Genome-Wide Identification, Drought-Responsive Expression, and EAR-Mediated Regulatory Network Construction of TOPLESS Genes in Populus ussuriensis Kom.
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of the TPL/TPR Genes in P. ussuriensis
2.2. Physicochemical Properties and Secondary Structures of TPL/TPR Proteins in P. ussuriensis
2.3. Gene Structure, Conserved Domains, and Cis-Regulatory Elements
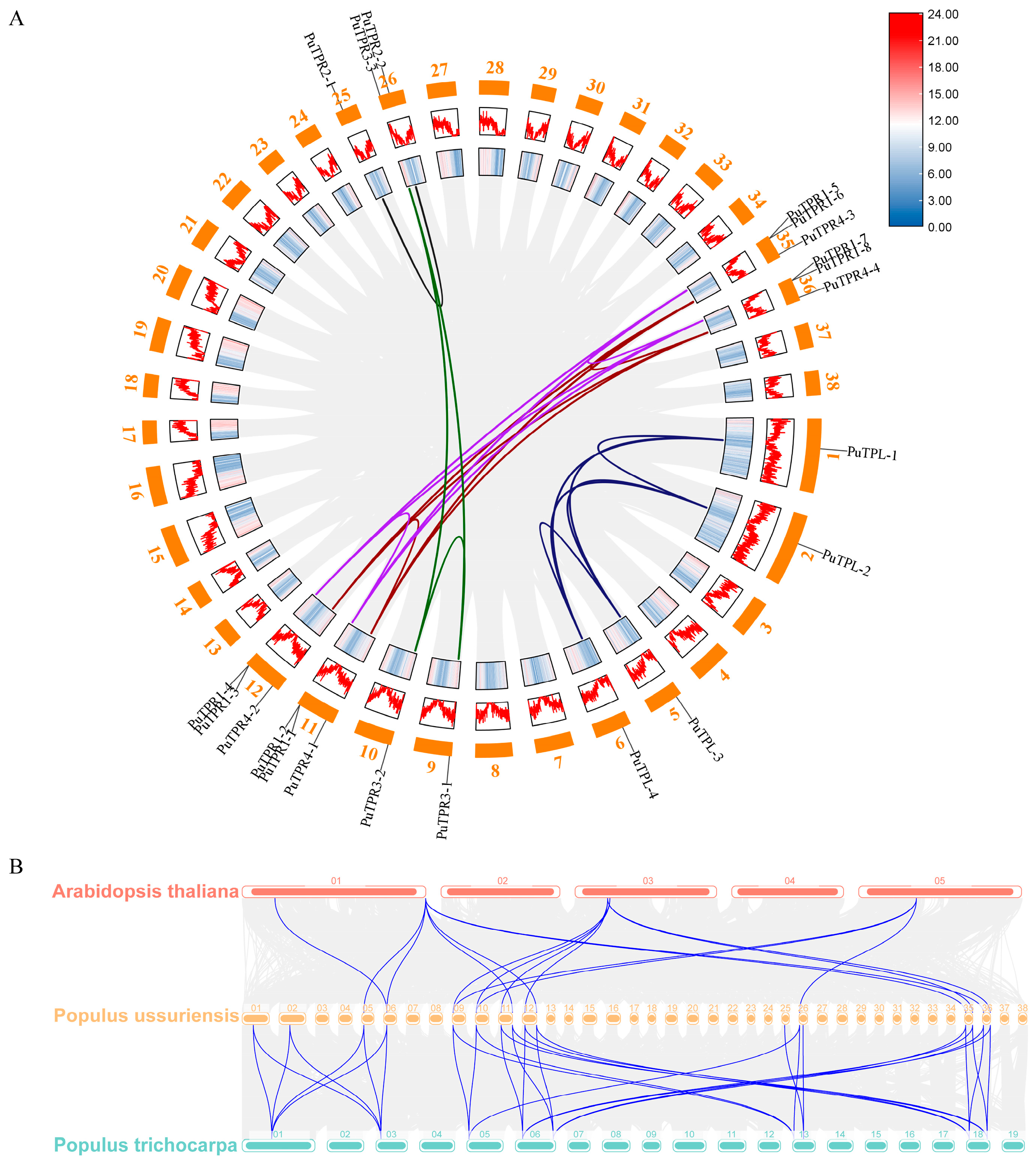
2.4. Chromosomal Localization and Synteny Analysis of PuTPLs
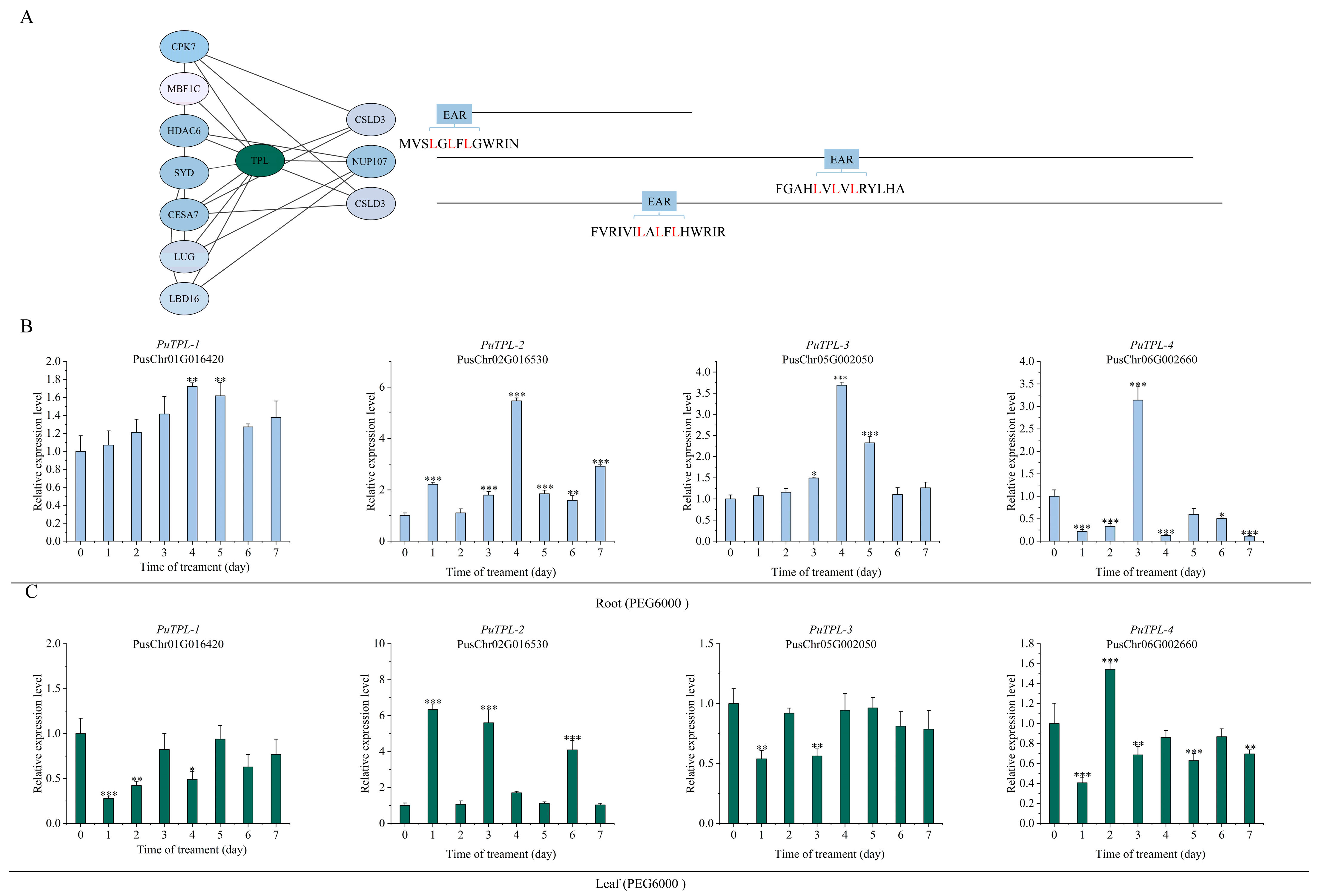
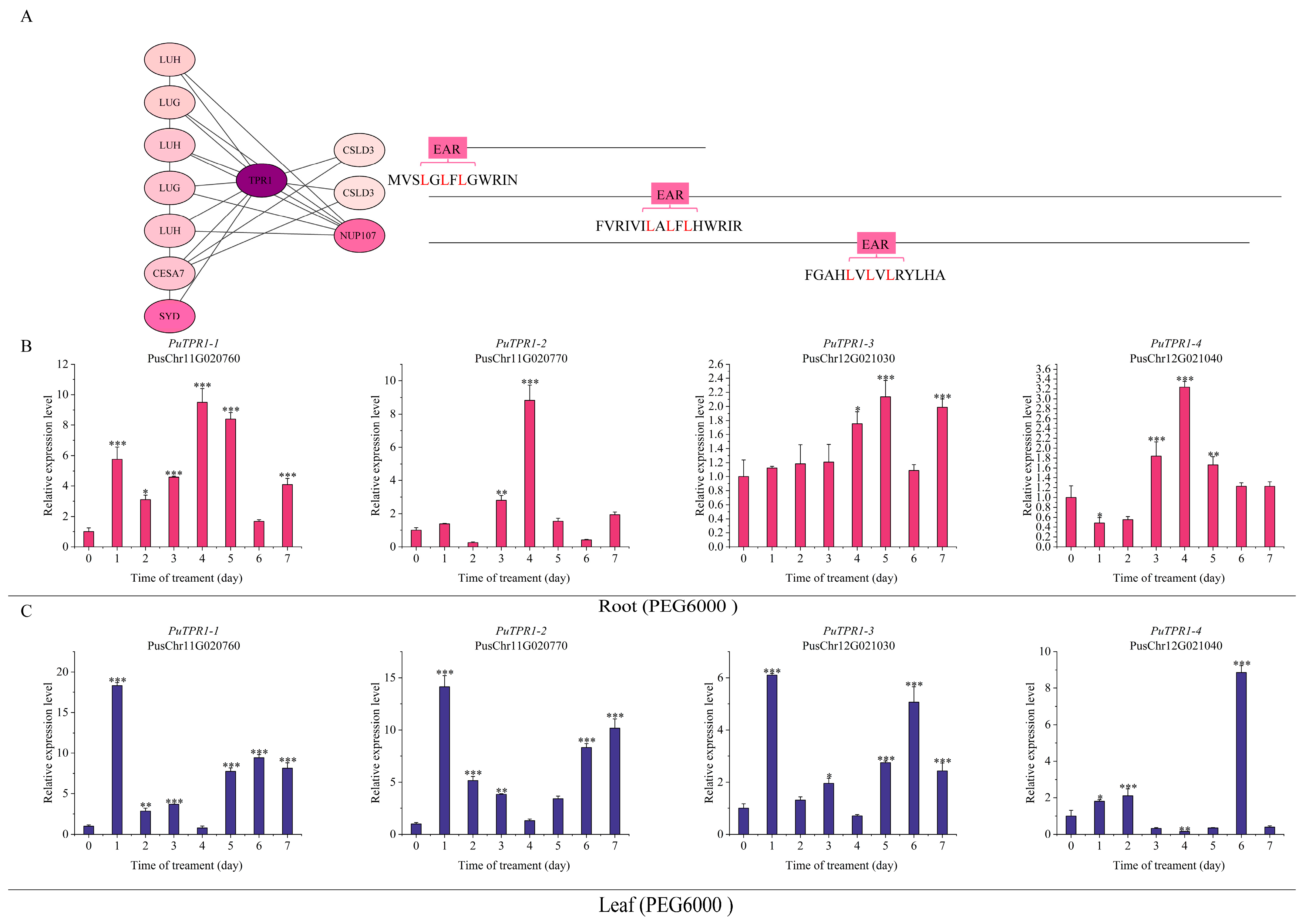
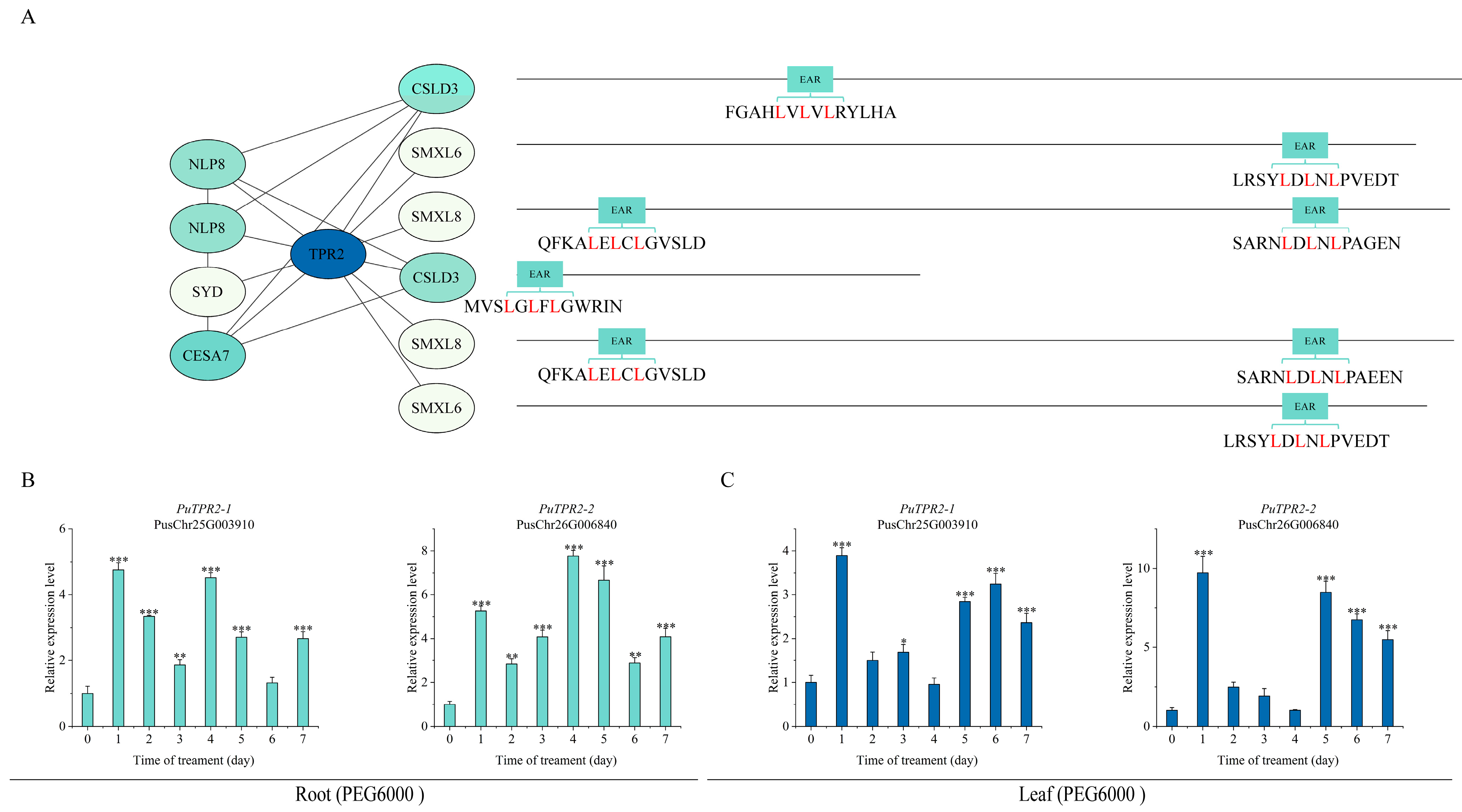
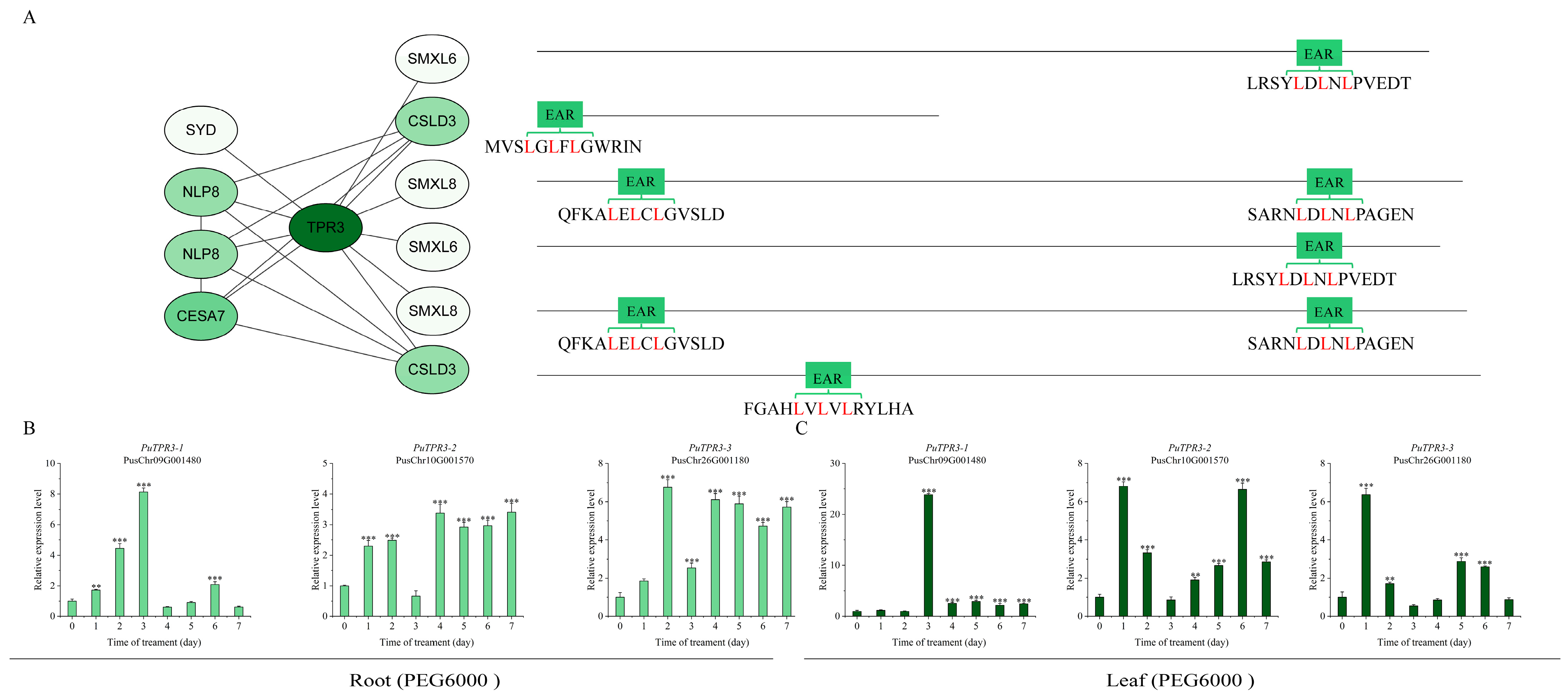
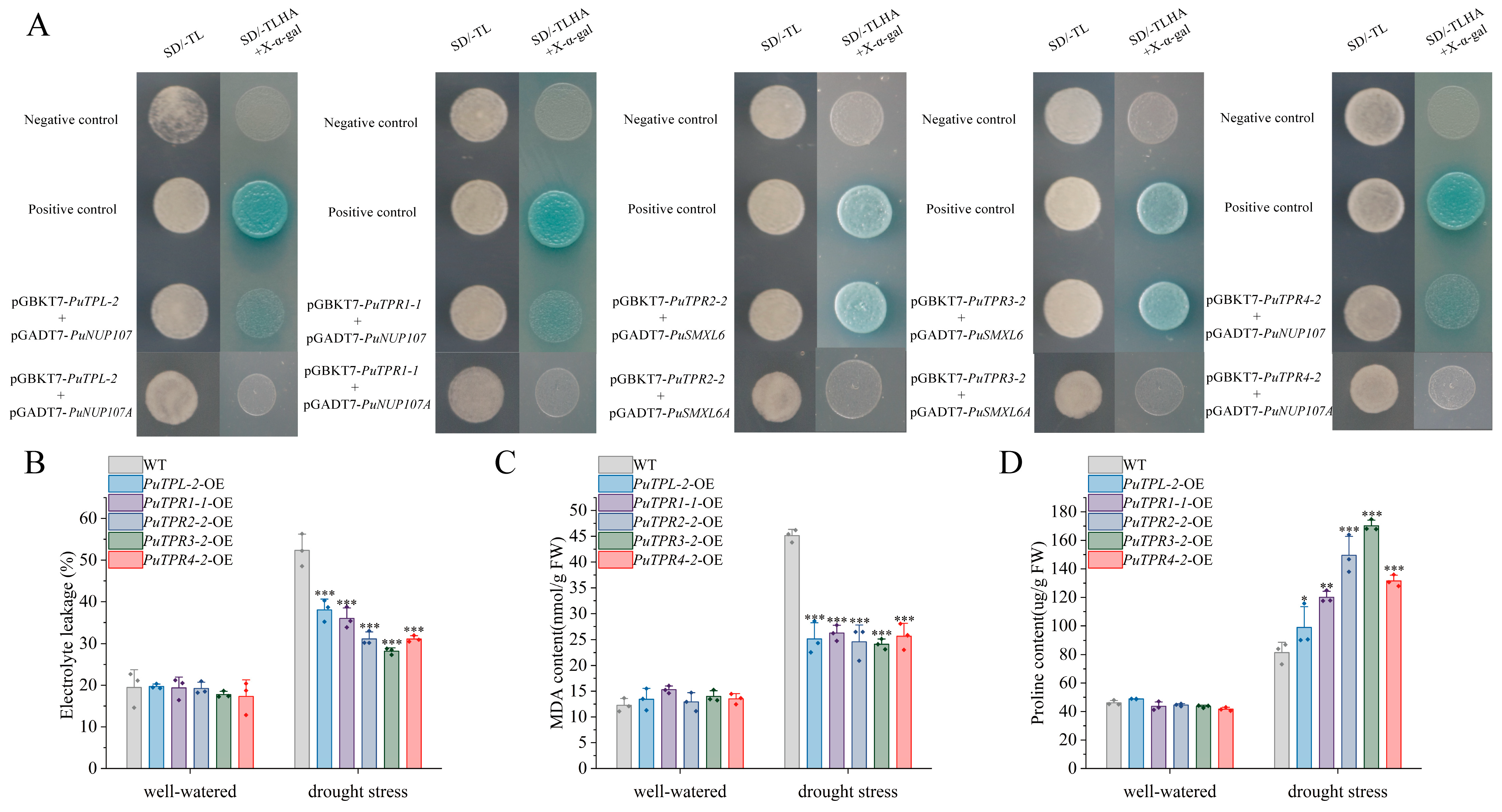
2.5. The TPL/TPR Co-Repressor Interaction Network Mediated by EAR Motifs
2.6. Expression Patterns of PuTPLs Under Drought Stress
2.7. Physiological Analysis of Drought Tolerance in PuTPLs-Overexpressing (OE) Lines
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization
4.2. Physicochemical Characterization and Secondary Structure Prediction
4.3. Phylogeny Reconstruction and Multiple Sequence Alignment
4.4. Analysis of Conserved Domains, Cis-Regulatory Elements and Gene Structure
4.5. Chromosomal Localization and Collinear Relationship
4.6. Plant Materials and Stress Treatment
4.7. Total RNA Extraction, Reverse Transcription, and RT-qPCR Analysis
4.8. Interaction Network Prediction
4.9. Cloning and Vector Construction
4.10. Yeast Two-Hybrid Assays
4.11. Transient Expression and Physiological Index Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Song, Q.; Kong, L.; Yang, J.; Lin, M.; Zhang, Y.; Yang, X.; Wang, X.; Zhao, Z.; Zhang, M.; Pan, J.; et al. The Transcription Factor PtoMYB142 Enhances Drought Tolerance in Populus tomentosa by Regulating Gibberellin Catabolism. Plant J. 2024, 118, 42–57. [Google Scholar] [CrossRef]
- Xu, W.; Tang, W.; Wang, C.; Ge, L.; Sun, J.; Qi, X.; He, Z.; Zhou, Y.; Chen, J.; Xu, Z.; et al. SiMYB56 Confers Drought Stress Tolerance in Transgenic Rice by Regulating Lignin Biosynthesis and ABA Signaling Pathway. Front. Plant Sci. 2020, 11, 785. [Google Scholar] [CrossRef]
- Zhao, H.; Fu, Y.; Lv, W.; Zhang, X.; Li, J.; Yang, D.; Shi, L.; Wang, H.; Li, W.; Huang, H.; et al. PuUBL5-Mediated ZINC FINGER PROTEIN 1 Stability Is Critical for Root Development under Drought Stress in Populus ussuriensis. Plant Physiol. 2025, 198, kiaf181. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhao, Y.; Zhao, Z.; Liu, W.; Jiang, C.; Li, J.; Zhang, Z.; Zhang, H.; Zhang, Y.; Wang, X.; et al. RRS1 Shapes Robust Root System to Enhance Drought Resistance in Rice. New Phytol. 2023, 238, 1146–1162. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Bao, X.; Sun, H.; Chen, J.; Zhu, L.; Zhang, J.; Zhang, H.; Zhang, Y.; Zhang, K.; Bai, Z.; et al. The Crucial Role of Lateral Root Angle in Enhancing Drought Resilience in Cotton. Front. Plant Sci. 2024, 15, 1358163. [Google Scholar] [CrossRef]
- Oh, J.E.; Kwon, Y.; Kim, J.H.; Noh, H.; Hong, S.-W.; Lee, H. A Dual Role for MYB60 in Stomatal Regulation and Root Growth of Arabidopsis Thaliana under Drought Stress. Plant Mol. Biol. 2011, 77, 91–103. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, T.; Li, X.; Song, C.-P.; Zhu, J.-K.; Chen, L.; Zhao, Y. Phosphorylation of SWEET Sucrose Transporters Regulates Plant Root:Shoot Ratio under Drought. Nat. Plants 2021, 8, 68–77. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Jin, X.; Niu, M.; Feng, C.; Liu, X.; Liu, C.; Wang, H.; Yin, W.; Xia, X. PeFUS3 Drives Lateral Root Growth Via Auxin and ABA Signalling Under Drought Stress in Populus. Plant Cell Environ. 2025, 48, 664–681. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF Transcription Factor PtoERF15 Confers Drought Tolerance via JA -mediated Signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wu, Y.; Li, M.; Ding, L.; Li, J.; Liu, Y.; Cao, Y.; Hua, Y.; Jia, Q.; Wang, D. The Auxin Response Factor (ARF) Gene Family in Cyclocarya Paliurus: Genome-Wide Identification and Their Expression Profiling under Heat and Drought Stresses. Physiol. Mol. Biol. Plants 2024, 30, 921–944. [Google Scholar] [CrossRef]
- Shiriga, K.; Sharma, R.; Kumar, K.; Yadav, S.K.; Hossain, F.; Thirunavukkarasu, N. Genome-Wide Identification and Expression Pattern of Drought-Responsive Members of the NAC Family in Maize. Meta Gene 2014, 2, 407–417. [Google Scholar] [CrossRef]
- Wang, X.-L.; Chen, X.; Yang, T.-B.; Cheng, Q.; Cheng, Z.-M. Genome-Wide Identification of BZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca). Int. J. Genom. 2017, 2017, 3981031. [Google Scholar] [CrossRef]
- Lee, J.E.; Golz, J.F. Diverse Roles of Groucho/Tup1 Co-Repressors in Plant Growth and Development. Plant Signal. Behav. 2012, 7, 86–92. [Google Scholar] [CrossRef]
- Plant, A.R.; Larrieu, A.; Causier, B. Repressor for Hire! The Vital Roles of TOPLESS-mediated Transcriptional Repression in Plants. New Phytol. 2021, 231, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K. Structural Basis for Recognition of Diverse Transcriptional Repressors by the TOPLESS Family of Corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef]
- Liu, Z.; Karmarkar, V. Groucho/Tup1 Family Co-Repressors in Plant Development. Trends Plant Sci. 2008, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Cheong, J.-J. AtMYB44 Interacts with TOPLESS-RELATED Corepressors to Suppress Protein Phosphatase 2C Gene Transcription. Biochem. Biophys. Res. Commun. 2018, 507, 437–442. [Google Scholar] [CrossRef]
- Griebel, T.; Lapin, D.; Locci, F.; Kracher, B.; Bautor, J.; Concia, L.; Benhamed, M.; Parker, J.E. Arabidopsis Topless-related 1 Mitigates Physiological Damage and Growth Penalties of Induced Immunity. New Phytol. 2023, 239, 1404–1419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Y.; Chen, H.; Guo, C.; Su, X.; Mu, T.; Feng, B.; Wang, Y.; Liu, Z.; Zhang, B.; et al. Genome-Wide Analysis of TOPLESS/TOPLESS-RELATED Co-Repressors and Functional Characterization of BnaA9.TPL Regulating the Embryogenesis and Leaf Morphology in Rapeseed. Plant Sci. 2024, 346, 112149. [Google Scholar] [CrossRef]
- Deng, H.; Chen, Y.; Liu, Z.; Liu, Z.; Shu, P.; Wang, R.; Hao, Y.; Su, D.; Pirrello, J.; Liu, Y.; et al. SlERF.F12 Modulates the Transition to Ripening in Tomato Fruit by Recruiting the Co-Repressor TOPLESS and Histone Deacetylases to Repress Key Ripening Genes. Plant Cell 2022, 34, 1250–1272. [Google Scholar] [CrossRef]
- Uhrig, J.F.; Huang, L.-J.; Barghahn, S.; Willmer, M.; Thurow, C.; Gatz, C. CC-Type Glutaredoxins Recruit the Transcriptional Co-Repressor TOPLESS to TGA-Dependent Target Promoters in Arabidopsis Thaliana. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 218–226. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA Connects the Co-Repressor TOPLESS to Jasmonate Signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, S.; Tao, W.; Zhang, X.; Liu, J.; Sun, J.; Zhang, H.; Pu, L.; Huang, R.; Chen, T. INDETERMINATE SPIKELET1 Recruits Histone Deacetylase and a Transcriptional Repression Complex to Regulate Rice Salt Tolerance. Plant Physiol. 2018, 178, 824–837. [Google Scholar] [CrossRef]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef]
- Martin-Arevalillo, R.; Nanao, M.H.; Larrieu, A.; Vinos-Poyo, T.; Mast, D.; Galvan-Ampudia, C.; Brunoud, G.; Vernoux, T.; Dumas, R.; Parcy, F. Structure of the Arabidopsis TOPLESS Corepressor Provides Insight into the Evolution of Transcriptional Repression. Proc. Natl. Acad. Sci. USA 2017, 114, 8107–8112. [Google Scholar] [CrossRef]
- Saini, R.; Nandi, A.K. TOPLESS in the Regulation of Plant Immunity. Plant Mol. Biol. 2022, 109, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xie, Q.; Wang, T.; Wang, X.; Xu, F.; Yan, Z.; Li, X.; Ouyang, S.; Chen, J.; Wang, Y.; et al. RALF33–FERONIA Signaling Orchestrates Postwounding Root-Tip Regeneration via TPR4–ERF115 Dynamics. Plant Cell 2025, 37, koaf098. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Gong, F.; Liu, G.; Deng, Y.; Liu, J.; Yang, W.; Sun, X.; Li, Y.; Gao, J.; Zhou, X.; et al. Petal Size Is Controlled by the MYB73/TPL/HDA19-miR159-CKX6 Module Regulating Cytokinin Catabolism in Rosa Hybrida. Nat. Commun. 2023, 14, 7106. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, W.; Li, Z.; Yang, L.; Zhuang, W.; Zhang, J.; Zhang, Z.; Fan, Z.; Wang, F.; Zhao, S.; et al. Integrated Multi-Omics Analysis Reveals Photosynthetic Acclimation and Metabolic Reprogramming in Populus Ussuriensis Kom. Under Cold Stress. Forests 2025, 16, 660. [Google Scholar] [CrossRef]
- Kagale, S.; Links, M.G.; Rozwadowski, K. Genome-Wide Analysis of Ethylene-Responsive Element Binding Factor-Associated Amphiphilic Repression Motif-Containing Transcriptional Regulators in Arabidopsis. Plant Physiol. 2010, 152, 1109–1134. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wang, X.; Li, X.; Bassa, C.; Mila, I.; Audran, C.; Maza, E.; Li, Z.; Bouzayen, M.; Van Der Rest, B.; et al. Genome-Wide Identification, Phylogenetic Analysis, Expression Profiling, and Protein–Protein Interaction Properties of TOPLESS Gene Family Members in Tomato. J. Exp. Bot. 2014, 65, 1013–1023. [Google Scholar] [CrossRef]
- Bartlett, M. Looking Back to Look Forward: Protein–Protein Interactions and the Evolution of Development. New Phytol. 2020, 225, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Guerois, R. Evolution of Protein Interactions: From Interactomes to Interfaces. Arch. Biochem. Biophys. 2014, 554, 65–75. [Google Scholar] [CrossRef]
- Wang, Q.; Gong, Z.; Zhu, Z. High Temperature-responsive DEAR4 Condensation Confers Thermotolerance through Recruiting TOPLESS in Arabidopsis Nucleus. Plant J. 2025, 122, e70172. [Google Scholar] [CrossRef]
- An, C.; Deng, L.; Zhai, H.; You, Y.; Wu, F.; Zhai, Q.; Goossens, A.; Li, C. Regulation of Jasmonate Signaling by Reversible Acetylation of TOPLESS in Arabidopsis. Mol. Plant 2022, 15, 1329–1346. [Google Scholar] [CrossRef]
- Navarrete, F.; Gallei, M.; Kornienko, A.E.; Saado, I.; Khan, M.; Chia, K.-S.; Darino, M.A.; Bindics, J.; Djamei, A. TOPLESS Promotes Plant Immunity by Repressing Auxin Signaling and Is Targeted by the Fungal Effector Naked1. Plant Commun. 2022, 3, 100269. [Google Scholar] [CrossRef]
- Garner, C.M.; Spears, B.J.; Su, J.; Cseke, L.J.; Smith, S.N.; Rogan, C.J.; Gassmann, W. Opposing Functions of the Plant TOPLESS Gene Family during SNC1-Mediated Autoimmunity. PLoS Genet. 2021, 17, e1009026. [Google Scholar] [CrossRef]
- Long, J.A.; Woody, S.; Poethig, S.; Meyerowitz, E.M.; Barton, M.K. Transformation of Shoots into Roots in Arabidopsis Embryos Mutant at the TOPLESS Locus. Development 2002, 129, 2797–2806. [Google Scholar] [CrossRef]
- Wang, W.; Ye, J.; Xu, H.; Liu, X.; Fu, Y.; Zhang, H.; Rouached, H.; Whelan, J.; Shen, Z.; Zheng, L. OsbHLH061 Links TOPLESS/TOPLESS-RELATED Repressor Proteins with POSITIVE REGULATOR OF IRON HOMEOSTASIS 1 to Maintain Iron Homeostasis in Rice. New Phytol. 2022, 234, 1753–1769. [Google Scholar] [CrossRef]
- Li, Y.; Yang, T.; Dai, D.; Hu, Y.; Guo, X.; Guo, H. Evolution, Gene Expression Profiling and 3D Modeling of CSLD Proteins in Cotton. BMC Plant Biol. 2017, 17, 119. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G.; et al. Transcriptional Regulation of Strigolactone Signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Lampugnani, E.R.; Bacic, A.; Golz, J.F. SEUSS and SEUSS-LIKE 2 Coordinate Auxin Distribution and KNOXI Activity during Embryogenesis. Plant J. 2014, 80, 122–135. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Chen, C.; Li, C.; Thapa, R.K.; Song, J.; Xie, X.; Nguyen, V.; Bian, S.; Liu, J.; Kohalmi, S.E.; et al. Genome-wide Occupancy of Arabidopsis SWI/SNF Chromatin Remodeler SPLAYED Provides Insights into Its Interplay with Its Close Homolog BRAHMA and Polycomb Proteins. Plant J. 2021, 106, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Lin, X.; Chan, T.-F.; Imtiaz, M.; Rehman, H.M.; Ali, M.A.; Baloch, F.S.; Atif, R.M.; Yang, S.H.; Chung, G. Characterization of Cellulose Synthase A (CESA) Gene Family in Eudicots. Biochem. Genet. 2019, 57, 248–272. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. EAR Motif-Mediated Transcriptional Repression in Plants: An Underlying Mechanism for Epigenetic Regulation of Gene Expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Liu, S.; Yang, C.; Wu, L.; Cai, H.; Li, H.; Xu, M. The Peu-miR160a−PeARF17.1/PeARF17.2 Module Participates in the Adventitious Root Development of Poplar. Plant Biotechnol. J. 2020, 18, 457–469. [Google Scholar] [CrossRef]
- Hamanishi, E.T.; Thomas, B.R.; Campbell, M.M. Drought Induces Alterations in the Stomatal Development Program in Populus. J. Exp. Bot. 2012, 63, 4959–4971. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. Transcriptional Regulatory Network of the Light Signaling Pathways. New Phytol. 2020, 227, 683–697. [Google Scholar] [CrossRef]
- Fang, Y.; Xiong, L. General Mechanisms of Drought Response and Their Application in Drought Resistance Improvement in Plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Kim, S.H.; Bahk, S.; Nguyen, N.T.; Pham, M.L.A.; Kadam, U.S.; Hong, J.C.; Chung, W.S. Phosphorylation of the Auxin Signaling Transcriptional Repressor IAA15 by MPKs Is Required for the Suppression of Root Development under Drought Stress in Arabidopsis. Nucleic Acids Res. 2022, 50, 10544–10561. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Ponting, C.P.; Schultz, J.; Milpetz, F.; Bork, P. SMART: Identification and Annotation of Domains from Signalling and Extracellular Protein Sequences. Nucleic Acids Res. 1999, 27, 229–232. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Richmond, T. A Simple Modular Architecture Research Tool for the Identification of Signaling Domains. Genome Biol. 2000, 1, reports234. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a Simple Modular Architecture Research Tool: Identification of Signaling Domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th Anniversary of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Zhao, S.; Li, D.; Wei, M.; Li, C. PuHSFA4a Enhances Tolerance to Excess Zinc by Regulating Reactive Oxygen Species Production and Root Development in Populus. Plant Physiol. 2019, 180, 2254–2271. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Ahmad, S.; Jose da Costa Gonzales, L.; Bowler-Barnett, E.H.; Rice, D.L.; Kim, M.; Wijerathne, S.; Luciani, A.; Kandasaamy, S.; Luo, J.; Watkins, X.; et al. The UniProt Website API: Facilitating Programmatic Access to Protein Knowledge. Nucleic Acids Res. 2025, 53, gkaf394. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Quick and Easy Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 35–37. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Wang, Y.; Liu, Z.; Gao, C. Comprehensive Analysis of MYB Gene Family and Their Expressions Under Abiotic Stresses and Hormone Treatments in Tamarix Hispida. Front. Plant Sci. 2018, 9, 1303. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Liu, Q.; Ren, J.; Fan, Z.; Gulfam, T.; Ma, Z.; Yang, J. Genome-Wide Identification, Drought-Responsive Expression, and EAR-Mediated Regulatory Network Construction of TOPLESS Genes in Populus ussuriensis Kom.. Plants 2025, 14, 3213. https://doi.org/10.3390/plants14203213
Li W, Liu Q, Ren J, Fan Z, Gulfam T, Ma Z, Yang J. Genome-Wide Identification, Drought-Responsive Expression, and EAR-Mediated Regulatory Network Construction of TOPLESS Genes in Populus ussuriensis Kom.. Plants. 2025; 14(20):3213. https://doi.org/10.3390/plants14203213
Chicago/Turabian StyleLi, Wanxin, Qianqian Liu, Jingru Ren, Zihan Fan, Tabeer Gulfam, Zhongzheng Ma, and Jingli Yang. 2025. "Genome-Wide Identification, Drought-Responsive Expression, and EAR-Mediated Regulatory Network Construction of TOPLESS Genes in Populus ussuriensis Kom." Plants 14, no. 20: 3213. https://doi.org/10.3390/plants14203213
APA StyleLi, W., Liu, Q., Ren, J., Fan, Z., Gulfam, T., Ma, Z., & Yang, J. (2025). Genome-Wide Identification, Drought-Responsive Expression, and EAR-Mediated Regulatory Network Construction of TOPLESS Genes in Populus ussuriensis Kom.. Plants, 14(20), 3213. https://doi.org/10.3390/plants14203213




