Genetic Diversity and Population Structure of Shanlan Upland Rice Germplasm Based on SSR Markers
Abstract
1. Introduction
2. Results and Analysis
2.1. Genetic Diversity of Shanlan Upland Rice
2.2. Genetic Diversity of Shanlan Upland Rice from Different Geographic Origins
2.3. Nei’s Genetic Distance Among Shanlan Upland Rice Populations from Different Geographical Origin
2.4. Analysis of Molecular Variance
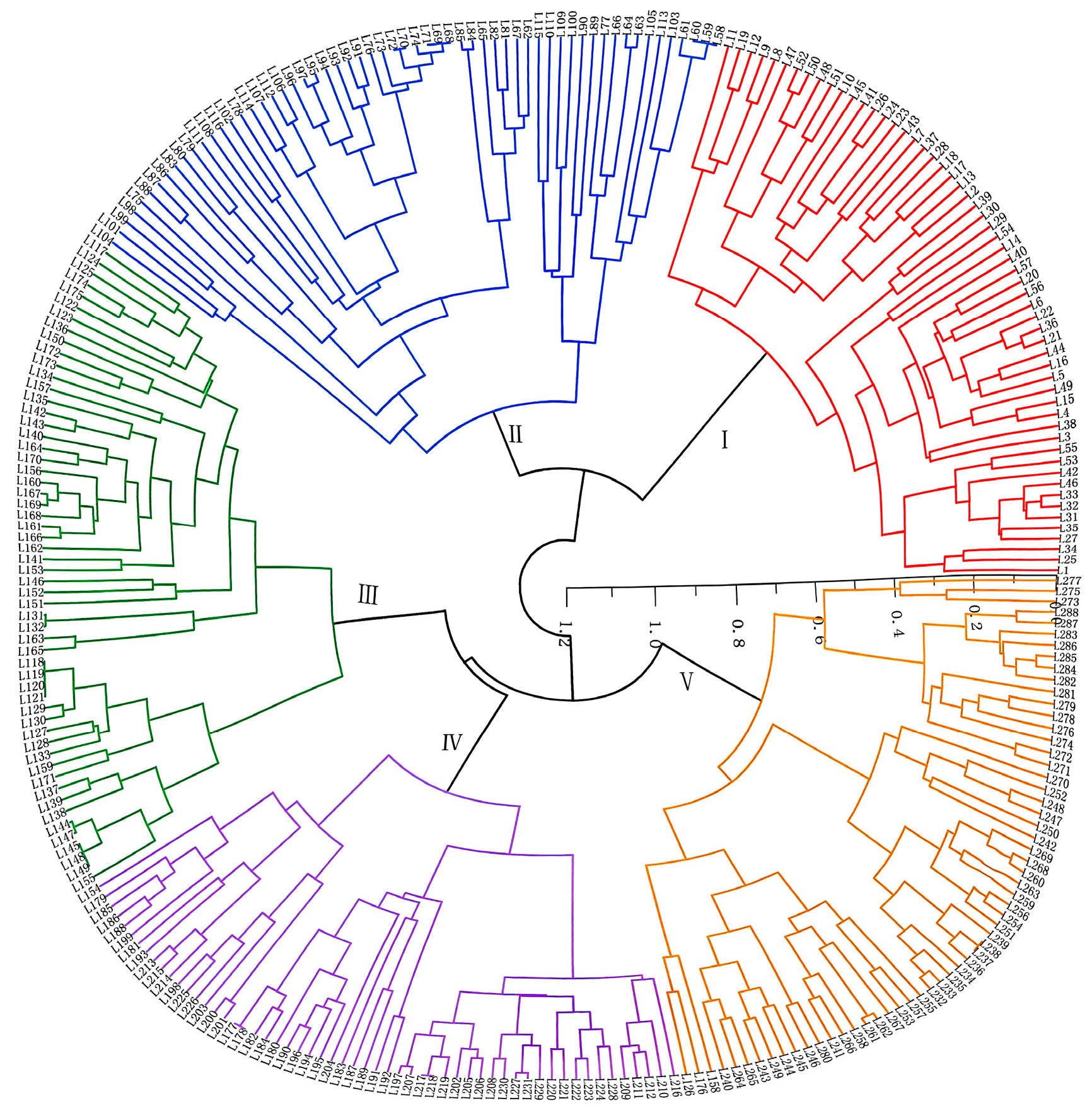
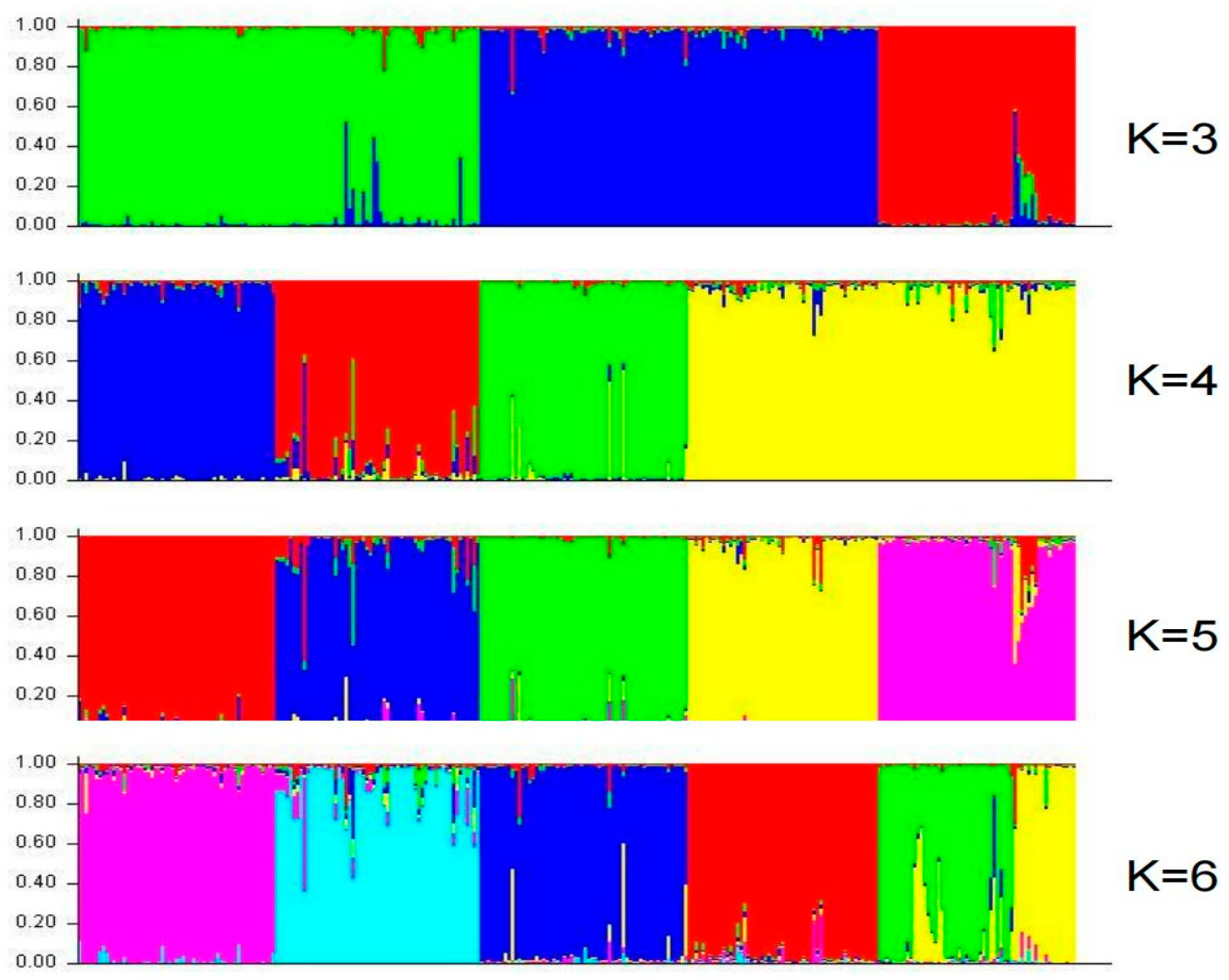
2.5. Cluster and Population Structure Analysis of Shanlan Upland Rice
3. Discussion
3.1. Moderate Genetic Diversity and Population Structure of Shanlan Upland Rice
3.2. Qiongzhong as a Potential Core Genetic Center and Patterns of Regional Differentiation
3.3. Phylogeography and Historical Dispersal Routes
3.4. The Genetically Distinct Dongfang Population: Implications for Conservation
3.5. Gene Introgression: A Double-Edged Sword
3.6. Limitations and Future Perspectives
4. Materials and Methods
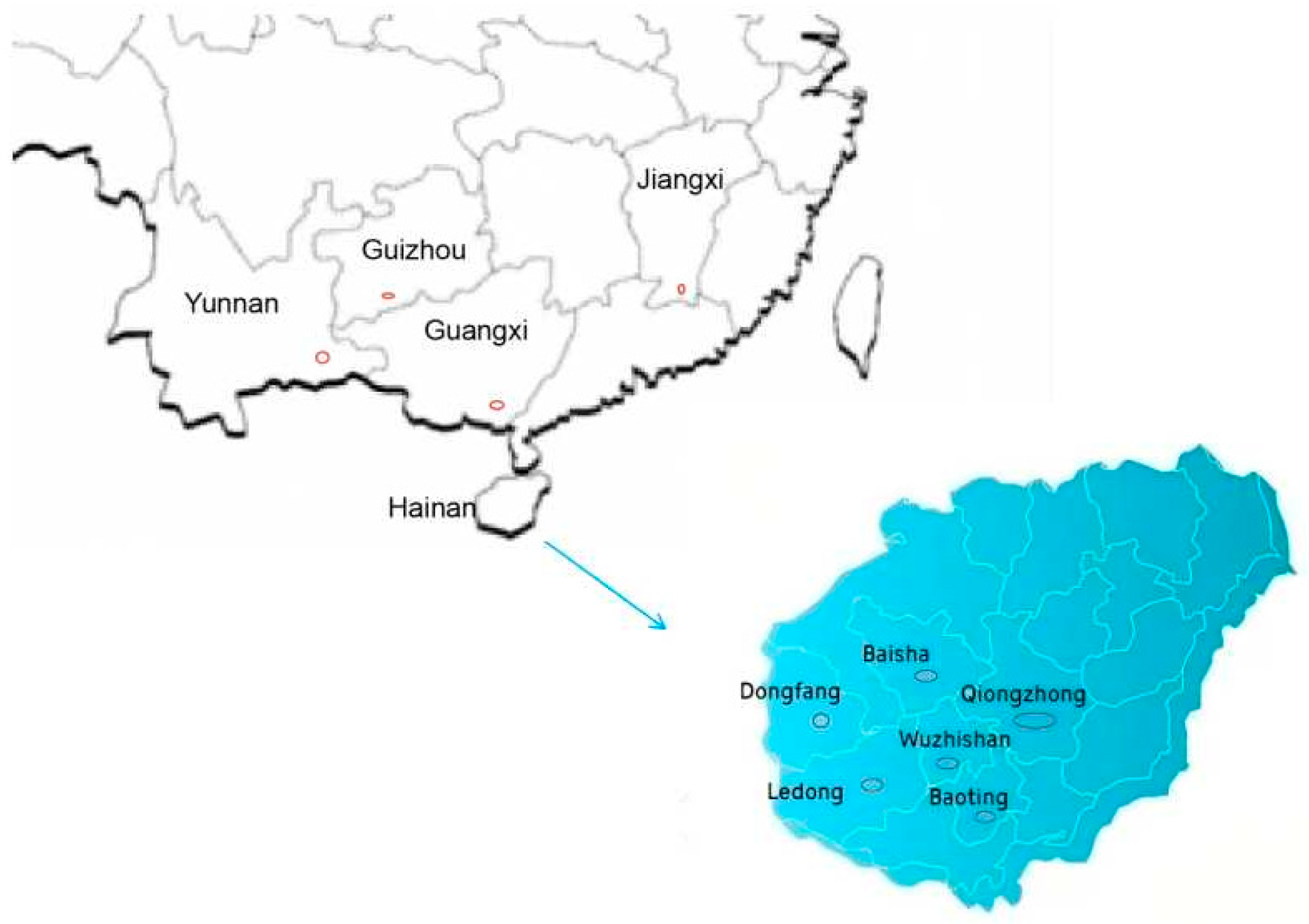
4.1. Plant Materials
4.2. DNA Extraction of Shanlan Upland Rice
4.3. Screening and Amplification of Molecular Marker Primers
4.4. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, X.; Liu, C.; Niu, X.; Wang, L.; Li, L.; Yuan, Q.; Pei, X. Research on lncRNA related to drought resistance of Shanlan upland rice. BMC Genom. 2022, 23, 336. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, X.; Niu, X.; Li, L.; Wang, L.; Liu, C.; Liu, J.; Yuan, Q.; Pei, X. Understanding the molecular mechanism of drought resistance in Shanlan upland rice by transcriptome and phenotype analyses. Int. J. Biol. Macromol. 2023, 231, 123387. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Yan, X.; Yang, G.; Zhong, Z.; Tang, L. Collection and survey of Shanlan upland rice resources in Hainan and recommendations for its development. Hybrid Rice. 2018, 33, 20–24, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Kumar, S.; Joshi, U.N.; Singh, V.; Singh, J.V.; Saini, M.L. Characterization of released and elite genotypes of guar [Cyamopsis tetragonoloba (L.) Taub.] from India proves unrelated to geographical origin. Genet. Resour. Crop Evol. 2013, 60, 2017–2032. [Google Scholar] [CrossRef]
- Li, R.; Huang, Y.; Yang, X.; Su, M.; Xiong, H.; Dai, Y.; Wu, W.; Pei, X.; Yuan, Q. Genetic Diversity and Relationship of Shanlan Upland Rice Were Revealed Based on 214 Upland Rice SSR Markers. Plants 2023, 12, 2876. [Google Scholar] [CrossRef]
- Vasumathy, S.; Alagu, M. SSR marker-based genetic diversity analysis and SNP haplotyping of genes associating abiotic and biotic stress tolerance, rice growth and development and yield across 93 rice landraces. Mol. Biol. Rep. 2021, 48, 5943–5953. [Google Scholar] [CrossRef]
- Nachimuthu, V.V.; Muthurajan, R.; Duraialaguraja, S.; Sivakami, R.; Pandian, B.A.; Ponniah, G.; Gunasekaran, K.; Swaminathan, M.; KK, S.; Sabariappan, R. Analysis of Population Structure and Genetic Diversity in Rice Germplasm Using SSR Markers: An Initiative Towards Association Mapping of Agronomic Traits in Oryza Sativa. Rice 2015, 8, 30. [Google Scholar] [CrossRef]
- Jasim, A.; Rafii, M.; Latif, M.; Sakimin, S.; Arolu, I.; Miah, G. Genetic Diversity of Aromatic Rice Germplasm Revealed By SSR Markers. Biomed. Res. Int. 2018, 2018, 7658032. [Google Scholar] [CrossRef]
- Temnykh, S.; Park, W.D.; Ayres, N.; Cartinhour, S.; Hauck, N.; Lipovich, L.; Cho, Y.G.; Ishii, T.; McCOUCH, S.R. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 697–712. [Google Scholar] [CrossRef]
- Ali, A.; Pan, Y.B.; Wang, Q.N.; Wang, J.D.; Chen, J.L.; Gao, S.J. Genetic diversity and population structure analysis of Saccharum and Erianthus genera using microsatellite (SSR) markers. Sci. Rep. 2019, 9, 395. [Google Scholar] [CrossRef]
- Diallo, S.; Badiane, F.A.; Kabkia, B.N.P.A.; Diédhiou, I.; Diouf, M.; Diouf, D. Genetic diversity and population structure of cowpea mutant collection using SSR and ISSR molecular markers. Sci. Rep. 2024, 14, 31833. [Google Scholar] [CrossRef]
- Zhu, Y.; Ma, T.; Lin, Y.; Peng, Y.; Huang, Y.; Jiang, J. SSR molecular marker developments and genetic diversity analysis of Zanthoxylum nitidum (Roxb.) DC. Sci. Rep. 2023, 13, 20767. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Yang, J.; Lv, Y.; Zhang, X.; Xia, C.; Zhao, H.; Wen, C. Genetic diversity analysis and variety identification using SSR and SNP markers in melon. BMC Plant Biol. 2023, 23, 39. [Google Scholar] [CrossRef]
- Mohan, L.; Sunita, M.; Sangeeta, B.; Samarjit, S.; Twahira, B.; Sudin, K. Molecular genetic diversity analysis using SSR marker amongst high solasodine content lines of Solanum khasianum C.B. Clarke, an industrially important plant. Ind. Crops Prod. Vol. 2022, 184, 115073. [Google Scholar] [CrossRef]
- Feng, S.; He, R.; Lu, J.; Jiang, M.; Shen, X.; Jiang, Y.; Wang, Z.; Wang, H. Development of SSR Markers and Assessment of Genetic Diversity in Medicinal Chrysanthemum morifolium Cultivars. Front. Genet. 2016, 7, 1664–8021. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yuan, N.; Wei, X.; Xue, D.; Yang, Q. The orogin and evolution of upland rice in Li ethnic communities in Hainan province. J. Plant Genet. Resour. 2013, 14, 202–207, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Yao, Y.; Wang, Y.; Zhuang, N.; Gao, H. Analysis on genetic diversity of Hainan upland rice local varieties and establishment of molecular ID with SRAP markers. Biotechnol. Bull. 2014, 11, 97–106, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Mills, L.; Allendorf, F. The One-Migrant-per-Generation Rule in Conservation and Management. Conserv. Biol. 1996, 10, 1509–1518. [Google Scholar] [CrossRef]
- Oliveira, M.M.; Sousa, L.B.; Reis, M.C.; Silva Junior, E.G.; Cardoso, D.B.O.; Hamawaki, O.T.; Nogueira, A.P.O. Evaluation of genetic diversity among soybean (Glycine max) genotypes using univariate and multivariate analysis. Genet. Mol. Res. 2017, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, S.; Kumar, N.; Rana, R.; Sharma, P.; Kumar, P.; Rani, M. Morpho-molecular genetic diversity and population structure analysis in garden pea (Pisum sativum L.) Genotypes Using Simple Sequence Repeat Markers. PLoS ONE 2022, 17, e0273499. [Google Scholar] [CrossRef]
- Zhai, L.; Tang, Q.; Zhou, B.; Zhou, S.; Wang, H.; Yun, Y.; Han, Y.; Wang, Q.; Yan, X.; Xing, F. Genetic diversity analysis and core germplasm construction of Oryza rufipogon Griff. Hainan J. Plant Genet. Resour. 2024, 25, 1624–1636, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Ma, M.; Lei, E.; Wang, T.; Meng, H.; Zhang, W.; Lu, B. Genetic Diversity and Association Mapping of Grain-Size Traits in Rice Landraces from the Honghe Hani Rice Terraces System in Yunnan Province. Plants 2023, 12, 1678. [Google Scholar] [CrossRef] [PubMed]
- Hour, A.L.; Hsieh, W.H.; Chang, S.H.; Wu, Y.P.; Chin, H.S.; Lin, Y.R. Genetic Diversity of Landraces and Improved Varieties of Rice (Oryza sativa L.) in Taiwan. Rice 2020, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yang, Y.; Guan, Y.; Xu, Z.; Wang, J.; Yun, Y.; Yan, X.; Tang, Q. Genetic Diversity of Shanlan Upland Rice (Oryza sativa L.) and Association Analysis of SSR Markers Linked to Agronomic Traits. BioMed. Res. Int. 2021, 7588652. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, X.; Zhang, F.; Wan, X.; Wang, C.; Liu, Y.; Xiao, B. Diversity of Rice Landraces Revealed by Molecular Markers and Phenotypic Traits. J. Plant Genet. Resour. 2023, 24, 636–647, (In Chinese with English Abstract). Available online: https://www.zwyczy.cn/zwyczyxb/article/abstract/202303005?st=article_issu (accessed on 1 May 2024).
- Zhang, H.; Sun, J.; Wang, M.; Liao, D.; Zeng, Y.; Shen, S.; Yu, P.; Mu, P.; Wang, X.; Li, Z. Genetic structure and phylogeography of rice landraces in Yunnan, China, revealed by SSR. Genome 2007, 50, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.; Septiningsih, E.; Suwardjo, F.; Santoso, T.; Silitonga, T.; McCouch, S. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2007, 114, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Cui, D.; Zhou, J.; Li, W.; Ma, X.; Han, B.; Guo, X.; Zhao, Z.; Han, L. Comparative analysis of genetic diversity of rice (Oryza sativa L.) varieties cultivated in different periods in China. Genet. Resour. Crop Evol. 2021, 68, 1439–1451. [Google Scholar] [CrossRef]
- Lin, H.; Wu, Y.; Hour, A.; Ho, S.; Wei, F.; Hsing, Y.; Lin, Y. Genetic diversity of rice germplasm used in Taiwan breeding programs. Bot. Stud. 2012, 53, 363–376. [Google Scholar]
- Zhang, Y.; He, Q.; Zhou, X.; Zheng, S.; Wang, Y.; Li, P.; Wang, Y. Genetic diversity and population structure of 93 rice cultivars (lines) (Oryza sativa Xian group) in Qinba in China by 3 types of genetic markers. BMC Genom. 2022, 23, 550. [Google Scholar] [CrossRef]
- Teshome, A.; Bryngelsson, T.; Dagne, K.; Geleta, M. Assessment of genetic diversity in Ethiopian field pea (Pisum sativum L.) accessions with newly developed EST-SSR markers. BMC Genet. 2015, 16, 102. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Z.; Zhang, J. Anthropological studies on Li nationality in Hainan island, China. Acta Anthropol. Sin. 1982, 1, 53–72, (In Chinese with English Abstract). Available online: https://www.anthropol.ac.cn/EN/abstract/abstract38.shtml (accessed on 21 April 2025).
- Lin, H. Chinese National History; Commercial Press: Beijing, China, 1984; p. 111, (In Chinese with English Abstract). [Google Scholar]
- Lv, S. Chinese National History; China Encyclopedia Press: Beijing, China, 1987; pp. 184–193, (In Chinese with English Abstract). [Google Scholar]
- Chen, G. A brief discussion on grain production in Hainan’s history. J. Guangdong Polytech. Norm. Univ. 2003, 1, 14–19, (In Chinese with English Abstract). Available online: https://kns.cnki.net/kcms2/article/abstract?v=i9XsIId0T12UY1tW586Y5CjSpVANWXUbTdVgqemVYIUGL-UU4mCLUgS1pij3Kl8ECh0buhXL1atpm51_KY9UrON2YFMKAKJrFI_leCA-pMzoD_djFaNGiq8mNQZ_VJZSb04D5NKIaWCOK-bJwADNncqtFfVy9Y0LxSl-OXdTX8oWjGqkE5k7Ug==&uniplatform=NZKPT&language=CHS (accessed on 21 April 2025).
- Adeyemo, O.; Menkir, A.; Melaku, G.; Omidiji, O. Genetic diversity assessment and relationship among tropicalyellow endosperm maize inbred lines using SSR markers. Maydica 2011, 56, 1703. Available online: https://www.semanticscholar.org/paper/Genetic-diversity-assessment-and-relationship-among-Adeyemo-Menkir/ddb588dd49ab4c3ff00f48ce1fcb8ff902e75a3e (accessed on 15 June 2025).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]


| Locus | Na | Ne | I | H | Ho | He | PIC (%) | Fis | Fit |
|---|---|---|---|---|---|---|---|---|---|
| RM7l | 2.800 | 2.035 | 0.758 | 0.246 | 0.196 | 0.490 | 70.00 | 0.600 | 0.663 |
| OSR28 | 2.000 | 1.913 | 0.668 | 0.333 | 0.043 | 0.475 | 93.33 | 0.909 | 0.912 |
| RM7102 | 2.800 | 2.300 | 0.883 | 0.321 | 0.577 | 0.550 | 90.00 | −0.050 | 0.130 |
| RM571 | 1.400 | 1.114 | 0.118 | 0.070 | 0.096 | 0.075 | 50.00 | −0.287 | −0.051 |
| RM336 | 1.640 | 1.483 | 0.369 | 0.257 | 0.099 | 0.081 | 64.00 | −0.216 | 0.012 |
| RM424 | 1.720 | 1.437 | 0.375 | 0.252 | 0.000 | 0.097 | 72.00 | 1.000 | 1.000 |
| RM85 | 1.500 | 1.430 | 0.326 | 0.230 | 0.019 | 0.100 | 50.00 | 0.812 | 0.893 |
| RM3331 | 2.000 | 1.857 | 0.652 | 0.454 | 0.058 | 0.459 | 100.00 | 0.873 | 0.880 |
| RM567 | 2.000 | 1.875 | 0.656 | 0.145 | 0.783 | 0.464 | 50.00 | −0.688 | −0.644 |
| RM551 | 1.300 | 1.081 | 0.099 | 0.059 | 0.004 | 0.056 | 30.00 | 0.937 | 0.987 |
| RM331 | 2.200 | 1.807 | 0.625 | 0.265 | 0.178 | 0.401 | 85.00 | 0.556 | 0.673 |
| RM17 | 2.000 | 1.784 | 0.626 | 0.135 | 0.021 | 0.435 | 100.00 | 0.952 | 0.959 |
| RM21 | 1.500 | 1.207 | 0.212 | 0.135 | 0.004 | 0.131 | 50.00 | 0.973 | 0.995 |
| RM2l9 | 2.000 | 1.718 | 0.580 | 0.397 | 0.004 | 0.397 | 100.00 | 0.991 | 0.992 |
| RM190 | 2.000 | 1.495 | 0.493 | 0.237 | 0.045 | 0.319 | 66.67 | 0.859 | 0.894 |
| RM231 | 1.200 | 1.021 | 0.139 | 0.019 | 0.200 | 0.100 | 20.00 | −1.000 | 0.778 |
| RM253 | 2.200 | 1.536 | 0.444 | 0.290 | 0.000 | 0.296 | 100.00 | 1.000 | 1.000 |
| RM267 | 2.000 | 1.565 | 0.533 | 0.351 | 0.007 | 0.351 | 100.00 | 0.980 | 0.981 |
| RM423 | 1.800 | 1.469 | 0.386 | 0.177 | 0.362 | 0.265 | 70.00 | −0.369 | 0.118 |
| RM481 | 2.800 | 2.093 | 0.816 | 0.330 | 0.011 | 0.497 | 93.33 | 0.979 | 0.984 |
| RM493 | 2.000 | 1.689 | 0.555 | 0.379 | 0.011 | 0.379 | 100.00 | 0.972 | 0.978 |
| mean | 1.946 | 1.615 | 0.491 | 0.242 | 0.129 | 0.306 | 74.02 | 0.513 | 0.673 |
| Sample Plot | Na | Ne | I | Ho | He | F | |
|---|---|---|---|---|---|---|---|
| Yunnan (China) | Mean | 1.667 | 1.471 | 0.373 | 0.079 | 0.246 | 0.609 |
| SE | 0.144 | 0.111 | 0.078 | 0.032 | 0.051 | 0.115 | |
| Baisha (China, Hainan) | Mean | 2.048 | 1.696 | 0.527 | 0.111 | 0.342 | 0.633 |
| SE | 0.129 | 0.126 | 0.073 | 0.045 | 0.048 | 0.108 | |
| Qiongzhong (China, Hainan) | Mean | 2.238 | 1.766 | 0.605 | 0.148 | 0.393 | 0.599 |
| SE | 0.095 | 0.109 | 0.052 | 0.051 | 0.035 | 0.117 | |
| Wuzhishan (China, Hainan) | Mean | 2.143 | 1.715 | 0.550 | 0.140 | 0.357 | 0.589 |
| SE | 0.125 | 0.118 | 0.070 | 0.049 | 0.046 | 0.116 | |
| Ledong (China, Hainan) | Mean | 2.048 | 1.779 | 0.589 | 0.196 | 0.390 | 0.507 |
| SE | 0.109 | 0.111 | 0.062 | 0.068 | 0.040 | 0.144 | |
| Dongfang (China, Hainan) | Mean | 1.762 | 1.426 | 0.389 | 0.123 | 0.253 | 0.543 |
| SE | 0.153 | 0.130 | 0.071 | 0.059 | 0.048 | 0.141 | |
| Guizhou (China) | Mean | 1.952 | 1.663 | 0.499 | 0.138 | 0.331 | 0.495 |
| SE | 0.129 | 0.121 | 0.073 | 0.060 | 0.049 | 0.142 | |
| Baoting (China, Hainan) | Mean | 1.905 | 1.528 | 0.473 | 0.151 | 0.305 | 0.526 |
| SE | 0.168 | 0.126 | 0.072 | 0.063 | 0.047 | 0.148 | |
| Jiangxi (China) | Mean | 1.286 | 1.198 | 0.224 | 0.111 | 0.151 | 0.309 |
| SE | 0.156 | 0.132 | 0.074 | 0.066 | 0.049 | 0.195 | |
| Guangxi (China) | Mean | 1.333 | 1.237 | 0.230 | 0.127 | 0.159 | 0.200 |
| SE | 0.126 | 0.106 | 0.067 | 0.067 | 0.047 | 0.198 | |
| Total | Mean | 1.838 | 1.548 | 0.446 | 0.132 | 0.293 | 0.532 |
| SE | 0.047 | 0.039 | 0.023 | 0.018 | 0.015 | 0.043 |
| Yunnan (China) | Baisha (China, Hainan) | Qiongzhong (China, Hainan) | Wuzhishan (China, Hainan) | Ledong (China, Hainan) | Dongfang (China, Hainan) | Guizhou (China) | Baoting (China, Hainan) | Jiangxi (China) | Guangxi (China) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Yunnan (China) | - | 0.069 | 0.112 | 0.134 | 0.125 | 0.284 | 0.272 | 0.201 | 0.241 | 0.329 |
| Baisha (China, Hainan) | 0.076 | - | 0.038 | 0.057 | 0.127 | 0.183 | 0.212 | 0.151 | 0.219 | 0.209 |
| Qiongzhong (China, Hainan) | 0.099 | 0.034 | - | 0.016 | 0.040 | 0.163 | 0.114 | 0.093 | 0.169 | 0.190 |
| Wuzhishan (China, Hainan) | 0.120 | 0.052 | 0.019 | - | 0.047 | 0.177 | 0.090 | 0.070 | 0.139 | 0.165 |
| Ledong (China, Hainan) | 0.165 | 0.096 | 0.033 | 0.039 | - | 0.176 | 0.086 | 0.103 | 0.189 | 0.274 |
| Dongfang (China, Hainan) | 0.245 | 0.165 | 0.134 | 0.145 | 0.150 | - | 0.345 | 0.212 | 0.285 | 0.182 |
| Guizhou (China) | 0.214 | 0.160 | 0.086 | 0.075 | 0.072 | 0.260 | - | 0.120 | 0.148 | 0.277 |
| Baoting (China, Hainan) | 0.192 | 0.140 | 0.091 | 0.076 | 0.099 | 0.212 | 0.122 | - | 0.211 | 0.139 |
| Jiangxi (China) | 0.307 | 0.248 | 0.183 | 0.175 | 0.209 | 0.322 | 0.212 | 0.273 | - | 0.255 |
| Guangxi (China) | 0.344 | 0.223 | 0.183 | 0.173 | 0.241 | 0.245 | 0.264 | 0.203 | 0.426 | - |
| Source | DF | SS | MS | Est. Var. | PV (%) | PhiPT |
|---|---|---|---|---|---|---|
| Among Pops | 9 | 435.303 | 48.367 | 1.419 | 8.390 | |
| In Pops | 278 | 4307.269 | 15.494 | 15.494 | 91.609 | |
| Total | 288 | 4742.573 | 16.913 | 100% | 0.084 (p < 0.01) |
| Origin | Total Number |
|---|---|
| Jiangxi | 3 |
| Guizhou | 14 |
| Guangxi | 3 |
| Yunnan | 3 |
| Baisha (Hainan) | 32 |
| Dongfang (Hainan) | 12 |
| Qiongzhong (Hainan) | 137 |
| Wuzhishan (Hainan) | 36 |
| Ledong (Hainan) | 36 |
| Baoting (Hainan) | 12 |
| Total | 288 |
| No. | Primer | Forward Sequences (5′-3′) | Invert the Sequence (5′-3′) |
|---|---|---|---|
| 1 | RM7l | agatccatccctgtggagag | gcgaactcgcgttgtaatc |
| 2 | OSR28 | agcagctatagcttagctgg | actgcacatgagcagagaca |
| 3 | RM7102 | taggagtgtttagagtgcca | tcggtttgcttatacatcag |
| 4 | RM571 | ggaggtgaaagcgaatcatg | cctgctgctctttcatcagc |
| 5 | RM336 | cttacagagaaacggcatcg | gctggtttgtttcaggttcg |
| 6 | RM424 | tttgtggctcaccagttgag | tggcgcattcatgtcatc |
| 7 | RM85 | ccaaagatgaaacctggattg | gcacaaggtgagcagtcc |
| 8 | RM3331 | cctcctccatgagctaatgc | aggaggagcggatttctctc |
| 9 | RM567 | atcagggaaatcctgaaggg | ggaaggagcaatcaccactg |
| 10 | RM551 | agcccagactagcatgattg | gaaggcgagaaggatcacag |
| 11 | RM331 | gaaccagaggacaaaaatgc | catcatacatttgcagccag |
| 12 | RM17 | tgccctgttattttcttctctc | ggtgatcctttcccatttca |
| 13 | RM21 | acagtattccgtaggcacgg | gctccatgagggtggtagag |
| 14 | RM2l9 | cgtcggatgatgtaaagcct | catatcggcattcgcctg |
| 15 | RM190 | ctttgtctatctcaagacac | ttgcagatgttcttcctgatg |
| 16 | RM231 | ccagattatttcctgaggtc | cacttgcatagttctgcattg |
| 17 | RM253 | tccttcaagagtgcaaaacc | gcattgtcatgtcgaagcc |
| 18 | RM267 | tgcagacatagagaaggaagtg | agcaacagcacaacttgatg |
| 19 | RM423 | agcacccatgccttatgttg | cctttttcagtagccctccc |
| 20 | RM481 | tagctagccgattgaatggc | ctccacctcctatgttgttg |
| 21 | RM493 | tagctccaacaggatcgacc | gtacgtaaacgcggaaggtg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, L.; Zhao, M.; Yan, X.; Li, Y.; Xiao, X.; Wang, Q.; Wang, H.; Zhou, B.; Yun, Y.; Xing, F.; et al. Genetic Diversity and Population Structure of Shanlan Upland Rice Germplasm Based on SSR Markers. Plants 2025, 14, 3233. https://doi.org/10.3390/plants14203233
Zhai L, Zhao M, Yan X, Li Y, Xiao X, Wang Q, Wang H, Zhou B, Yun Y, Xing F, et al. Genetic Diversity and Population Structure of Shanlan Upland Rice Germplasm Based on SSR Markers. Plants. 2025; 14(20):3233. https://doi.org/10.3390/plants14203233
Chicago/Turabian StyleZhai, Linan, Mingchao Zhao, Xiaowei Yan, Yapeng Li, Xiaorong Xiao, Qingyu Wang, Huijian Wang, Bangji Zhou, Yong Yun, Funeng Xing, and et al. 2025. "Genetic Diversity and Population Structure of Shanlan Upland Rice Germplasm Based on SSR Markers" Plants 14, no. 20: 3233. https://doi.org/10.3390/plants14203233
APA StyleZhai, L., Zhao, M., Yan, X., Li, Y., Xiao, X., Wang, Q., Wang, H., Zhou, B., Yun, Y., Xing, F., & Tang, Q. (2025). Genetic Diversity and Population Structure of Shanlan Upland Rice Germplasm Based on SSR Markers. Plants, 14(20), 3233. https://doi.org/10.3390/plants14203233






