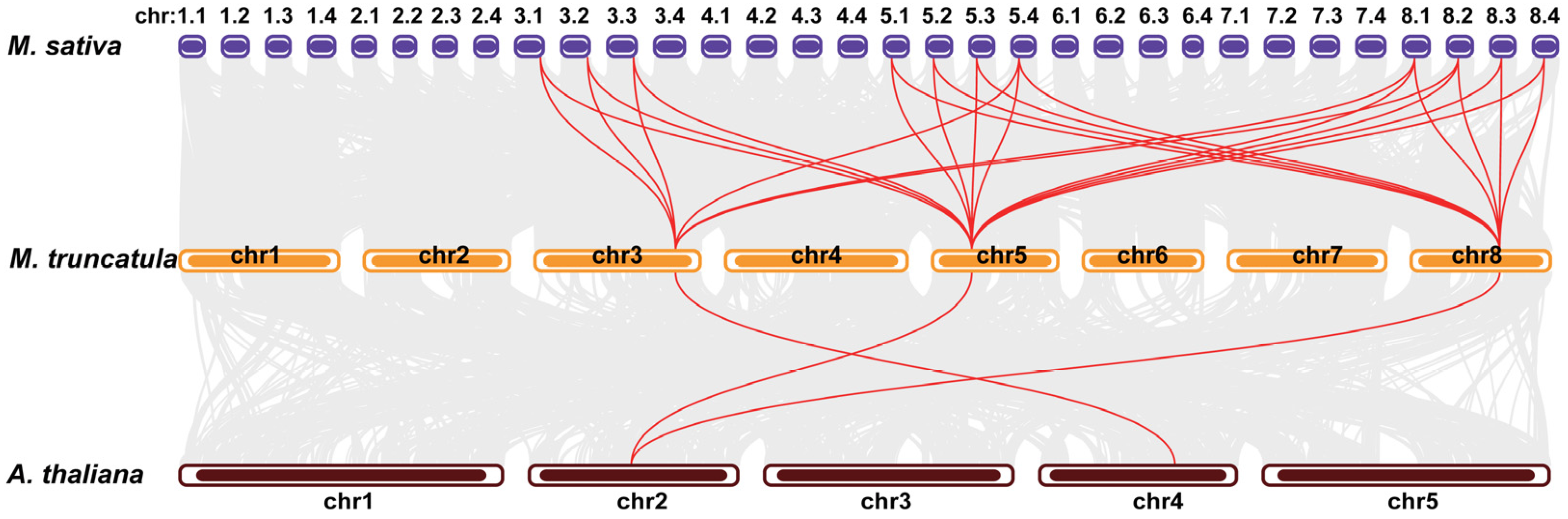
2.2. Synteny Analysis of DICs in the Genome of Medicago
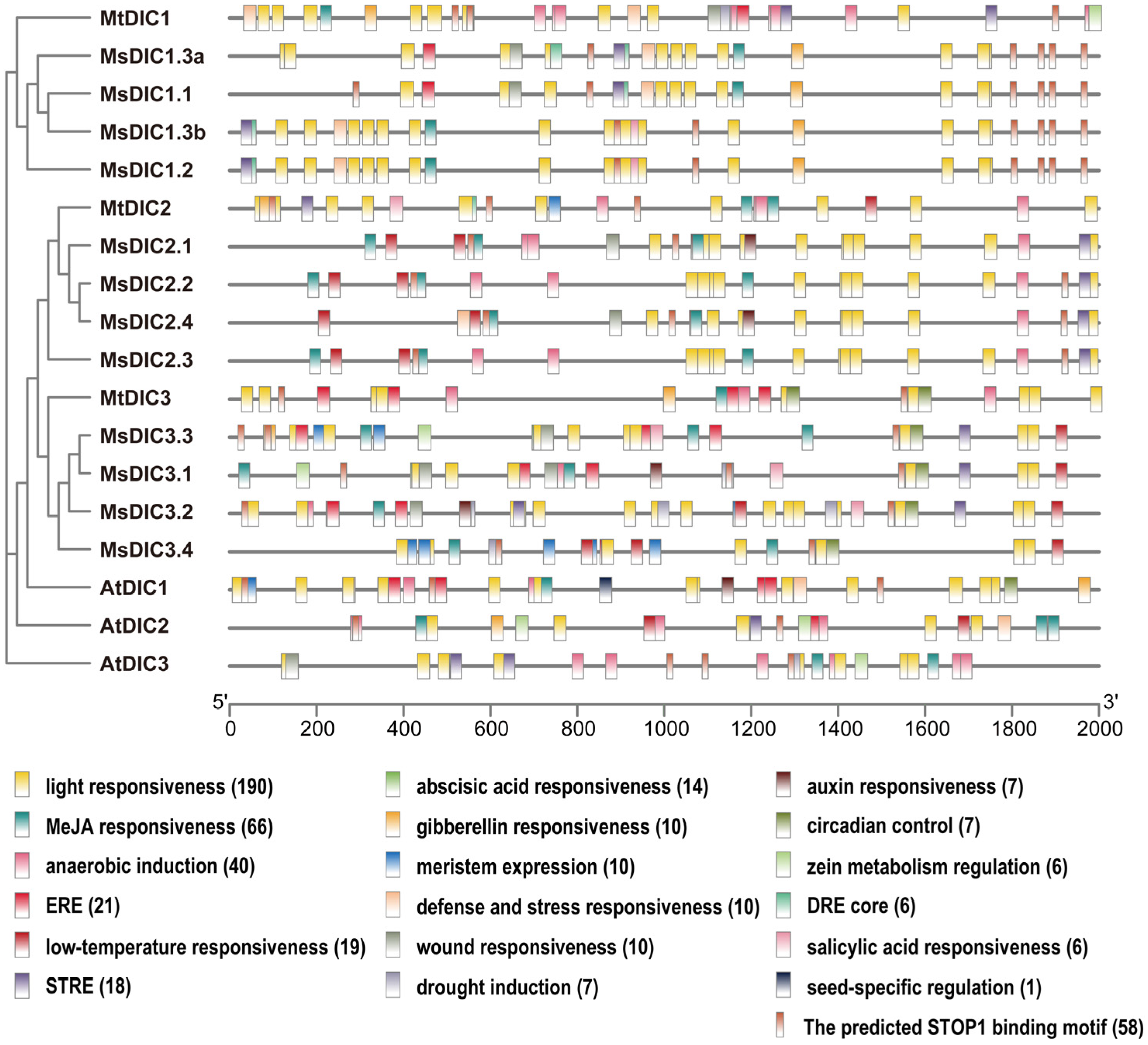
2.3. Protein Motifs, Gene Structures and Cis-Elements in DIC Promoters
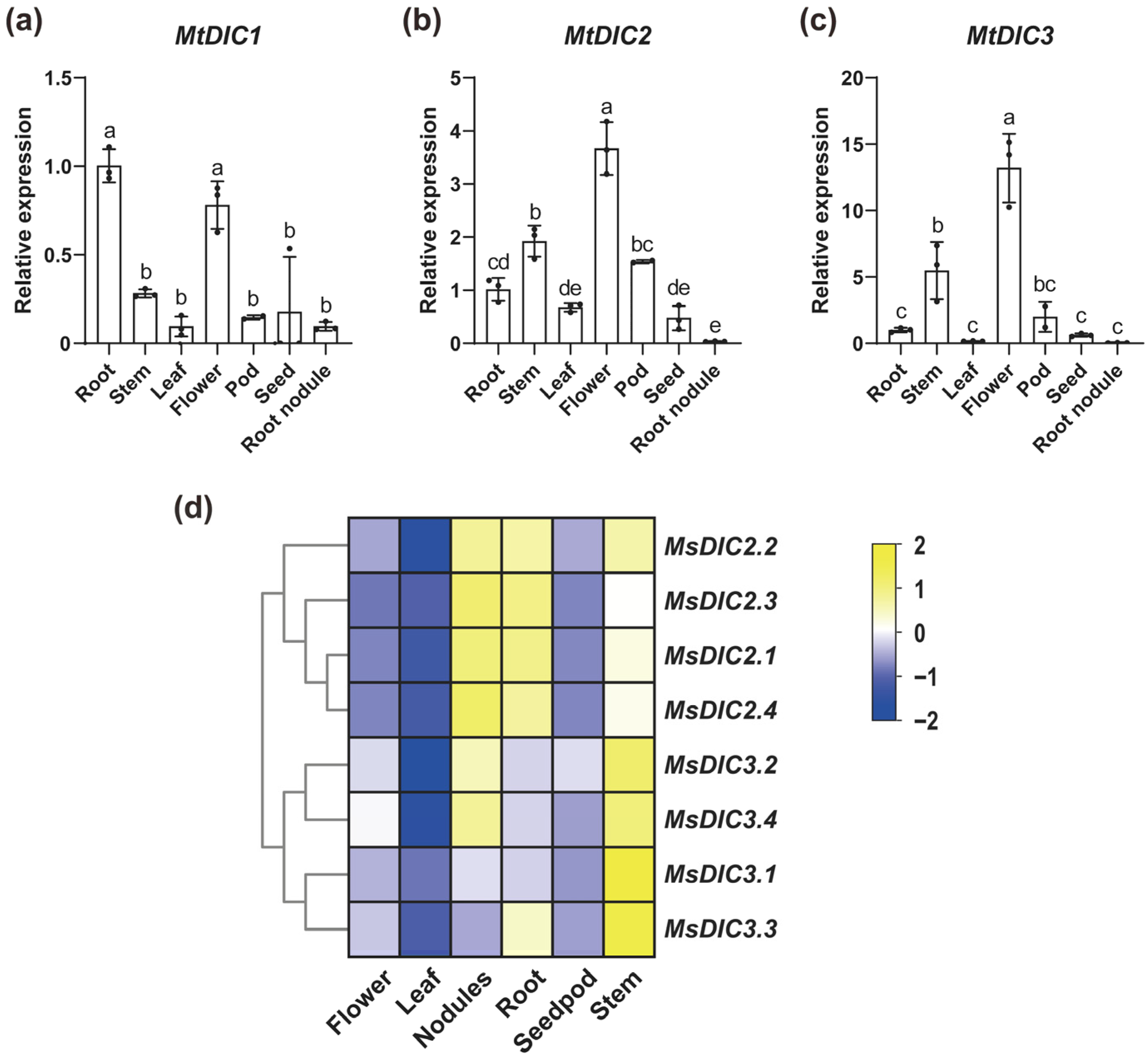
2.4. Expression Analysis of DIC Genes in Medicago Under Al Treatment
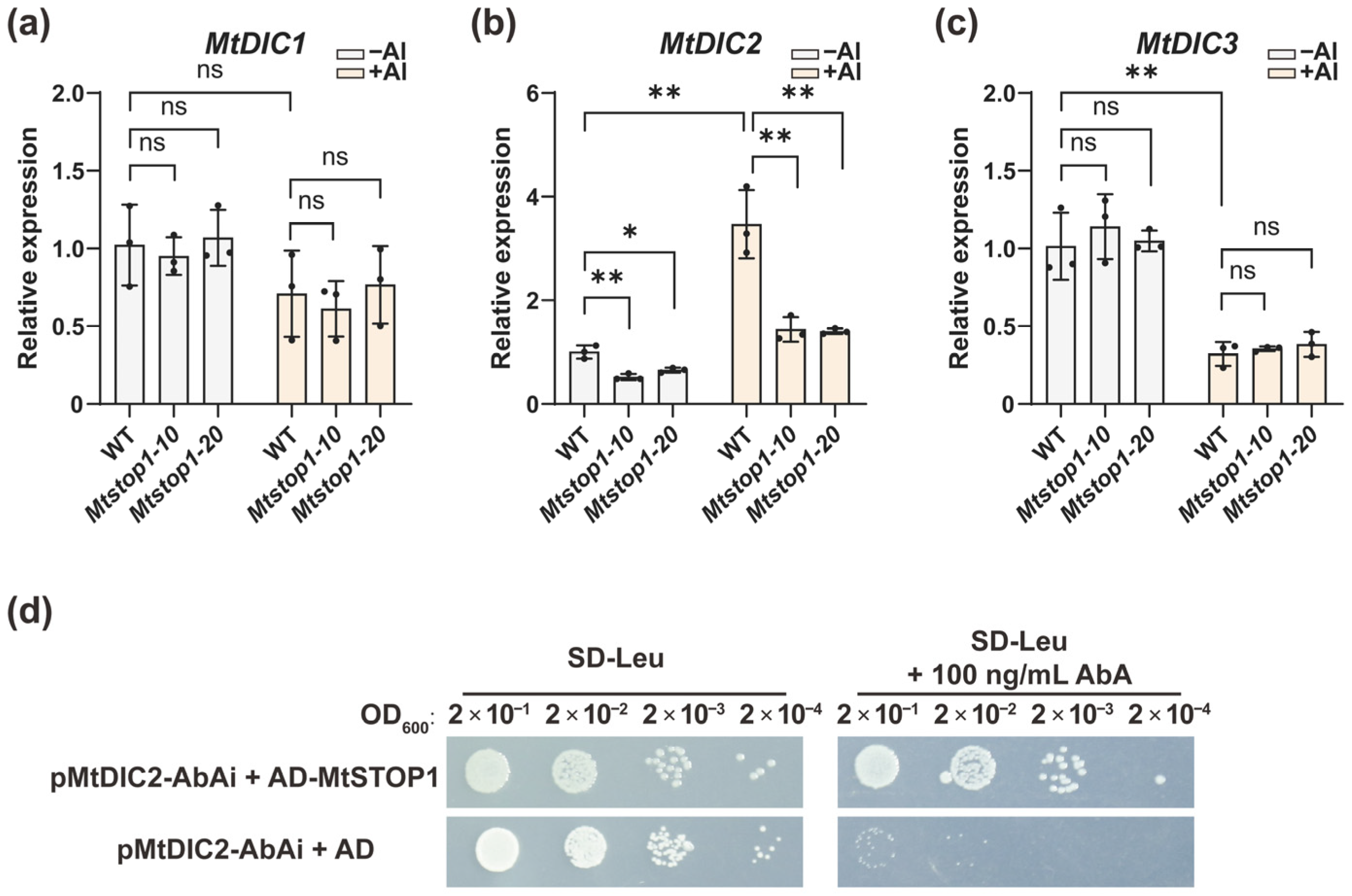
2.5. MtSTOP1 Upregulates MtDIC2 Expression in Roots Under Al Stress
3. Discussion
4. Materials and Methods
4.1. Identification of DIC Gene Family in Medicago
4.2. Phylogenetic and Synteny Analysis
4.3. Protein Motif Prediction and Gene Structure Survey
4.4. Cis-Element Profiling
4.5. Plant Materials and Growth Conditions
4.6. RNA-Seq Data Analysis
4.7. Real-Time Quantitative Reverse-Transcription PCR (qRT-PCR) Analysis
4.8. Yeast One-Hybrid Assay
4.9. Aluminum Speciation Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| chr | Chromosome |
| RNA-seq | RNA sequencing |
| qRT-PCR | Real-time quantitative reverse transcription PCR |
| FC | Fold change |
| FDR | False discovery rate |
| DAP-seq | DNA affinity purification sequencing |
References
- Giegé, P.; Heazlewood, J.L.; Roessner-Tunali, U.; Millar, A.H.; Fernie, A.R.; Leaver, C.J.; Sweetlove, L.J. Enzymes of Glycolysis Are Functionally Associated with the Mitochondrion in Arabidopsis Cells. Plant Cell 2003, 15, 2140–2151. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory Metabolism: Glycolysis, the TCA Cycle and Mitochondrial Electron Transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Palmieri, F. The Mitochondrial Transporter Family SLC25: Identification, Properties and Physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef]
- Fernie, A.R.; Cavalcanti, J.H.F.; Nunes-Nesi, A. Metabolic Roles of Plant Mitochondrial Carriers. Biomolecules 2020, 10, 1013. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Cavalcanti, J.H.F.; Fernie, A.R. Characterization of In Vivo Function(s) of Members of the Plant Mitochondrial Carrier Family. Biomolecules 2020, 10, 1226. [Google Scholar] [CrossRef]
- Picault, N.; Hodges, M.; Palmieri, L.; Palmieri, F. The Growing Family of Mitochondrial Carriers in Arabidopsis. Trends Plant Sci. 2004, 9, 138–146. [Google Scholar] [CrossRef]
- Palmieri, L.; Picault, N.; Arrigoni, R.; Besin, E.; Palmieri, F.; Hodges, M. Molecular Identification of Three Arabidopsis thaliana Mitochondrial Dicarboxylate Carrier Isoforms: Organ Distribution, Bacterial Expression, Reconstitution into Liposomes and Functional Characterization. Biochem. J. 2008, 410, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Elsässer, M.; Fuchs, P.; Fenske, R.; Schwarzländer, M.; Millar, A.H. The Versatility of Plant Organic Acid Metabolism in Leaves Is Underpinned by Mitochondrial Malate–Citrate Exchange. Plant Cell 2021, 33, 3700–3720. [Google Scholar] [CrossRef] [PubMed]
- Toleco, M.R.; Naake, T.; Zhang, Y.; Heazlewood, J.L.; Fernie, A.R. Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism. Plants 2020, 9, 117. [Google Scholar] [CrossRef]
- Barreto, P.; Koltun, A.; Nonato, J.; Yassitepe, J.; Maia, I.d.G.; Arruda, P. Metabolism and Signaling of Plant Mitochondria in Adaptation to Environmental Stresses. Int. J. Mol. Sci. 2022, 23, 11176. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-Batista, R.d.C.; Siqueira, J.A.; da Fonseca-Pereira, P.; Barreto, P.; Feitosa-Araujo, E.; Araújo, W.L.; Nunes-Nesi, A. Potential Roles of Mitochondrial Carrier Proteins in Plant Responses to Abiotic Stress. J. Exp. Bot. 2025, eraf032. [Google Scholar] [CrossRef]
- Barreto, P.; Arcuri, M.L.C.; Lima, R.P.M.; Marino, C.L.; Maia, I.G. Comprehensive In Silico Analysis and Transcriptional Profiles Highlight the Importance of Mitochondrial Dicarboxylate Carriers (DICs) on Hypoxia Response in Both Arabidopsis thaliana and Eucalyptus grandis. Plants 2022, 11, 181. [Google Scholar] [CrossRef]
- van Dongen, J.T.; Licausi, F. Oxygen Sensing and Signaling. Annu. Rev. Plant Biol. 2015, 66, 345–367. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.R.; Weits, D.A.; Feulner, C.F.J.; van Dongen, J.T. Oxygen Sensing and Integrative Stress Signaling in Plants. Plant Physiol. 2018, 176, 1131–1142. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular Mechanisms of Aluminum Toxicity and Resistance in Plants. Annu. Rev. Plant Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F. Syndrome of Aluminum Toxicity and Diversity of Aluminum Resistance in Higher Plants. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 2007; Volume 264, pp. 225–252. [Google Scholar]
- Poschenrieder, C.; Gunsé, B.; Corrales, I.; Barceló, J. A Glance into Aluminum Toxicity and Resistance in Plants. Sci. Total Environ. 2008, 400, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.A.; Good, A.G.; Taylor, G.J. Induction of Vacuolar ATPase and Mitochondrial ATP Synthase by Aluminum in an Aluminum-Resistant Cultivar of Wheat. Plant Physiol. 2001, 125, 2068–2077. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Aluminum Toxicity Is Associated with Mitochondrial Dysfunction and the Production of Reactive Oxygen Species in Plant Cells. Plant Physiol. 2002, 128, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium Tolerance in Plants and the Complexing Role of Organic Acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Katsuhara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A Wheat Gene Encoding an Aluminum-activated Malate Transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef]
- Magalhaes, J.V.; Liu, J.; Guimarães, C.T.; Lana, U.G.P.; Alves, V.M.C.; Wang, Y.-H.; Schaffert, R.E.; Hoekenga, O.A.; Piñeros, M.A.; Shaff, J.E.; et al. A Gene in the Multidrug and Toxic Compound Extrusion (MATE) Family Confers Aluminum Tolerance in Sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, Which Encodes a Malate Transporter, Is Identified as One of Several Genes Critical for Aluminum Tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc Finger Protein STOP1 Is Critical for Proton Tolerance in Arabidopsis and Coregulates a Key Gene in Aluminum Tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated Citrate and Malate Transporters from the MATE and ALMT Families Function Independently to Confer Arabidopsis Aluminum Tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef]
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 Encodes a Phloem-Localized ABC Transporter-like Protein That Is Required for Aluminum Tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A Bacterial-Type ABC Transporter Is Involved in Aluminum Tolerance in Rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Piñeros, M.A.; Li, X.; Yang, H.; Liu, Y.; Murphy, A.S.; Kochian, L.V.; Liu, D. An Arabidopsis ABC Transporter Mediates Phosphate Deficiency-Induced Remodeling of Root Architecture by Modulating Iron Homeostasis in Roots. Mol. Plant 2017, 10, 244–259. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Chen, M.; Li, L.; Wang, L.; Huang, C.F. The Cell Cycle Checkpoint Regulator ATR Is Required for Internal Aluminum Toxicity-Mediated Root Growth Inhibition in Arabidopsis. Front. Plant Sci. 2018, 9, 118. [Google Scholar] [CrossRef]
- Fan, N.; Li, X.; Xie, W.; Wei, X.; Fang, Q.; Xu, J.; Huang, C.F. Modulation of External and Internal Aluminum Resistance by ALS3-Dependent STAR1-Mediated Promotion of STOP1 Degradation. New Phytol. 2024, 244, 511–527. [Google Scholar] [CrossRef]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, T.; Sakurai, N.; Fujita, M.; Shinozaki, K.; Shibata, D.; Kobayashi, M.; et al. STOP1 Regulates Multiple Genes That Protect Arabidopsis from Proton and Aluminum Toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef]
- He, Q.; Jin, J.; Li, P.; Zhu, H.; Wang, Z.; Fan, W.; Yang, J.L. Involvement of SlSTOP1 Regulated SlFDH Expression in Aluminum Tolerance by Reducing NAD+ to NADH in the Tomato Root Apex. Plant J. 2023, 113, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Tokizawa, M.; Enomoto, T.; Ito, H.; Wu, L.; Kobayashi, Y.; Mora-Macías, J.; Armenta-Medina, D.; Iuchi, S.; Kobayashi, M.; Nomoto, M.; et al. High Affinity Promoter Binding of STOP1 Is Essential for Early Expression of Novel Aluminum-Induced Resistance Genes GDH1 and GDH2 in Arabidopsis. J. Exp. Bot. 2021, 72, 2769–2789. [Google Scholar] [CrossRef]
- Chandran, D.; Sharopova, N.; VandenBosch, K.A.; Garvin, D.F.; Samac, D.A. Physiological and Molecular Characterization of Aluminum Resistance in Medicago truncatula. BMC Plant Biol. 2008, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Zhang, J.; Xuan, W.; Xie, Y. Up-Regulation of Heme Oxygenase-1 Contributes to the Amelioration of Aluminum-Induced Oxidative Stress in Medicago sativa. J. Plant Physiol. 2013, 170, 1328–1336. [Google Scholar] [CrossRef]
- Jin, D.; Chen, J.; Kang, Y.; Yang, F.; Yu, D.; Liu, X.; Yan, C.; Guo, Z.; Zhang, Y. Genome-Wide Characterization, Transcriptome Profiling, and Functional Analysis of the ALMT Gene Family in Medicago for Aluminum Resistance. J. Plant Physiol. 2024, 297, 154262. [Google Scholar] [CrossRef]
- Jin, D.; Chen, J.; Yan, C.; Liu, X.; Lin, Y.; Li, Z.; Guo, Z.; Zhang, Y. The Zinc Finger Transcription Factor MtSTOP1 Modulates Aluminum Resistance and Low pH Tolerance in Medicago truncatula. J. Exp. Bot. 2025, 76, eraf112. [Google Scholar] [CrossRef]
- Wang, J.; Hou, Q.; Li, P.; Yang, L.; Sun, X.; Benedito, V.A.; Wen, J.; Chen, B.; Mysore, K.S.; Zhao, J. Diverse Functions of Multidrug and Toxin Extrusion (MATE) Transporters in Citric Acid Efflux and Metal Homeostasis in Medicago truncatula. Plant J. 2017, 90, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Lv, A.; Wen, W.; Fan, N.; Su, L.; Zhou, P.; An, Y. Dehydrin MsDHN1 Improves Aluminum Tolerance of Alfalfa (Medicago sativa L.) by Affecting Oxalate Exudation from Root Tips. Plant J. 2021, 108, 441–458. [Google Scholar] [CrossRef]
- Su, L.; Lv, A.; Wen, W.; Fan, N.; Li, J.; Gao, L.; Zhou, P.; An, Y. MsMYB741 Is Involved in Alfalfa Resistance to Aluminum Stress by Regulating Flavonoid Biosynthesis. Plant J. 2022, 112, 756–771. [Google Scholar] [CrossRef]
- Tesfaye, M.; Temple, S.J.; Allan, D.L.; Vance, C.P.; Samac, D.A. Overexpression of Malate Dehydrogenase in Transgenic Alfalfa Enhances Organic Acid Synthesis and Confers Tolerance to Aluminum. Plant Physiol. 2001, 127, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Barone, P.; Rosellini, D.; LaFayette, P.; Bouton, J.; Veronesi, F.; Parrott, W. Bacterial Citrate Synthase Expression and Soil Aluminum Tolerance in Transgenic Alfalfa. Plant Cell Rep. 2008, 27, 893–901. [Google Scholar] [CrossRef]
- Kang, Y.; Chen, L. Structural Basis for the Binding of DNP and Purine Nucleotides onto UCP1. Nature 2023, 620, 226–231. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L.; De Grassi, A.; Nunes-Nesi, A.; Fernie, A.R. Evolution, Structure and Function of Mitochondrial Carriers: A Review with New Insights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Monné, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial Carriers for Aspartate, Glutamate and Other Amino Acids: A Review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Miller, S.S.; Dornbusch, M.R.; Farmer, A.D.; Huertas, R.; Gutierrez-Gonzalez, J.J.; Young, N.D.; Samac, D.A.; Curtin, S.J. Alfalfa (Medicago sativa L.) Pho2 Mutant Plants Hyperaccumulate Phosphate. G3 Genes|Genomes|Genet. 2022, 12, jkac096. [Google Scholar] [CrossRef]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-Box Family Genes in Medicago truncatula: Key Elements in Response to Salt, Cold, and Drought Stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef]
- Ojeda-Rivera, J.O.; Oropeza-Aburto, A.; Herrera-Estrella, L. Dissection of Root Transcriptional Responses to Low pH, Aluminum Toxicity and Iron Excess Under Pi-Limiting Conditions in Arabidopsis Wild-Type and Stop1 Seedlings. Front. Plant Sci. 2020, 11, 01200. [Google Scholar] [CrossRef]
- Monné, M.; Daddabbo, L.; Gagneul, D.; Obata, T.; Hielscher, B.; Palmieri, L.; Miniero, D.V.; Fernie, A.R.; Weber, A.P.M.; Palmieri, F. Uncoupling Proteins 1 and 2 (UCP1 and UCP2) from Arabidopsis thaliana are Mitochondrial Transporters of Aspartate, Glutamate, and Dicarboxylates. J. Biol. Chem. 2018, 293, 4213–4227. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. Mitochondrial Carrier Proteins. FEBS Lett. 1994, 346, 48–54. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G.J.-M.; Brandolin, G. Structure of Mitochondrial ADP/ATP Carrier in Complex with Carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhu, Y.; Xie, W.; Ren, W.; Zhang, Y.; Zhang, H.; Dai, S.; Huang, C.F. H2O2 Negatively Regulates Aluminum Resistance via Oxidation and Degradation of the Transcription Factor STOP1. Plant Cell 2024, 36, 688–708. [Google Scholar] [CrossRef]
- Koyama, H.; Kawamura, A.; Kihara, T.; Hara, T.; Takita, E.; Shibata, D. Overexpression of Mitochondrial Citrate Synthase in Arabidopsis thaliana Improved Growth on a Phosphorus-Limited Soil. Plant Cell Physiol. 2000, 41, 1030–1037. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Lytovchenko, A.; Morgan, M.; Nunes-Nesi, A.; Taylor, N.L.; Baxter, C.J.; Eickmeier, I.; Fernie, A.R. Mitochondrial Uncoupling Protein Is Required for Efficient Photosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19587–19592. [Google Scholar] [CrossRef]
- Agrahari, R.K.; Kobayashi, Y.; Enomoto, T.; Miyachi, T.; Sakuma, M.; Fujita, M.; Ogata, T.; Fujita, Y.; Iuchi, S.; Kobayashi, M.; et al. STOP1-Regulated SMALL AUXIN UP RNA55 (SAUR55) Is Involved in Proton/Malate Co-Secretion for Al Tolerance in Arabidopsis. Plant Direct 2024, 8, e557. [Google Scholar] [CrossRef]
- Huang, C.F.; Ma, Y. Aluminum Resistance in Plants: A Critical Review Focusing on STOP1. Plant Commun. 2024, 101200. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.F. F-Box Protein RAE1 Regulates the Stability of the Aluminum-Resistance Transcription Factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef]
- Fang, Q.; Zhou, F.; Zhang, Y.; Singh, S.; Huang, C.F. Degradation of STOP1 Mediated by the F-box Proteins RAH1 and RAE1 Balances Aluminum Resistance and Plant Growth in Arabidopsis thaliana. Plant J. 2021, 106, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.; Huang, C.F. Mutation of HPR1 Encoding a Component of the THO/TREX Complex Reduces STOP1 Accumulation and Aluminium Resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193. [Google Scholar] [CrossRef]
- Zhu, Y.-F.; Guo, J.; Zhang, Y.; Huang, C.F. The THO/TREX Complex Component RAE2/TEX1 Is Involved in the Regulation of Aluminum Resistance and Low Phosphate Response in Arabidopsis. Front. Plant Sci. 2021, 12, 698443. [Google Scholar] [CrossRef]
- Zhou, F.; Singh, S.; Zhang, J.; Fang, Q.; Li, C.; Wang, J.; Zhao, C.; Wang, P.; Huang, C.F. The MEKK1-MKK1/2-MPK4 Cascade Phosphorylates and Stabilizes STOP1 to Confer Aluminum Resistance in Arabidopsis. Mol. Plant 2023, 16, 337–353. [Google Scholar] [CrossRef]
- Cao, H.; Cui, R.; Guo, H.; Xia, Q.; Zhang, J.; Liu, W.; Yang, Z.-B. Ca2+-Dependent Cytoplasmic and Nuclear Phosphorylation of STOP1 by CPK21 and CPK23 Confers ALMT1-Dependent Aluminum Resistance. Nat. Commun. 2025, 16, 5225. [Google Scholar] [CrossRef]
- Ma, Y.; Zheng, H.; Schmitz-Thom, I.; Wang, J.; Zhou, F.; Li, C.; Zhang, Y.; Cheng, Y.; Miki, D.; Kudla, J.; et al. CPK28-Mediated Ca2+ Signaling Regulates STOP1 Localization and Accumulation to Facilitate Plant Aluminum Resistance. Nat. Commun. 2025, 16, 5224. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; Van Den Burg, H.A.; Huang, C.F. Regulation of Aluminum Resistance in Arabidopsis Involves the SUMOylation of the Zinc Finger Transcription Factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef] [PubMed]
- Le Poder, L.; Mercier, C.; Février, L.; Duong, N.; David, P.; Pluchon, S.; Nussaume, L.; Desnos, T. Uncoupling Aluminum Toxicity from Aluminum Signals in the STOP1 Pathway. Front. Plant Sci. 2022, 13, 785791. [Google Scholar] [CrossRef] [PubMed]
- Godon, C.; Mercier, C.; Wang, X.; David, P.; Richaud, P.; Nussaume, L.; Liu, D.; Desnos, T. Under Phosphate Starvation Conditions, Fe and Al Trigger Accumulation of the Transcription Factor STOP1 in the Nucleus of Arabidopsis Root Cells. Plant J. 2019, 99, 937–949. [Google Scholar] [CrossRef]
- Enomoto, T.; Tokizawa, M.; Ito, H.; Iuchi, S.; Kobayashi, M.; Yamamoto, Y.Y.; Kobayashi, Y.; Koyama, H. STOP1 Regulates the Expression of HsfA2 and GDHs That Are Critical for Low-Oxygen Tolerance in Arabidopsis. J. Exp. Bot. 2019, 70, 3297–3311. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Enomoto, T.; Kobayashi, Y.; Watanabe, T.; Iuchi, S.; Kobayashi, M.; Sahoo, L.; Yamamoto, Y.Y.; Koyama, H. Sensitive to Proton Rhizotoxicity1 Regulates Salt and Drought Tolerance of Arabidopsis thaliana through Transcriptional Regulation of CIPK23. Plant Cell Physiol. 2019, 60, 2113–2126. [Google Scholar] [CrossRef]
- Li, X.; Tian, Y. STOP1 and STOP1-like Proteins, Key Transcription Factors to Cope with Acid Soil Syndrome. Front. Plant Sci. 2023, 14, 1200139. [Google Scholar] [CrossRef]
- Balzergue, C.; Dartevelle, T.; Godon, C.; Laugier, E.; Meisrimler, C.; Teulon, J.-M.; Creff, A.; Bissler, M.; Brouchoud, C.; Hagège, A.; et al. Low Phosphate Activates STOP1-ALMT1 to Rapidly Inhibit Root Cell Elongation. Nat. Commun. 2017, 8, 15300. [Google Scholar] [CrossRef]
- Ullah, F.; Saqib, S.; Zaman, W.; Khan, W.; Zhao, L.; Khan, A.; Khan, W.; Xiong, Y.-C. Mitigating Drought and Heavy Metal Stress in Maize Using Melatonin and Sodium Nitroprusside. Plant Soil 2025, 511, 1597–1619. [Google Scholar] [CrossRef]
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Brière, C.; Moreau, S.; Carrère, S.; Blein, T.; Jardinaud, M.-F.; Latrasse, D.; Zouine, M.; et al. Whole-Genome Landscape of Medicago truncatula Symbiotic Genes. Nat. Plants 2018, 4, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-Aware Chromosome-Level Genome Assembly and Efficient Transgene-Free Genome Editing for the Autotetraploid Cultivated Alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, C.; Liu, X.; Li, Z.; Lin, Y.; Guo, Z.; Zhang, Y. Genome-Wide Identification and Expression Analysis of Mitochondrial Dicarboxylate Carriers (DICs) in Medicago Under Aluminum Stress. Plants 2025, 14, 3250. https://doi.org/10.3390/plants14213250
Yan C, Liu X, Li Z, Lin Y, Guo Z, Zhang Y. Genome-Wide Identification and Expression Analysis of Mitochondrial Dicarboxylate Carriers (DICs) in Medicago Under Aluminum Stress. Plants. 2025; 14(21):3250. https://doi.org/10.3390/plants14213250
Chicago/Turabian StyleYan, Chengcheng, Xiaoqing Liu, Zhen Li, Yujie Lin, Zhenfei Guo, and Yang Zhang. 2025. "Genome-Wide Identification and Expression Analysis of Mitochondrial Dicarboxylate Carriers (DICs) in Medicago Under Aluminum Stress" Plants 14, no. 21: 3250. https://doi.org/10.3390/plants14213250
APA StyleYan, C., Liu, X., Li, Z., Lin, Y., Guo, Z., & Zhang, Y. (2025). Genome-Wide Identification and Expression Analysis of Mitochondrial Dicarboxylate Carriers (DICs) in Medicago Under Aluminum Stress. Plants, 14(21), 3250. https://doi.org/10.3390/plants14213250







