Transcription Factor Analysis of Rhodophytes Suggests Trihelix Transcription Factors Across the Florideophyceae
Abstract
1. Introduction
2. Results
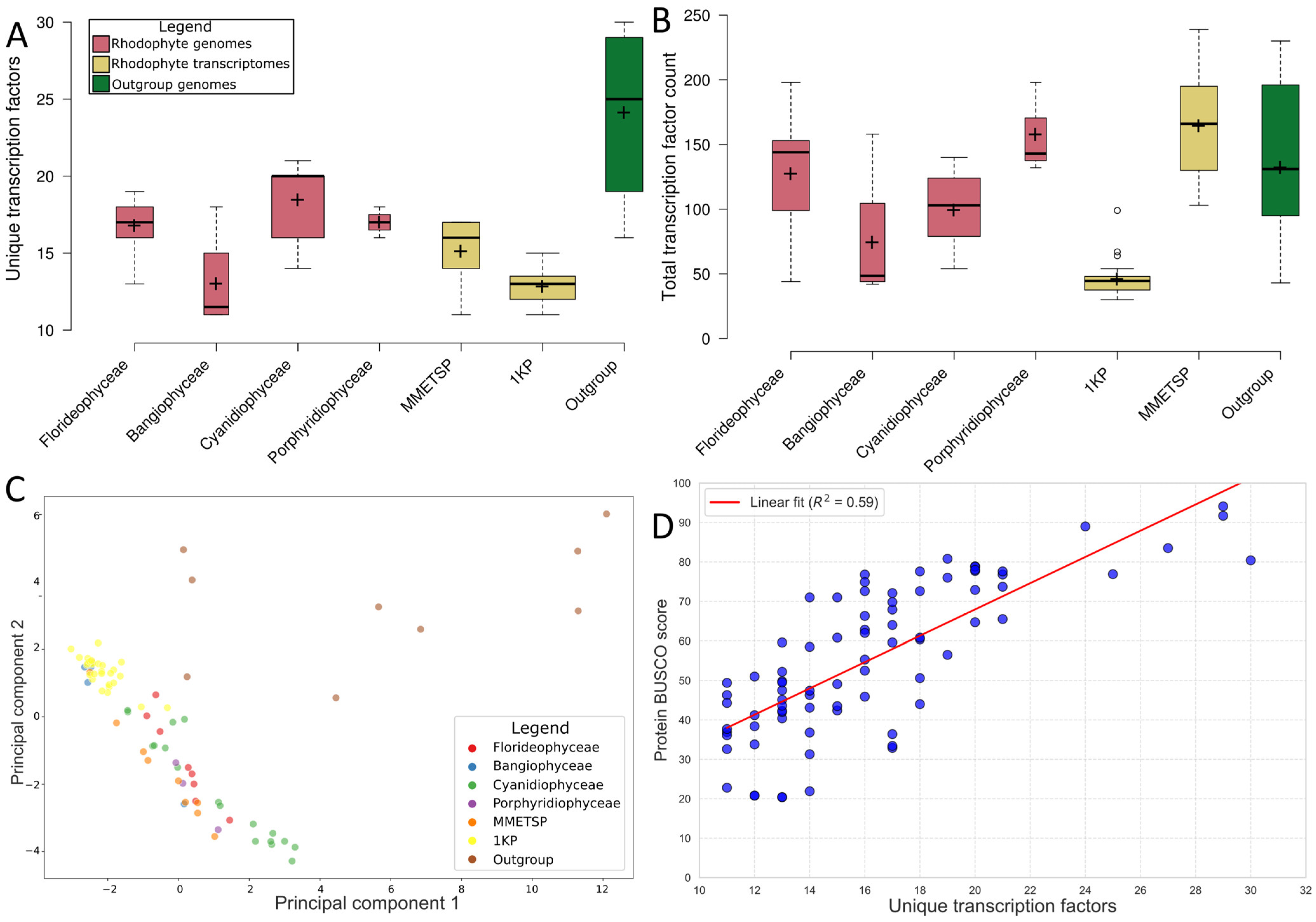
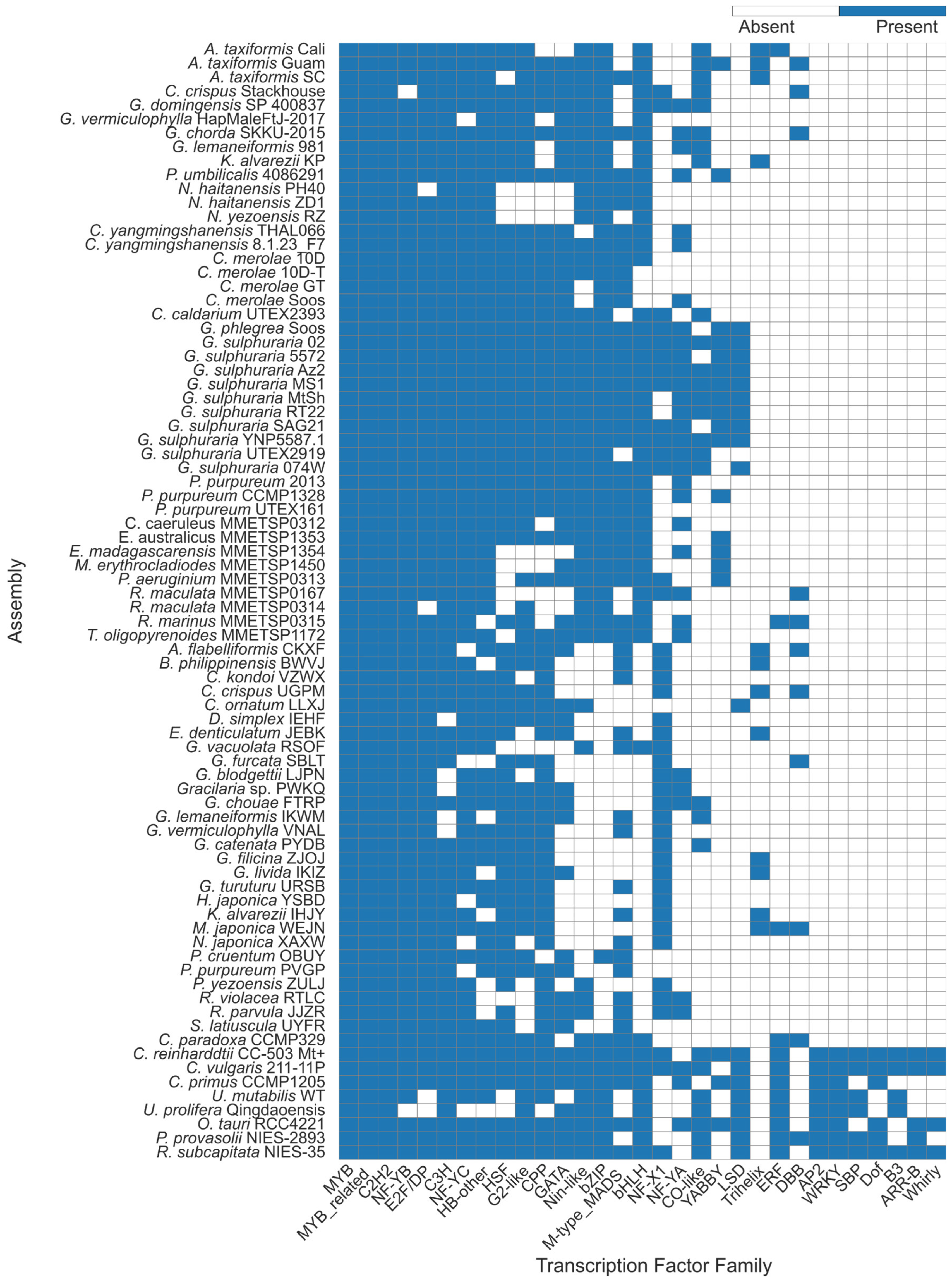
2.1. Transcription Factor Predictions Were Consistent with Prior Studies
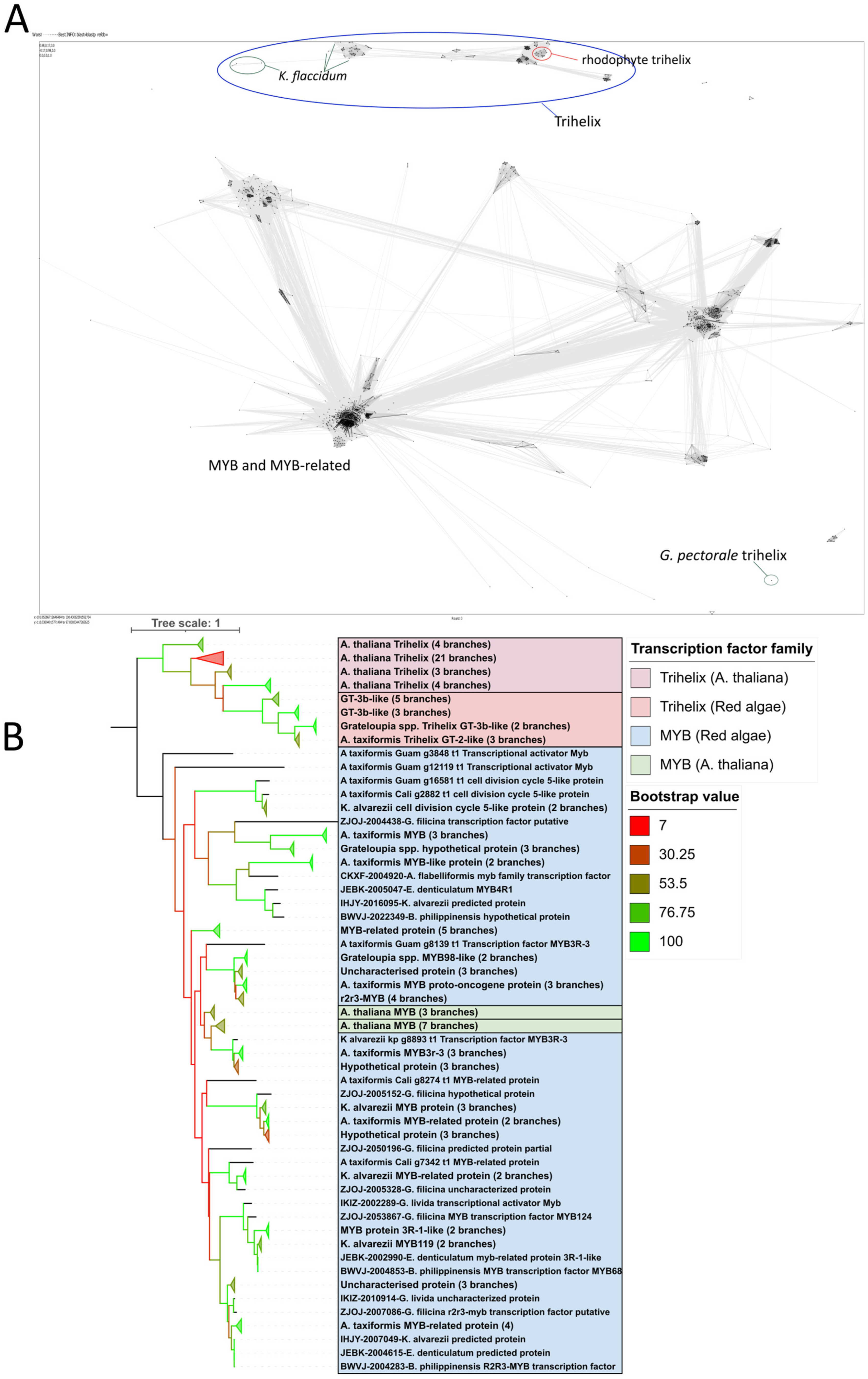
2.2. Putative Trihelix Factor Identified in Florideophyte
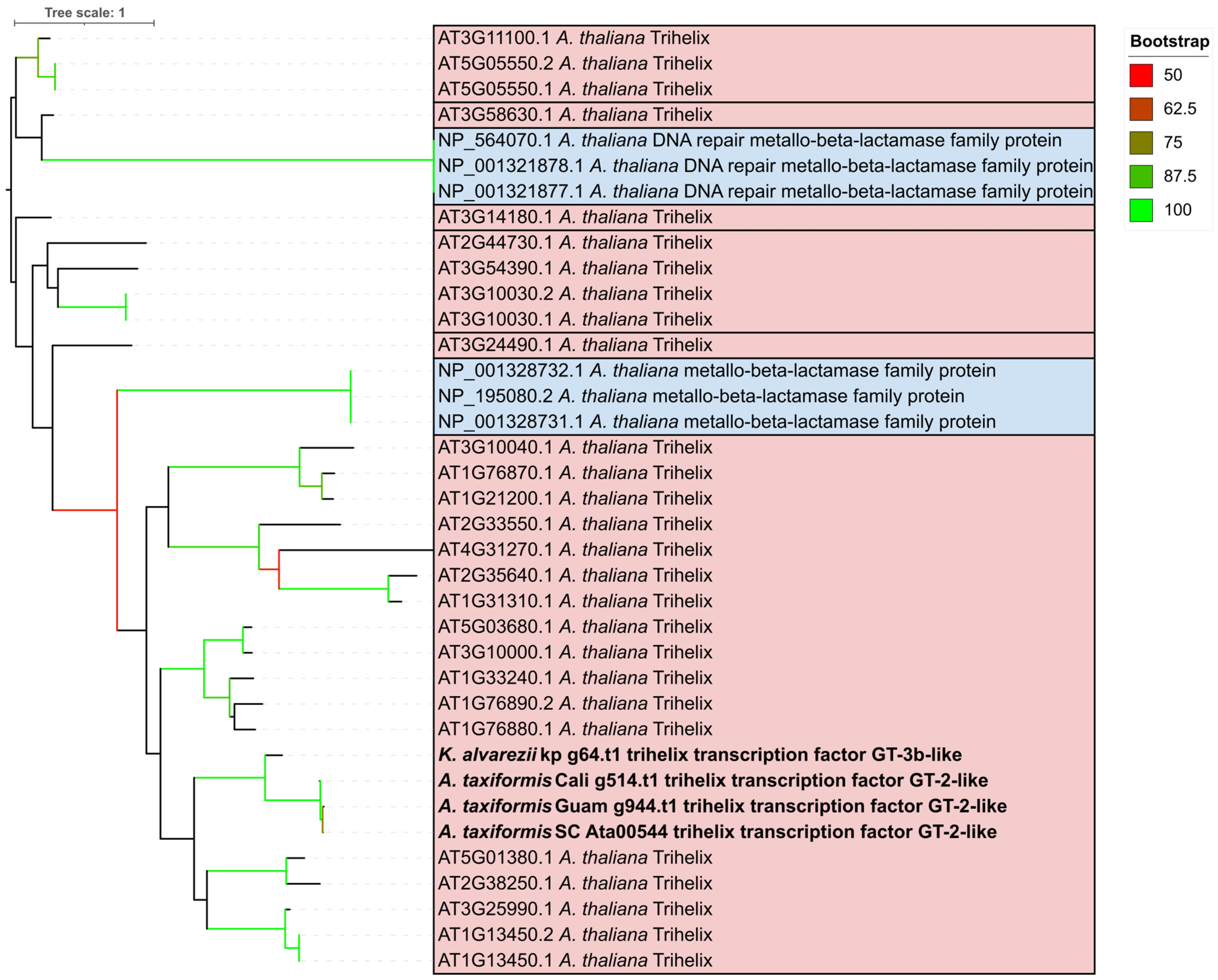
2.3. Trihelix Annotation Conserved Across Florideophyte Algae
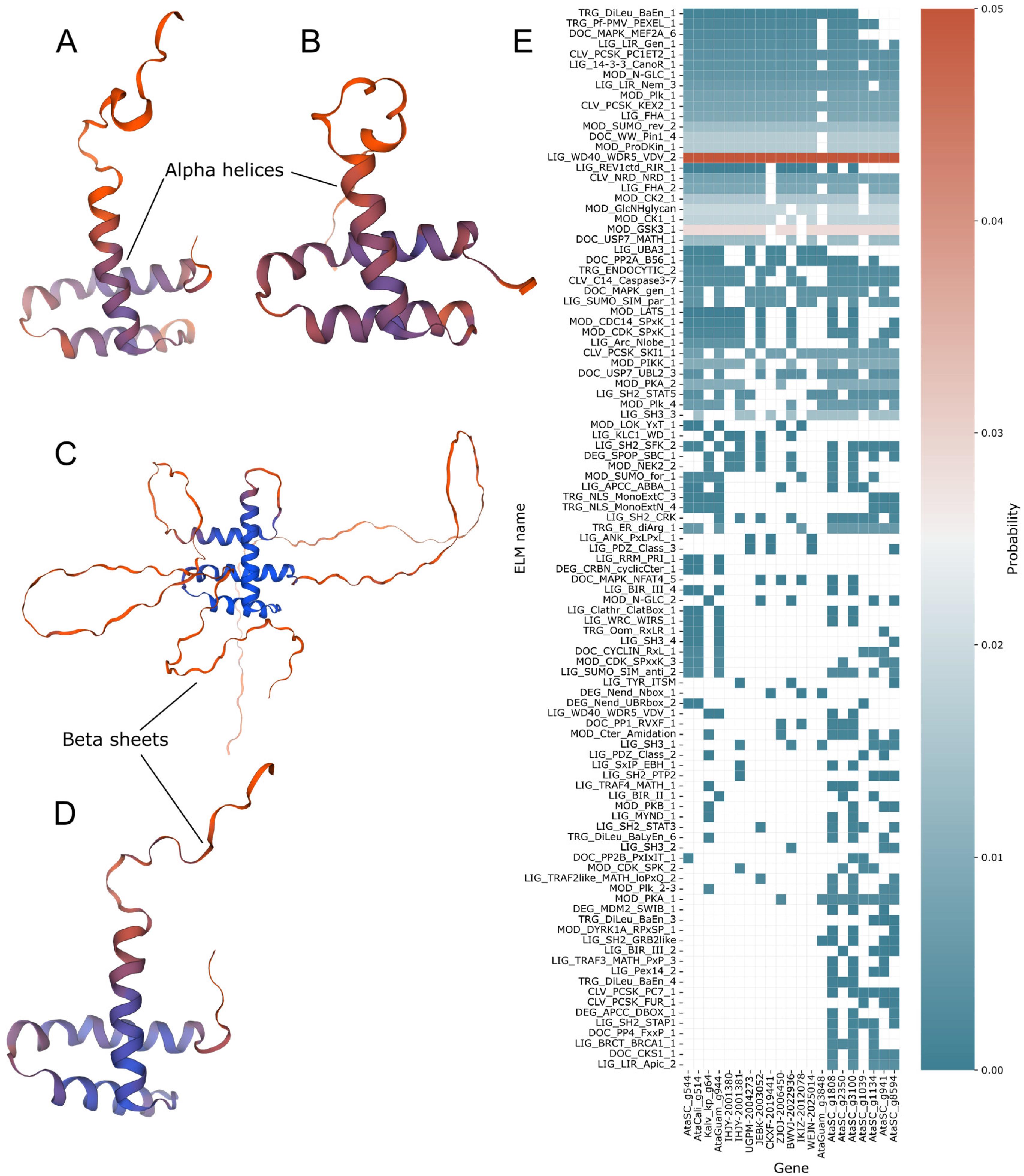
2.4. Protein Modelling and Motif Prediction
3. Discussion
3.1. Overall Transcription Factor Proportions Supported Previous Studies
3.2. Trihelix and YABBY TFs Were Predicted Contrary to Previous Evidence
3.3. Putative TF Predictions May Have Impacts on Secondary Metabolism
4. Materials and Methods
4.1. Identification of TFs Across Rhodophyte Assemblies
4.2. Phylogenomic Analysis of Putative Trihelix Transcription Factors
4.3. Protein Modelling and Motif Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1KP | One Thousand Plant Transcriptomes Project |
| AtaCali | Asparagopsis taxiformis Cali |
| AtaGuam | Asparagopsis taxiformis Guam |
| AtaSC | Asparagopsis taxiformis Sunshine Coast |
| BLAST | Basic Local Alignment Search Tool |
| BUSCO | Benchmarking Universal Single Copy Orthologs |
| CLANS | Cluster ANalysis Sequences |
| KW | Kruskal–Wallis |
| MAPK | Mitogen-activated protein kinase |
| MBL | Metallo- β-lactamase |
| MMETSP | Marine Microbial Eukaryotic Transcriptome Sequencing Project |
| PlantTFDB | Plant Transcription Factor Database |
| TF | Transcription factor |
| TTF | Trihelix transcription factor |
| WOX | WUSCHEL-related homeobox |
References
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Warren, A.J. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 2002, 12, 107–114. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Panchy, N.; Wang, P.; Uygun, S.; Shiu, S.-H. Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2017, 1860, 3–20. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Chénais, B.; Trottier, C.; Bougaran, G.; Cadoret, J.-P.; Schoefs, B.; Saint-Jean, B. Transcription factors in microalgae: Genome-wide prediction and comparative analysis. BMC Genom. 2016, 17, 282. [Google Scholar] [CrossRef]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Catarino, B.; Hetherington, A.J.; Emms, D.M.; Kelly, S.; Dolan, L. The Stepwise Increase in the Number of Transcription Factor Families in the Precambrian Predated the Diversification of Plants On Land. Mol. Biol. Evol. 2016, 33, 2815–2819. [Google Scholar] [CrossRef]
- Petroll, R.; Schreiber, M.; Finke, H.; Cock, J.M.; Gould, S.B.; Rensing, S.A. Signatures of Transcription Factor Evolution and the Secondary Gain of Red Algae Complexity. Genes 2021, 12, 1055. [Google Scholar] [CrossRef]
- Wilhelmsson, P.K.I.; Mühlich, C.; Ullrich, K.K.; Rensing, S.A. Comprehensive Genome-Wide Classification Reveals That Many Plant-Specific Transcription Factors Evolved in Streptophyte Algae. Genome Biol. Evol. 2017, 9, 3384–3397. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Khaldun, A.B.M.; Chen, J.; Zhang, C.; Lv, H.; Yuan, L.; Wang, Y. A R2R3-MYB Transcription Factor Regulates the Flavonol Biosynthetic Pathway in a Traditional Chinese Medicinal Plant, Epimedium sagittatum. Front. Plant Sci. 2016, 7, 1089. [Google Scholar] [CrossRef]
- Yang, J.; Tang, Z.; Yang, W.; Huang, Q.; Wang, Y.; Huang, M.; Wei, H.; Liu, G.; Lian, B.; Chen, Y.; et al. Genome-wide characterization and identification of Trihelix transcription factors and expression profiling in response to abiotic stresses in Chinese Willow (Salix matsudana Koidz). Front. Plant Sci. 2023, 14, 1125519. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors–light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef]
- Smalle, J.; Kurepa, J.; Haegman, M.; Gielen, J.; Van Montagu, M.; Van Der Straeten, D. The trihelix DNA-binding motif in higher plants is not restricted to the transcription factors GT-1 and GT-2. Proc. Natl. Acad. Sci. USA 1998, 95, 3318–3322. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.-J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of Seed Maturation Genes by a Trihelix Transcriptional Repressor in Arabidopsis Seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999, 4, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Jiang, S.; Yu, X.; Cheng, C.; Chen, S.; Cheng, Y.; Yuan, J.S.; Jiang, D.; He, P.; Shan, L. Phosphorylation of Trihelix Transcriptional Repressor ASR3 by MAP KINASE4 Negatively Regulates Arabidopsis Immunity. Plant Cell 2015, 27, 839–856. [Google Scholar] [CrossRef]
- Völz, R.; Kim, S.-K.; Mi, J.; Mariappan, K.G.; Guo, X.; Bigeard, J.; Alejandro, S.; Pflieger, D.; Rayapuram, N.; Al-Babili, S.; et al. The Trihelix transcription factor GT2-like 1 (GTL1) promotes salicylic acid metabolism, and regulates bacterial-triggered immunity. PLoS Genet. 2018, 14, e1007708. [Google Scholar] [CrossRef]
- Wu, T.; Yang, Q.; Zhou, R.; Yu, T.; Shen, S.; Cao, R.; Ma, X.; Song, X. Large-scale analysis of trihelix transcription factors reveals their expansion and evolutionary footprint in plants. Physiol. Plant. 2023, 175, e14039. [Google Scholar] [CrossRef]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Li, F.-W.; Kramer, E.M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef] [PubMed]
- Romanova, M.A.; Maksimova, A.I.; Pawlowski, K.; Voitsekhovskaja, O.V. YABBY Genes in the Development and Evolution of Land Plants. Int. J. Mol. Sci. 2021, 22, 4139. [Google Scholar] [CrossRef] [PubMed]
- Kayani, S.-I.; Shen, Q.; Rahman, S.-U.; Fu, X.; Li, Y.; Wang, C.; Hassani, D.; Tang, K. Transcriptional regulation of flavonoid biosynthesis in Artemisia annua by AaYABBY5. Hortic. Res. 2021, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-W.; Nishiyama, T.; Waller, M.; Frangedakis, E.; Keller, J.; Li, Z.; Fernandez-Pozo, N.; Barker, M.S.; Bennett, T.; Blázquez, M.A.; et al. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 2020, 6, 259–272. [Google Scholar] [CrossRef]
- Worden, A.Z.; Lee, J.-H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green Evolution and Dynamic Adaptations Revealed by Genomes of the Marine Picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef]
- McKinnie, L.J.; Cummins, S.F.; Zhao, M. Identification of Incomplete Annotations of Biosynthesis Pathways in Rhodophytes Using a Multi-Omics Approach. Mar. Drugs 2024, 22, 3. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2013, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An Arabidopsis Transcriptional Regulatory Map Reveals Distinct Functional and Evolutionary Features of Novel Transcription Factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Zeke, A.; Lazar, T.; Glavina, J.; Nagy-Kanta, E.; Donagh, J.M.; Kalman, Z.E.; Pascarelli, S.; et al. ELM-the Eukaryotic Linear Motif resource-2024 update. Nucleic Acids Res. 2024, 52, D442–D455. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Mészáros, B.; Sámano-Sánchez, H.; Zeke, A.; Dobson, L.; Lazar, T.; Örd, M.; Nagpal, A.; et al. The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Res. 2022, 50, D497–D508. [Google Scholar] [CrossRef]
- Dinkel, H.; Van Roey, K.; Michael, S.; Kumar, M.; Uyar, B.; Altenberg, B.; Milchevskaya, V.; Schneider, M.; Kühn, H.; Behrendt, A.; et al. ELM 2016—Data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016, 44, D294–D300. [Google Scholar] [CrossRef]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef]
- Wang, L.; Mo, Z.; Yu, X.; Mao, Y. Characterization of the basic leucine zipper transcription factor family of Neoporphyra haitanensis and its role in acclimation to dehydration stress. BMC Plant Biol. 2023, 23, 617. [Google Scholar] [CrossRef]
- Collén, J.; Porcel, B.; Carré, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K.; et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, D.; Li, J.; Jing, G.; Ning, K.; Xu, J. Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 2014, 4, 5454. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Hayes, R.D.; Calhoun, S.; Kamel, B.; Wang, A.; Ahrendt, S.; Dusheyko, S.; Nikitin, R.; Mondo Stephen, J.; Salamov, A.; et al. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res. 2020, 49, D1004–D1011. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Wan, C.; Li, C.; Ma, X.; Wang, Y.; Sun, C.; Huang, R.; Zhong, P.; Gao, Z.; Chen, D.; Xu, Z.; et al. GRY79 encoding a putative metallo-β-lactamase-trihelix chimera is involved in chloroplast development at early seedling stage of rice. Plant Cell Rep. 2015, 34, 1353–1363. [Google Scholar] [CrossRef]
- Han, H.Q.; Liu, Y.; Jiang, M.M.; Ge, H.Y.; Chen, H.Y. Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum L.). Genet. Mol. Res. 2015, 14, 7079–7091. [Google Scholar] [CrossRef]
- Rossoni, A.W.; Price, D.C.; Seger, M.; Lyska, D.; Lammers, P.; Bhattacharya, D.; Weber, A.P.M. The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. eLife 2019, 8, e45017. [Google Scholar] [CrossRef]
- Qiu, H.; Price, D.C.; Yang, E.C.; Yoon, H.S.; Bhattacharya, D. Evidence of ancient genome reduction in red algae (Rhodophyta). J. Phycol. 2015, 51, 624–636. [Google Scholar] [CrossRef]
- Brawley, S.H.; Blouin, N.A.; Ficko-Blean, E.; Wheeler, G.L.; Lohr, M.; Goodson, H.V.; Jenkins, J.W.; Blaby-Haas, C.E.; Helliwell, K.E.; Chan, C.X.; et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. USA 2017, 114, E6361–E6370. [Google Scholar] [CrossRef]
- Malgapo, M.I.P.; Safadi, J.M.; Linder, M.E. Metallo-β-lactamase domain-containing protein 2 is S-palmitoylated and exhibits acyl-CoA hydrolase activity. J. Biol. Chem. 2021, 296, 100106. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Zhou, D. ADAM metallopeptidase domain 10 knockdown enables podocytes to resist high glucose stimulation by inhibiting pyroptosis via MAPK pathway. Exp. Ther. Med. 2023, 25, 260. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Zhou, X.; Huang, H.; Wu, D.; Shao, J.; Zhan, R.; Chen, L. Identification of trihelix transcription factors in Pogostemon cablin reveals PatGT-1 negatively regulates patchoulol biosynthesis. Ind. Crops Prod. 2021, 161, 113182. [Google Scholar] [CrossRef]
- Ambrosino, A.; Chianese, A.; Zannella, C.; Piccolella, S.; Pacifico, S.; Giugliano, R.; Franci, G.; De Natale, A.; Pollio, A.; Pinto, G.; et al. Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds. Mar. Drugs 2023, 21, 383. [Google Scholar] [CrossRef]
- Gürlek, C.; Yarkent, Ç.; Köse, A.; Tuğcu, B.; Gebeloğlu, I.K.; Öncel, S.Ş.; Elibol, M. Screening of antioxidant and cytotoxic activities of several microalgal extracts with pharmaceutical potential. Health Technol. 2020, 10, 111–117. [Google Scholar] [CrossRef]
- Goiris, K.; Muylaert, K.; Voorspoels, S.; Noten, B.; De Paepe, D.; E Baart, G.J.; De Cooman, L. Detection of flavonoids in microalgae from different evolutionary lineages. J. Phycol. 2014, 50, 483–492. [Google Scholar] [CrossRef]
- Del Mondo, A.; Sansone, C.; Brunet, C. Insights into the biosynthesis pathway of phenolic compounds in microalgae. Comput. Struct. Biotechnol. J. 2022, 20, 1901–1913. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.-Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. Leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. STAG: Species Tree Inference from All Genes. bioRxiv 2018. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. STRIDE: Species Tree Root Inference from Gene Duplication Events. Mol. Biol. Evol. 2017, 34, 3267–3278. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kelly, S.; Maini, P.K. DendroBLAST: Approximate Phylogenetic Trees in the Absence of Multiple Sequence Alignments. PLoS ONE 2013, 8, e58537. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
| Query | Name | Accession | Interval | E-Value | Coverage % |
|---|---|---|---|---|---|
| A_taxiformis_SC_g544.t1 | GT1 | cd12203 | 9–75 | 1.12 × 10−18 | 14.44201 |
| A_taxiformis_SC_g544.t1 | 2A1904 super family | cl36772 | 255–443 | 4.59 × 10−3 | 41.13786 |
| A_taxiformis_Cali_g514.t1 | GT1 | cd12203 | 9–75 | 7.88 × 10−19 | 14.44201 |
| A_taxiformis_Cali_g514.t1 | 2A1904 super family | cl36772 | 255–443 | 1.90 × 10−4 | 41.13786 |
| K_alvarezii_kp_g64.t1 | Myb_DNA-bind_4 | pfam13837 | 9–94 | 8.37 × 10−16 | 24.14773 |
| K_alvarezii_kp_g64.t1 | 2A1904 super family | cl36772 | 202–342 | 3.46 × 10−5 | 39.77273 |
| A_taxiformis_Guam_g944.t1 | GT1 | cd12203 | 6–72 | 1.11 × 10−18 | 14.53744 |
| scaffold-IHJY-2001380-Kappaphycus_alvarezii | Myb_DNA-bind_4 | pfam13837 | 11–96 | 1.98 × 10−15 | 81.73077 |
| scaffold-IHJY-2001381-Kappaphycus_alvarezii | Myb_DNA-bind_4 | pfam13837 | 79–164 | 4.60 × 10−18 | 49.4186 |
| scaffold-UGPM-2004273-Chondrus_crispus | GT1 | cd12203 | 18–82 | 3.39 × 10−17 | 60.37736 |
| scaffold-JEBK-2003052-Eucheuma_denticulatum | GT1 | cd12203 | 81–145 | 2.81 × 10−17 | 60.37736 |
| scaffold-CKXF-2019441-Ahnfeltiopsis_flabelliformis | GT1 | cd12203 | 1–65 | 4.54 × 10−16 | 71.91011 |
| scaffold-ZJOJ-2006450-Grateloupia_filicina | GT1 | cd12203 | 38–104 | 2.21 × 10−15 | 51.9685 |
| scaffold-BWVJ-2022936-Betaphycus_philippinensis | Myb_DNA-bind_4 | pfam13837 | 73–158 | 1.35 × 10−18 | 51.20482 |
| scaffold-IKIZ-2012078-Grateloupia_livida | GT1 super family | cl23759 | 1–65 | 7.35 × 10−15 | 72.72727 |
| scaffold-WEJN-2025014-Mazzaella_japonica | GT1 | cd12203 | 18–82 | 3.39 × 10−17 | 60.37736 |
| A_taxiformis_Guam_g3848.t1 | SANT super family | cl21498 | 17–95 | 8.67 × 10−6 | 65.54622 |
| A_taxiformis_Guam_g3848.t1 | SANT | smart00717 | 84–119 | 4.10 × 10−3 | 29.41176 |
| Query | Name | Accession # | Interval | E-Value |
|---|---|---|---|---|
| Ata00544 | GT1 | cd12203 | 9–75 | 1.12 × 10−18 |
| Ata00544 | 2A1904 super family | cl36772 | 255–443 | 4.59 × 10−3 |
| A_taxiformis_Cali_g514.t1 | GT1 | cd12203 | 9–75 | 7.88 × 10−19 |
| A_taxiformis_Cali_g514.t1 | 2A1904 super family | cl36772 | 255–443 | 1.90 × 10−4 |
| K_alvarezii_kp_g64.t1 | Myb_DNA-bind_4 | pfam13837 | 9–94 | 8.37 × 10−16 |
| K_alvarezii_kp_g64.t1 | 2A1904 super family | cl36772 | 202–342 | 3.46 × 10−5 |
| A_taxiformis_Guam_g944.t1 | GT1 | cd12203 | 6–72 | 1.11 × 10−18 |
| scaffold-IHJY-2001380-Kappaphycus_alvarezii | Myb_DNA-bind_4 | pfam13837 | 11–96 | 1.98 × 10−15 |
| scaffold-IHJY-2001381-Kappaphycus_alvarezii | Myb_DNA-bind_4 | pfam13837 | 79–164 | 4.60 × 10−18 |
| scaffold-UGPM-2004273-Chondrus_crispus | GT1 | cd12203 | 18–82 | 3.39 × 10−17 |
| scaffold-JEBK-2003052-Eucheuma_denticulatum | GT1 | cd12203 | 81–145 | 2.81 × 10−17 |
| scaffold-CKXF-2019441-Ahnfeltiopsis_flabelliformis | GT1 | cd12203 | 1–65 | 4.54 × 10−16 |
| scaffold-ZJOJ-2006450-Grateloupia_filicina | GT1 | cd12203 | 38–104 | 2.21 × 10−15 |
| scaffold-BWVJ-2022936-Betaphycus_philippinensis | Myb_DNA-bind_4 | pfam13837 | 73–158 | 1.35 × 10−18 |
| scaffold-IKIZ-2012078-Grateloupia_livida | GT1 super family | cl23759 | 1–65 | 7.35 × 10−15 |
| scaffold-WEJN-2025014-Mazzaella_japonica | GT1 | cd12203 | 18–82 | 3.39 × 10−17 |
| A_taxiformis_Guam_g3848.t1 | SANT super family | cl21498 | 17–95 | 8.67 × 10−6 |
| A_taxiformis_Guam_g3848.t1 | SANT | smart00717 | 84–119 | 4.10 × 10−3 |
| Orthologue | Description | Tree Length | Phylogenetic Diversity | Mean Entropy |
|---|---|---|---|---|
| OG0001075 | 20S core proteasome subunit alpha 1 | 2.04459 | 3.81446 | 0.712415 |
| OG0002371 | Trihelix | 1.737687 | 4.060484 | 0.789495 |
| OG0001145 | CCR4-NOT transcription complex subunit | 1.822181 | 4.154696 | 0.965758 |
| OG0003140 | T-complex protein 1 subunit | 2.460099 | 4.209469 | 0.589745 |
| OG0003182 | DNA-directed RNA polymerase | 1.94762 | 4.402117 | 0.682362 |
| OG0003030 | GTP-binding family protein | 2.570532 | 4.42165 | 0.765911 |
| OG0001141 | Pyruvate dehydrogenase E1 component alpha subunit | 2.500867 | 4.43349 | 0.842321 |
| OG0001163 | Signal recognition particle, SRP54 subunit protein | 2.582355 | 4.488274 | 0.940031 |
| OG0001156 | vacuolar ATP synthase subunit A | 2.093928 | 4.580014 | 0.893024 |
| OG0002704 | carbamoyl phosphate synthetase B | 2.904098 | 4.607873 | 0.80962 |
| OG0001079 | Ubiquitin thioesterase OTU1 | 2.502898 | 4.654698 | 0.874793 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McKinnie, L.J.; Cummins, S.F.; Subramanian, S.; Zhao, M. Transcription Factor Analysis of Rhodophytes Suggests Trihelix Transcription Factors Across the Florideophyceae. Plants 2025, 14, 3143. https://doi.org/10.3390/plants14203143
McKinnie LJ, Cummins SF, Subramanian S, Zhao M. Transcription Factor Analysis of Rhodophytes Suggests Trihelix Transcription Factors Across the Florideophyceae. Plants. 2025; 14(20):3143. https://doi.org/10.3390/plants14203143
Chicago/Turabian StyleMcKinnie, Lachlan J., Scott F. Cummins, Sankar Subramanian, and Min Zhao. 2025. "Transcription Factor Analysis of Rhodophytes Suggests Trihelix Transcription Factors Across the Florideophyceae" Plants 14, no. 20: 3143. https://doi.org/10.3390/plants14203143
APA StyleMcKinnie, L. J., Cummins, S. F., Subramanian, S., & Zhao, M. (2025). Transcription Factor Analysis of Rhodophytes Suggests Trihelix Transcription Factors Across the Florideophyceae. Plants, 14(20), 3143. https://doi.org/10.3390/plants14203143







