Study of Stress Granule Core Protein AtUBP1b Phosphorylation In Vitro
Abstract
1. Introduction
2. Results
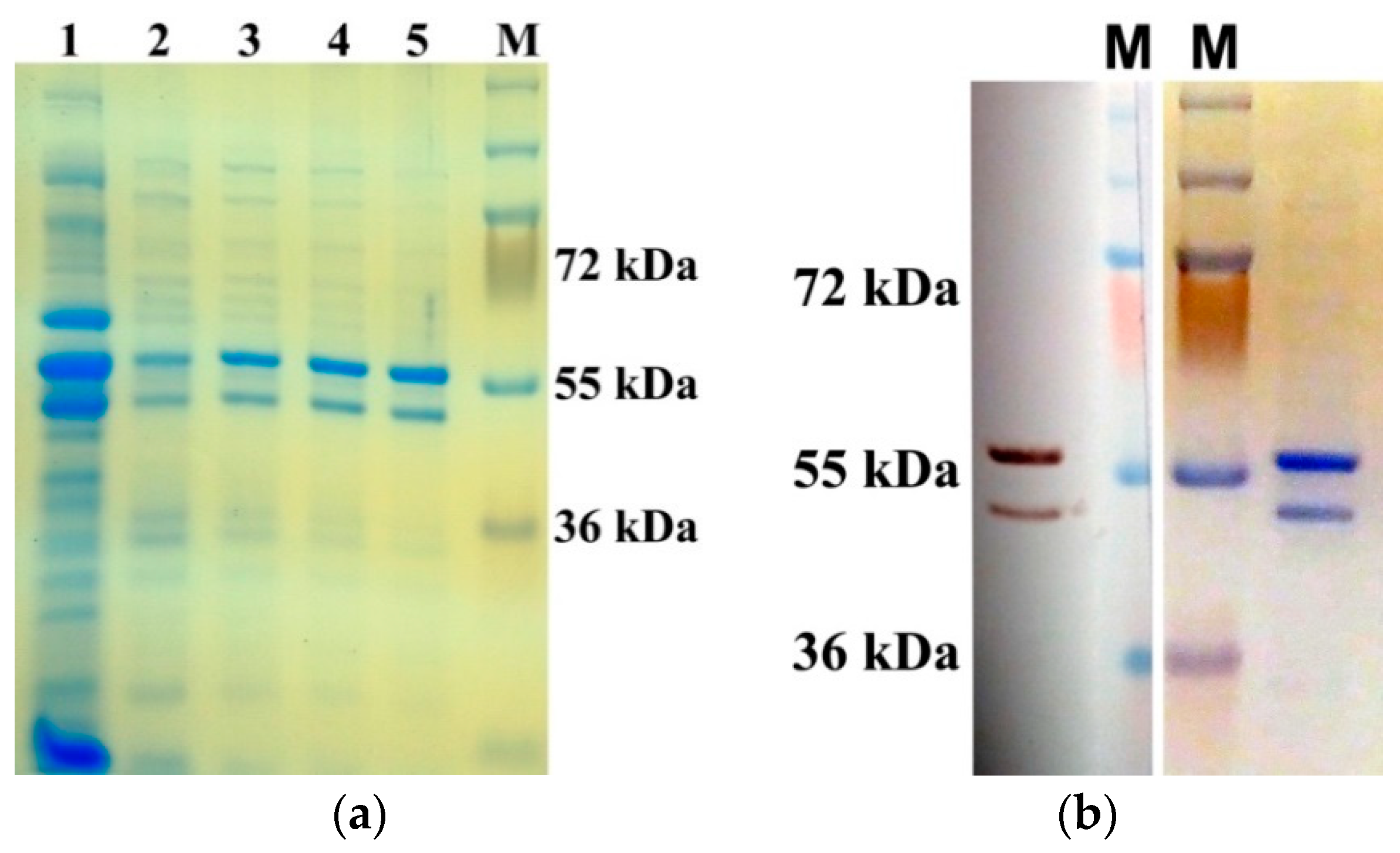
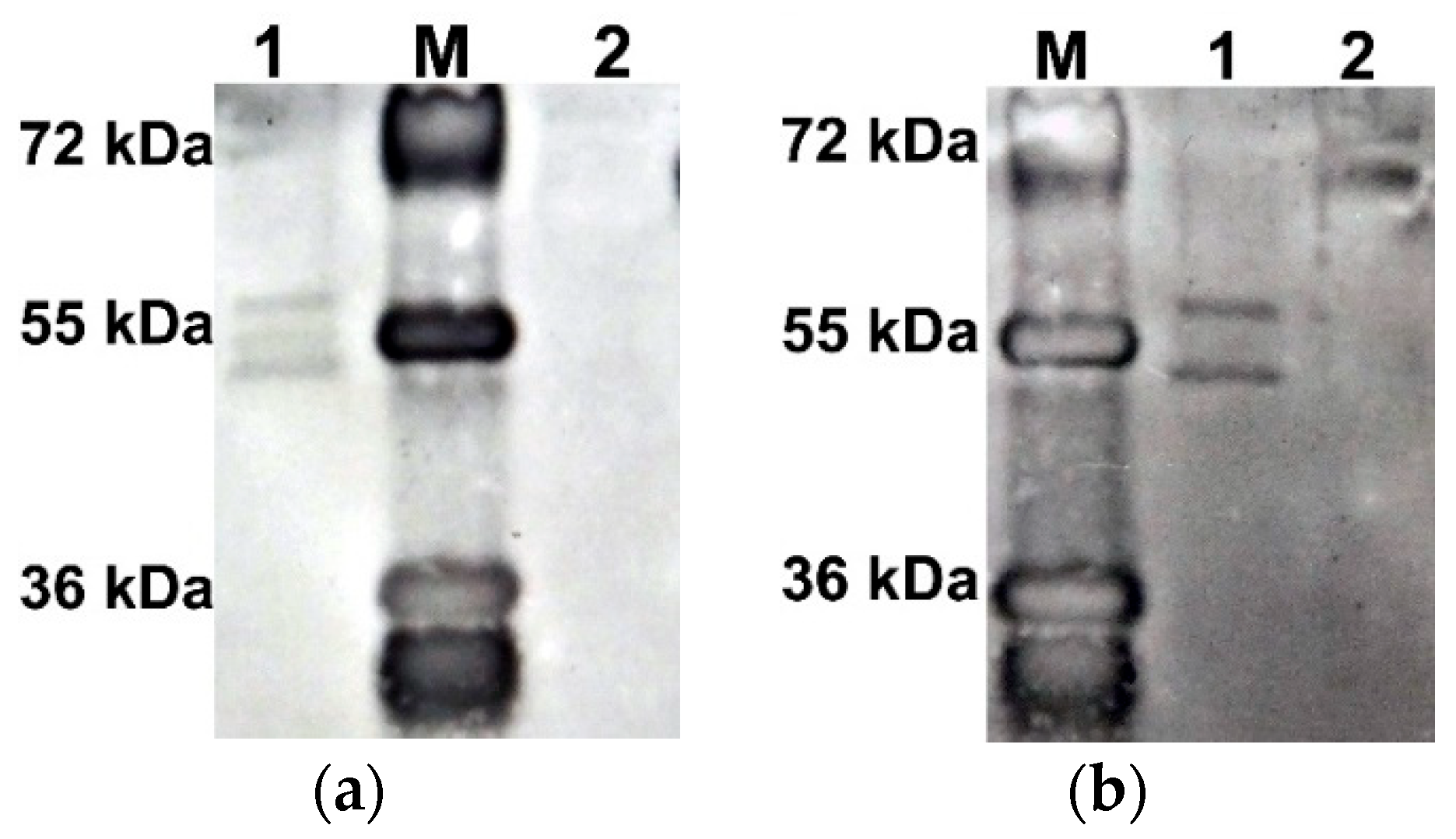
2.1. Expression and Purification of AtUBP1b from E. coli
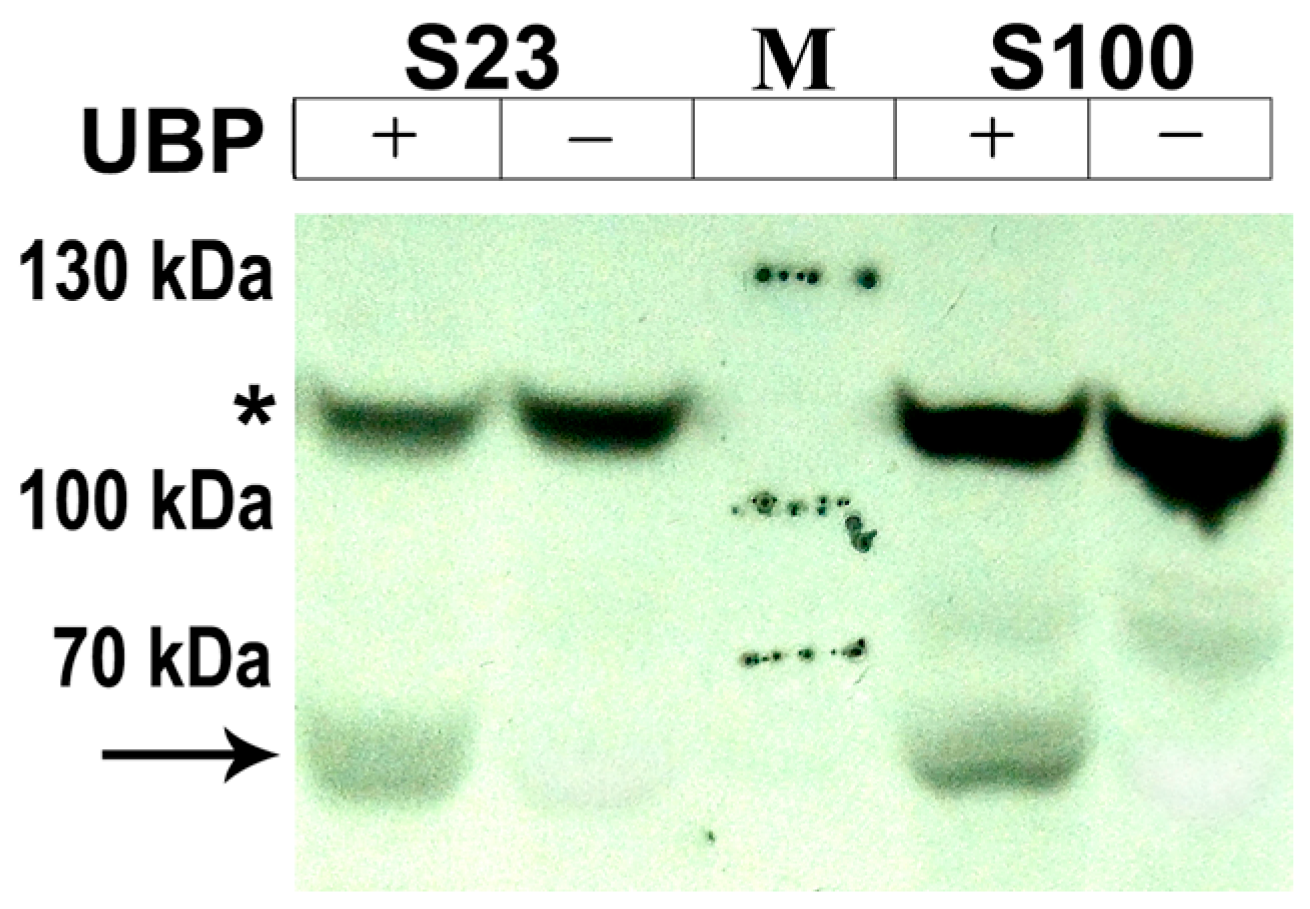
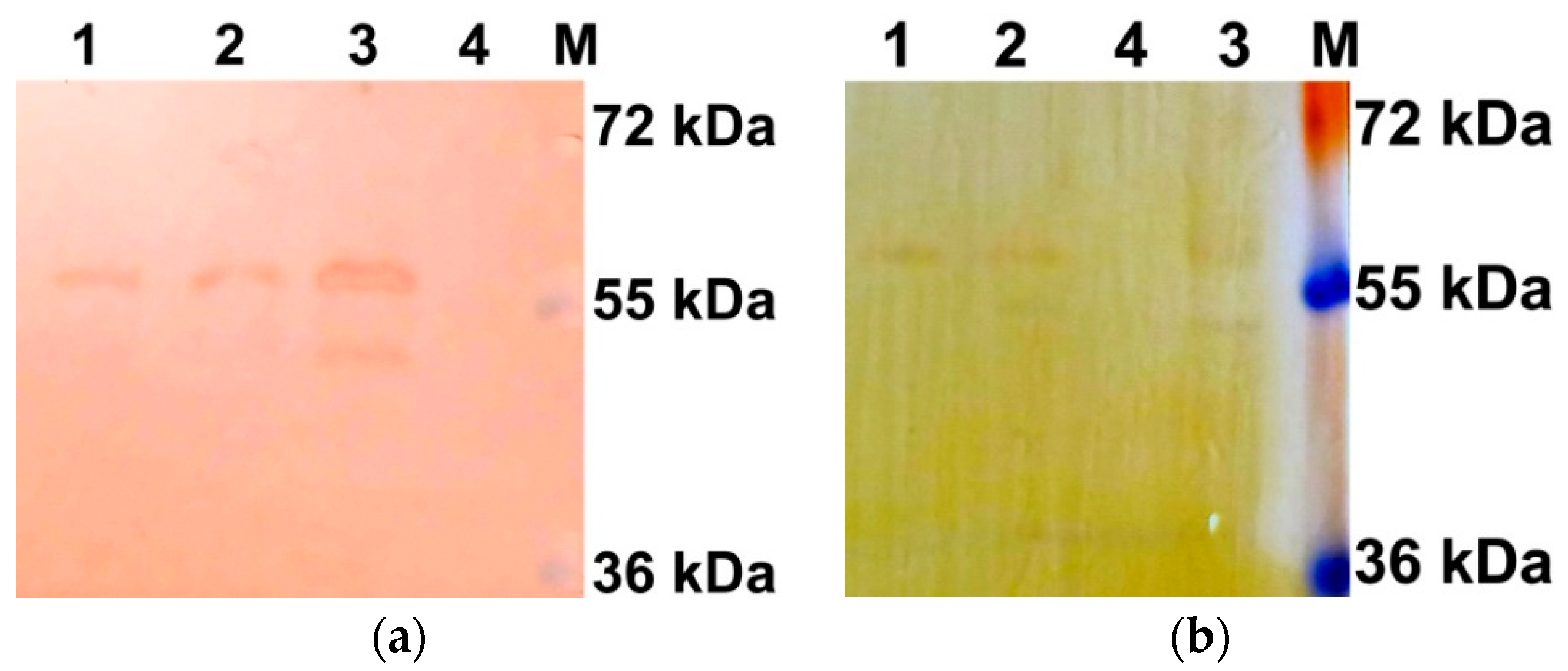
2.2. Determination of AtUBP1b Phosphorylation
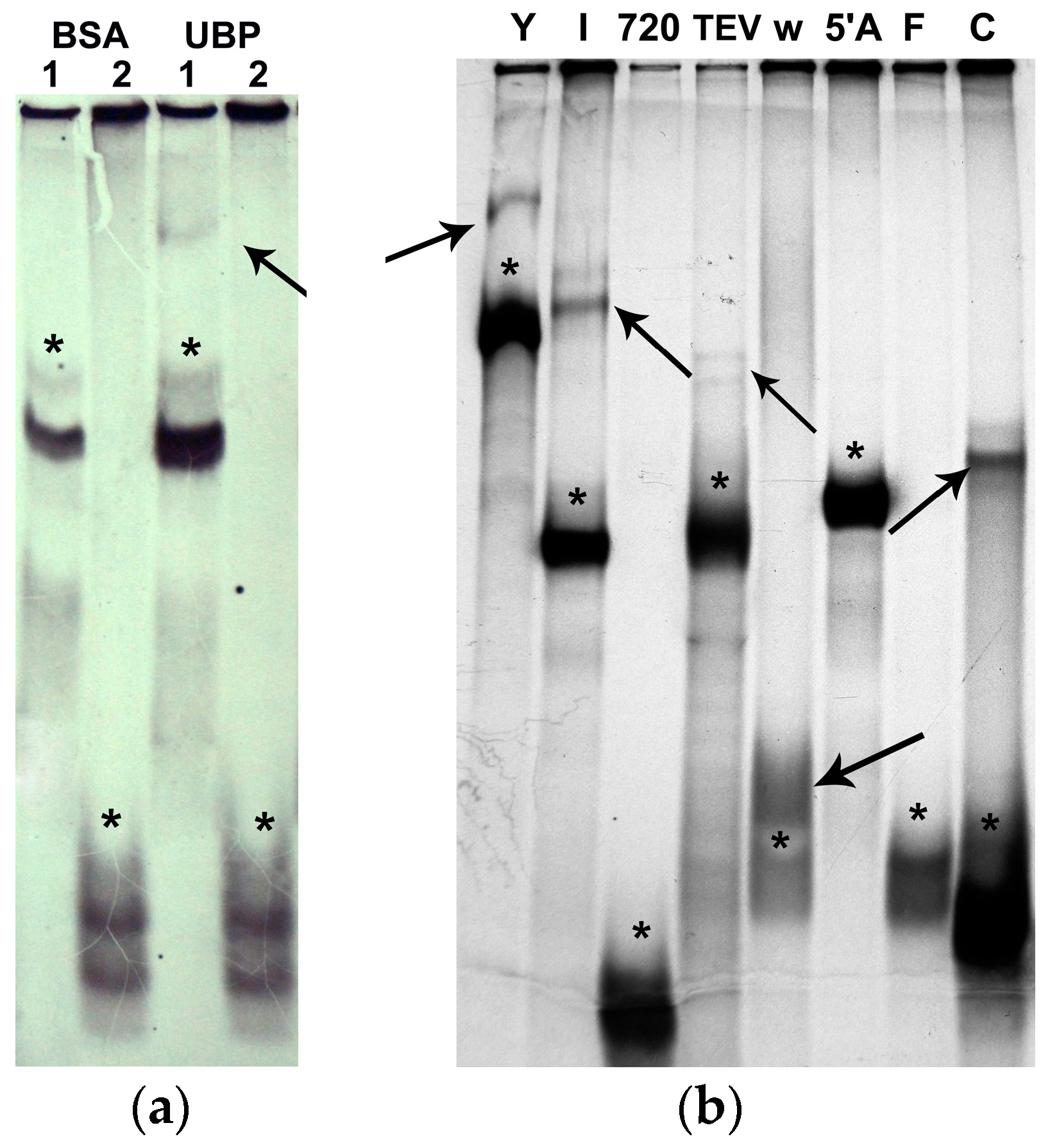
2.3. AtUBP1b Induced Sequence-Specific Gel Shifting of RNA
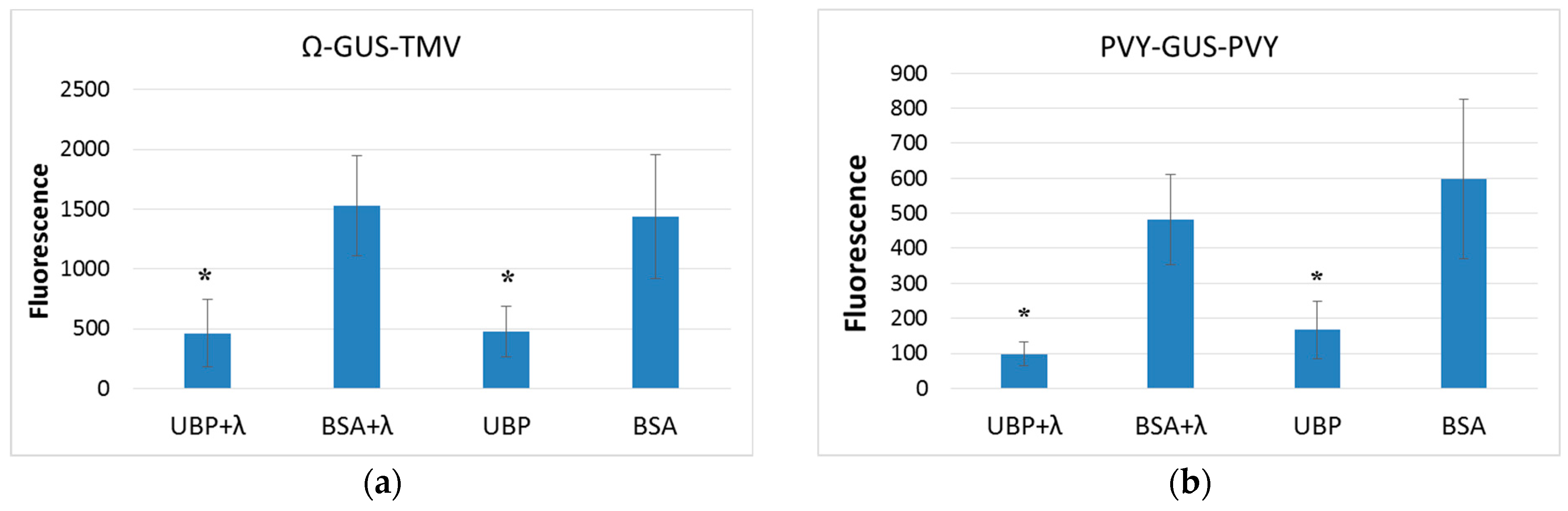
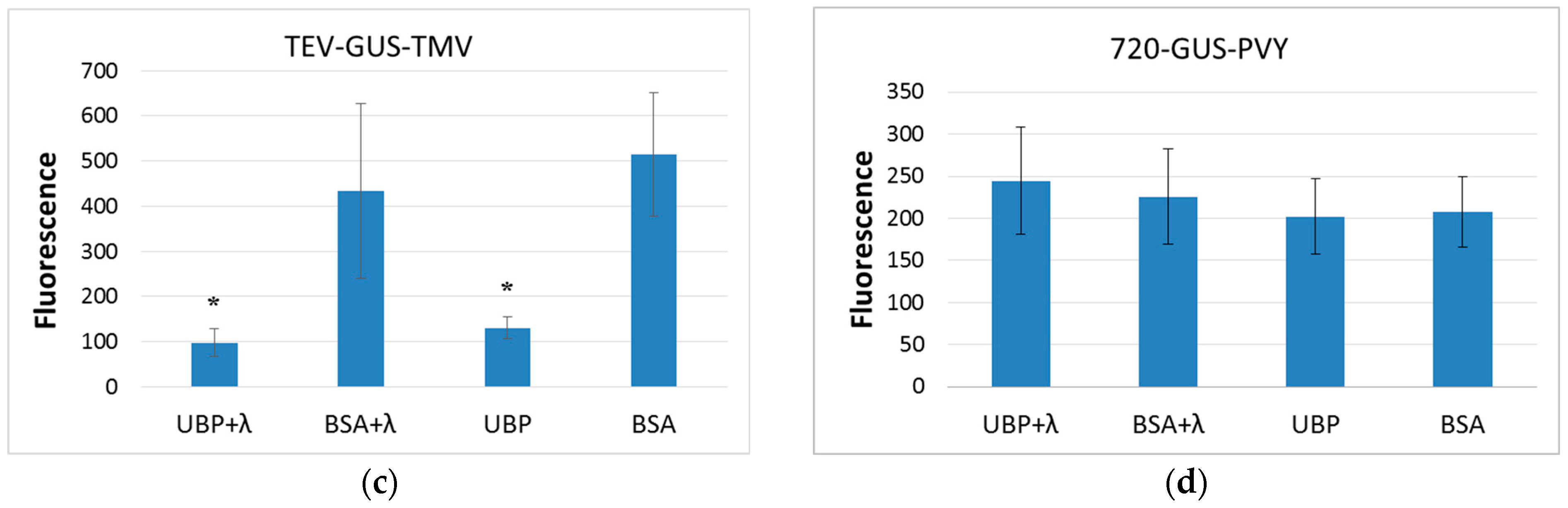
2.4. AtUBP1b Influence on In Vitro Translation
3. Discussion
4. Materials and Methods
4.1. Cloning of the AtUbp1b cDNA Gene, Expression, and Protein Purification from E. coli
4.2. Cell-Free In Vitro Translation
4.3. Protein Phosphorylation In Vitro
4.4. Radio-Labeled mRNA Synthesis
4.5. EMSA (Electrophoretic Mobility Shift Assay)
4.6. Western Blotting for Determination of Protein Phosphorylation
4.7. Protein Dephosphorylation by λ-Phosphatase
4.8. Liquid Chromatography (LC)-MS Analysis
4.9. Proteomics Database Search
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SGs | Stress granules |
| 5′TOP | 5′-terminal oligopyrimidine tracts |
| AtUBP1b | Oligouridylate binding protein |
| UTR | untranslated region |
| EMSA | electrophoretic mobility shift assay |
| BSA | bovine serum albumin |
| GUS | β-glucuronidase |
| PVY | potato virus Y |
| PVM | potato virus M |
| TMV | tobacco mosaic virus |
| EcCspA | cold-shock protein A of E. coli |
| TEV | tobacco etch virus |
| RRM | RNA recognition motifs |
References
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 2017, 68, 808–820. [Google Scholar] [CrossRef]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016, 212, 845–860, Erratum in J. Cell Biol. 2020, 219, e20150802809202019c. [Google Scholar] [CrossRef]
- Mateju, D.; Eichenberger, B.; Voigt, F.; Eglinger, J.; Roth, G.; Chao, J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 2020, 183, 1801–1812. [Google Scholar] [CrossRef]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by Oligouridylate-Binding Protein 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef] [PubMed]
- Mazan-Mamczarz, K.; Lal, A.; Martindale, J.L.; Kawai, T.; Gorospe, M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006, 26, 2716–2727. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, C.K.; Lykke-Andersen, J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011, 25, 2057–2068. [Google Scholar] [CrossRef] [PubMed]
- Maruri-López, I.; Figueroa, N.E.; Hernández-Sánchez, I.E.; Chodasiewicz, M. Plant Stress Granules: Trends and Beyond. Front. Plant Sci. 2021, 12, 722643. [Google Scholar] [CrossRef]
- Tian, S.; Curnutte, H.A.; Trcek, T. RNA Granules: A View from the RNA Perspective. Molecules 2020, 25, 3130. [Google Scholar] [CrossRef]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar] [CrossRef]
- Réthoré, E.; Pelletier, S.; Balliau, T.; Zivy, M.; Avelange-Macherel, M.-H.; Macherel, D. Multi-scale analysis of heat stress acclimation in Arabidopsis seedlings highlights the primordial contribution of energy-transducing organelles. Plant J. 2024, 119, 300–331. [Google Scholar] [CrossRef]
- Hamada, T.; Yako, M.; Minegishi, M.; Sato, M.; Kamei, Y.; Yanagawa, Y.; Toyooka, K.; Watanabe, Y.; Hara-Nishimura, I. Stress granule formation is induced by a threshold temperature rather than a temperature difference in Arabidopsis. J. Cell Sci. 2018, 131, jcs216051. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 2016, 5, e18413. [Google Scholar] [CrossRef]
- Marmor-Kollet, H.; Siany, A.; Kedersha, N.; Knafo, N.; Rivkin, N.; Danino, Y.M. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell 2020, 80, 876–891. [Google Scholar] [CrossRef]
- Maxwell, B.A.; Gwon, Y.; Mishra, A.; Peng, J.; Nakamura, H.; Zhang, K.; Kim, H.J.; Taylor, J.P. Ubiquitination is essential for recovery of cellular activities after heat shock. Science 2021, 372, eabc3593. [Google Scholar] [CrossRef] [PubMed]
- Loughlin, F.E.; West, D.L.; Gunzburg, M.J.; Waris, S.; Crawford, S.A.; Wilce, M.C.J.; Wilce, J.A. Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1. Nucleic Acids Res. 2021, 49, 2403–2417. [Google Scholar] [CrossRef] [PubMed]
- Lambermon, M.H.; Simpson, G.G.; Wieczorek, K.D.A.; Hemmings-Mieszczak, M.; Klahre, U.; Filipowicz, W. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 2000, 19, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Taupin, J.; Elledge, S.; Robertson, M.; Anderson, P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med. 1995, 182, 865–874. [Google Scholar] [CrossRef]
- England, W.E.; Wang, J.; Chen, S.; Baldi, P.; Flynn, R.A.; Spitale, R.C. An atlas of posttranslational modifications on RNA binding proteins. Nucleic Acids Res. 2022, 50, 4329–4339. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Madin, K.; Sawasaki, T.; Ogasawara, T.; Endo, Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 1997, 2, 559–564. [Google Scholar] [CrossRef]
- Cozzone, A.J. Diversity and specificity of protein-phosphorylating systems in bacteria. Folia Microbiol. 1997, 42, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Baldwin, T.J.; Sweeney, S.T.; Williams, P.H.; Leach, K.L. A protein kinase C-like activity in Escherichia coli. Mol. Microbiol. 1991, 5, 2977–2981. [Google Scholar] [CrossRef]
- Gradowski, M.; Baranowski, B.; Pawłowski, K. The expanding world of protein kinase-like families in bacteria: Forty families and counting. Biochem. Soc. Trans. 2020, 48, 1337–1352. [Google Scholar] [CrossRef]
- Gniadkowski, M.; Hemmings-Mieszczak, M.; Klahre, U.; Liu, H.X.; Filipowicz, W. Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res. 1996, 24, 619–627. [Google Scholar] [CrossRef]
- Golshani, A.; Golomehova, V.; Mironova, R.; Ivanov, I.G.; AbouHaidar, M.G. Does the Epsilon Sequence of phage T7 function as an initiator for the translation of CAT mRNA in Escherichia coli? Biochem. Biophys. Res. Commun. 1997, 236, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Cervera, M. Histochemical and fluorometric assays for uidA (GUS) gene detection. Methods Mol. Biol. 2005, 286, 203–214. [Google Scholar] [CrossRef]
- Izquierdo, J.M.; Valcárcel, J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J. Biol. Chem. 2007, 282, 1539–1543. [Google Scholar] [CrossRef]
- Velázquez-Cruz, A.; Baños-Jaime, B.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation. Front. Mol. Biosci. 2021, 8, 658852. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Grant, S.; Freestone, P.; Canvin, J.; Sheikh, F.N.; Toth, I.; Trinei, M.; Modha, K.; Norman, R.I. Calcium signalling in bacteria. J. Bacteriol. 1996, 178, 3677–3682. [Google Scholar] [CrossRef]
- Vincent, C.; Doublet, P.; Grangeasse, C.; Vaganay, E.; Cozzone, A.J.; Duclos, B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 1999, 181, 3472–3477. [Google Scholar] [CrossRef]
- Yao, Q.; Gao, J.; Bollinger, C.; Thelen, J.J.; Xu, D. Predicting and analyzing protein phosphorylation sites in plants using musite. Front. Plant Sci. 2012, 3, 186. [Google Scholar] [CrossRef]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.; Hennig, J.; Jagtap, P.K.A.; Sonntag, M.; Valcárcel, J.; Sattler, M. Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res. 2014, 42, 5949–5966. [Google Scholar] [CrossRef] [PubMed]
- Waris, S.; García-Mauriño, S.M.; Sivakumaran, A.; Beckham, S.A.; Loughlin, F.E.; Gorospe, M.; Díaz-Moreno, I.; Wilce, M.C.J.; Wilce, J.A. TIA-1 RRM23 binding and recognition of target oligonucleotides. Nucleic Acids Res. 2017, 45, 4944–4957. [Google Scholar] [CrossRef]
- Cruz-Gallardo, I.; Aroca, Á.; Gunzburg, M.J.; Sivakumaran, A.; Yoon, J.H.; Angulo, J.; Persson, C.; Gorospe, M.; Karlsson, B.G.; Wilce, J.A.; et al. The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain. RNA Biol. 2014, 11, 766–776. [Google Scholar] [CrossRef]
- Mustoe, A.M.; Corley, M.; Laederach, A.; Weeks, K.M. Messenger RNA Structure Regulates Translation Initiation: A Mechanism Exploited from Bacteria to Humans. Biochemistry 2018, 57, 3537–3539. [Google Scholar] [CrossRef] [PubMed]
- Grayeski, P.J.; Weidmann, C.A.; Kumar, J.; Lackey, L.; Mustoe, A.M.; Busan, S.; Laederach, A.; Weeks, K.M. Global 5′-UTR RNA structure regulates translation of a SERPINA1 mRNA. Nucleic Acids Res. 2022, 50, 9689–9704. [Google Scholar] [CrossRef]
- Akbergenov, R.Z.; Zhanybekova, S.S.; Kryldakov, R.V.; Zhigailov, A.; Polimbetova, N.S.; Hohn, T.; Iskakov, B.K. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res. 2004, 32, 239–247. [Google Scholar] [CrossRef]
- Gallie, D.R.; Sleat, D.E.; Watts, J.W.; Turner, P.C.; Wilson, T.M. A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 1987, 15, 8693–8711. [Google Scholar] [CrossRef]
- Kim, S.H.; Heo, M.A.; Kim, Y.J.; Kim, S.Y.; Neelamegam, R.; Lee, S.G. The effect of the cspA 5′-untranslated region on recombinant protein production at low temperature. Biotechnol. Bioproc. 2008, 13, 366–371. [Google Scholar] [CrossRef]
- Gallie, D.R.; Tanguay, R.L.; Leathers, V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 1995, 165, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Tanguay, R.L.; Gallie, D.R. Isolation and characterization of the 102-kilodalton RNA-binding protein that binds to the 5′ and 3′ translational enhancers of tobacco mosaic virus RNA. J. Biol. Chem. 1996, 271, 14316–14322. [Google Scholar] [CrossRef]
- Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Cuthill, I.C. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. Camb. Philos. Soc. 2007, 82, 591–605. [Google Scholar] [CrossRef] [PubMed]


| UTR | Sequence | U Content,% |
|---|---|---|
| 5′PVY | AAUUAAAACAACUCAAUACAACAUAAGAAAAACAACGCAAAAACACUCAUAAACGCUUAUUCUCACUCAAGCAACUUGCUAAGUUUCAGUUUAAAUCAUUUCCUUGCAACUCUCUUAAACGAUAUUGGAAACCAUUUCAACUCAACAAGUAAUUUCAUCACUUCCAACCAAUUUCAGAUCCU 1 (182 nt) | 28% |
| 5′CspA | GGUUUGACGUACAGACCAUUAAAGCAGUGUAGUAAGGCAAGUCCCUUCAAGAGUUAUCGUUGAUACCCCUCGUAGUGCACAUUCCUUUAACGCUUCAAAAUCUGUAAAGCACGCCAUAUCGCCGAAAGGCACACUUAAUUAUUAAAGGUAAUACAC (156 nt) | 26% |
| 5′TEV | AAAUAACAAAUCUCAACACAACAUAUACAAAACAAACGAAUCUCAAGCAAUCAAGCAUUCUACUUCUAUUGCAGCAAUUUAAAUCAUUUCUUUUAAAGCAAAAGCAAUUUUCUGAAAAUUUUCACCAUUUACGAACGAUAGCA (143 nt) | 28% |
| 5′I | UAAUUAAAACAACUCAAUACAACAUAAGAAAAACAACGCAAAAACACUCAUAAACGCUUAUUCUCACUCAAGCAACUUGCUAAGUUUCAGUUUAAAUCAUUUCCUUGCAACUCUCUUAAACGAUAUUGGAAA (132 nt) | 27% |
| 5′Ω | GACCAUGAUUAGGGGGGAGAUUUAUUUUUACAACAAUUACCAACAACAACAAACAACAAACAACAUUACAAUUACUAUUUACAAUUACAGUCGACGAU (98 nt) | 26% |
| 5′PVM | AUUAAACAAACAUACAAUAUCUGGACUUACACUACAAUAUACUACCAGGAAAUACUAUAUUUGGUCUAAGUCAGC (75 nt) | 28% |
| 5′F | UAAUUAAAACAACUCAAUACAACAUAAGAAAAACAACGCAAAAACACUCAUAAACGCUUAUUCUCACUCA (70 nt) | 20% |
| 5′C | CUCAAGCAACUUGCUAAGUUUCAGUUUAAAUCAUUUCCUUGCAACUCUCUUAAACGAUAUUGGAAAC (67 nt) | 34% |
| 5′720 | CAUUUCAACUCAACAAGUAAUUUCAUCACUUCCAACCAAUUUCAGAUCCU (50 nt) | 32% |
| 5′εII | UCUAGAUUUAACUUUAUUUACCUUAUCUCUAUUCCAUGG (39 nt) | 49% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nizkorodova, A.S.; Kislitsin, V.Y.; Zhigailov, A.V.; Kulyyassov, A.T.; Nadirova, L.M.; Stanbekova, G.E.; Iskakov, B.K. Study of Stress Granule Core Protein AtUBP1b Phosphorylation In Vitro. Plants 2025, 14, 3191. https://doi.org/10.3390/plants14203191
Nizkorodova AS, Kislitsin VY, Zhigailov AV, Kulyyassov AT, Nadirova LM, Stanbekova GE, Iskakov BK. Study of Stress Granule Core Protein AtUBP1b Phosphorylation In Vitro. Plants. 2025; 14(20):3191. https://doi.org/10.3390/plants14203191
Chicago/Turabian StyleNizkorodova, Anna S., Valeriy Y. Kislitsin, Andrey V. Zhigailov, Arman T. Kulyyassov, Leila M. Nadirova, Gulshan E. Stanbekova, and Bulat K. Iskakov. 2025. "Study of Stress Granule Core Protein AtUBP1b Phosphorylation In Vitro" Plants 14, no. 20: 3191. https://doi.org/10.3390/plants14203191
APA StyleNizkorodova, A. S., Kislitsin, V. Y., Zhigailov, A. V., Kulyyassov, A. T., Nadirova, L. M., Stanbekova, G. E., & Iskakov, B. K. (2025). Study of Stress Granule Core Protein AtUBP1b Phosphorylation In Vitro. Plants, 14(20), 3191. https://doi.org/10.3390/plants14203191








