Integrated Multi-Omics Analysis Reveals the Survival Strategy of Dongxiang Wild Rice (DXWR, Oryza rufipogon Griff.) Under Low-Temperature and Anaerobic Stress
Abstract
1. Introduction
2. Results
2.1. Germination Characteristics of DXWR Under Low-Temperature and Anaerobic Conditions
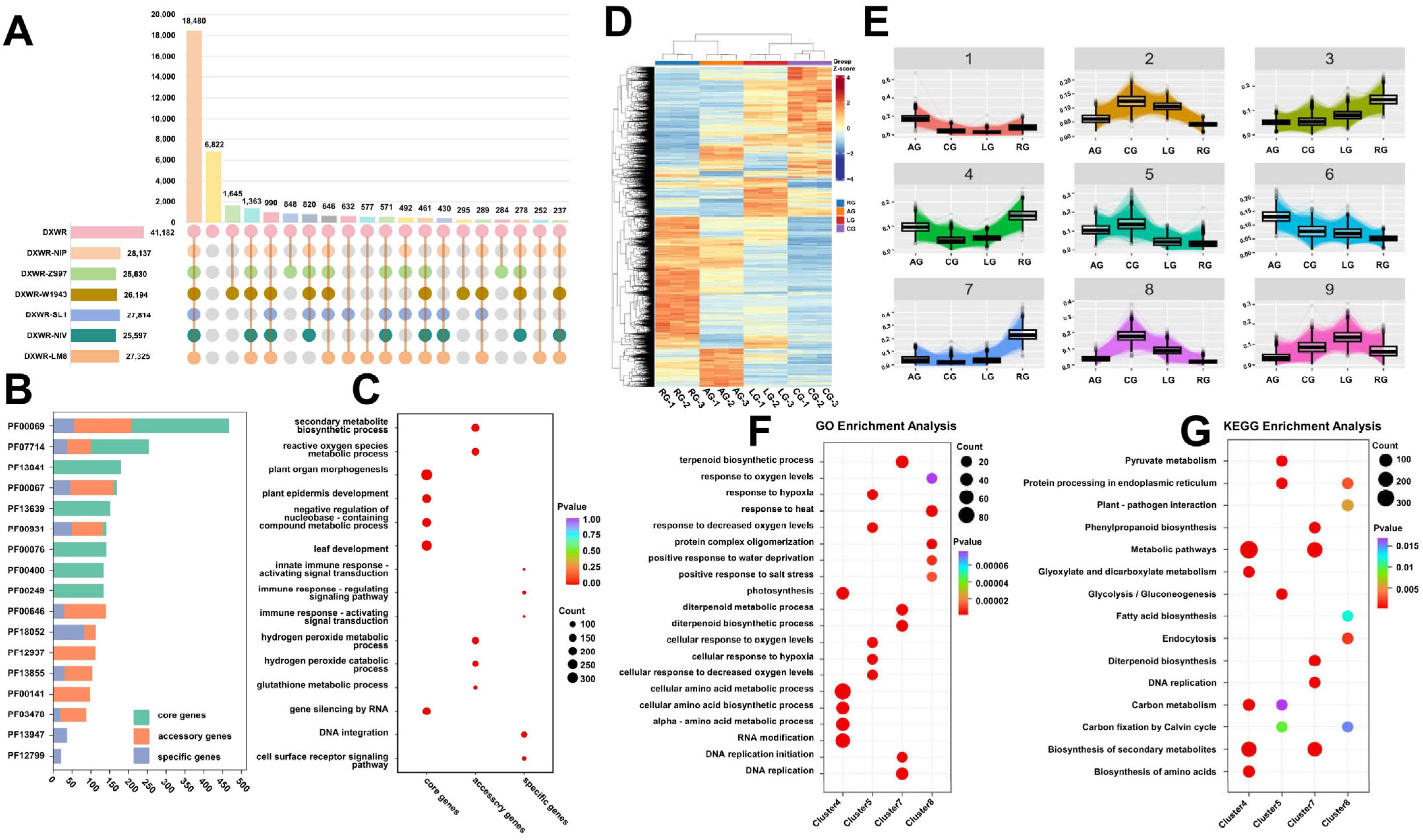
2.2. Gene Characteristics and Transcriptomic Responses of DXWR Under Stress
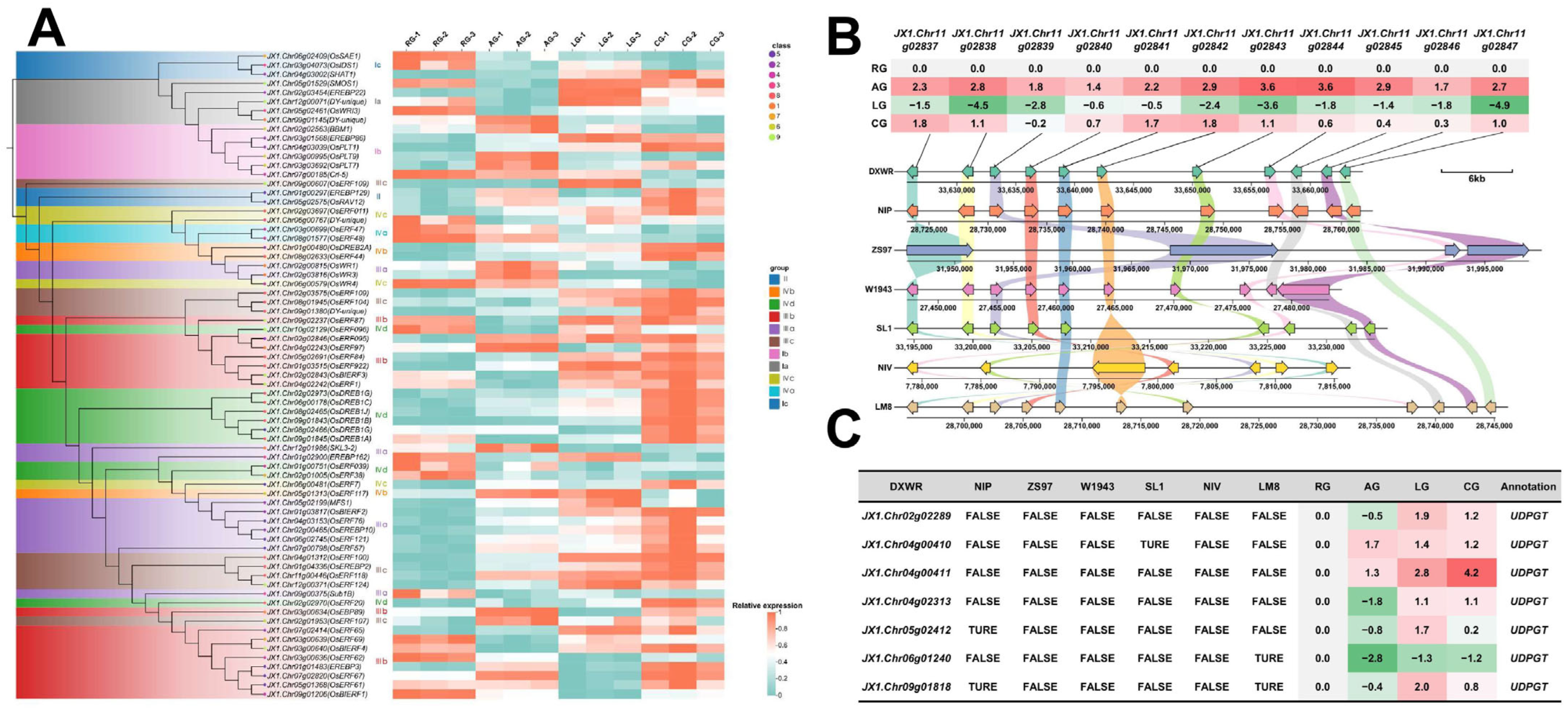
2.3. Multi-Layered Gene Co-Regulation Mechanism of Low-Temperature and Anaerobic Composite Stress During Germination of DXWR
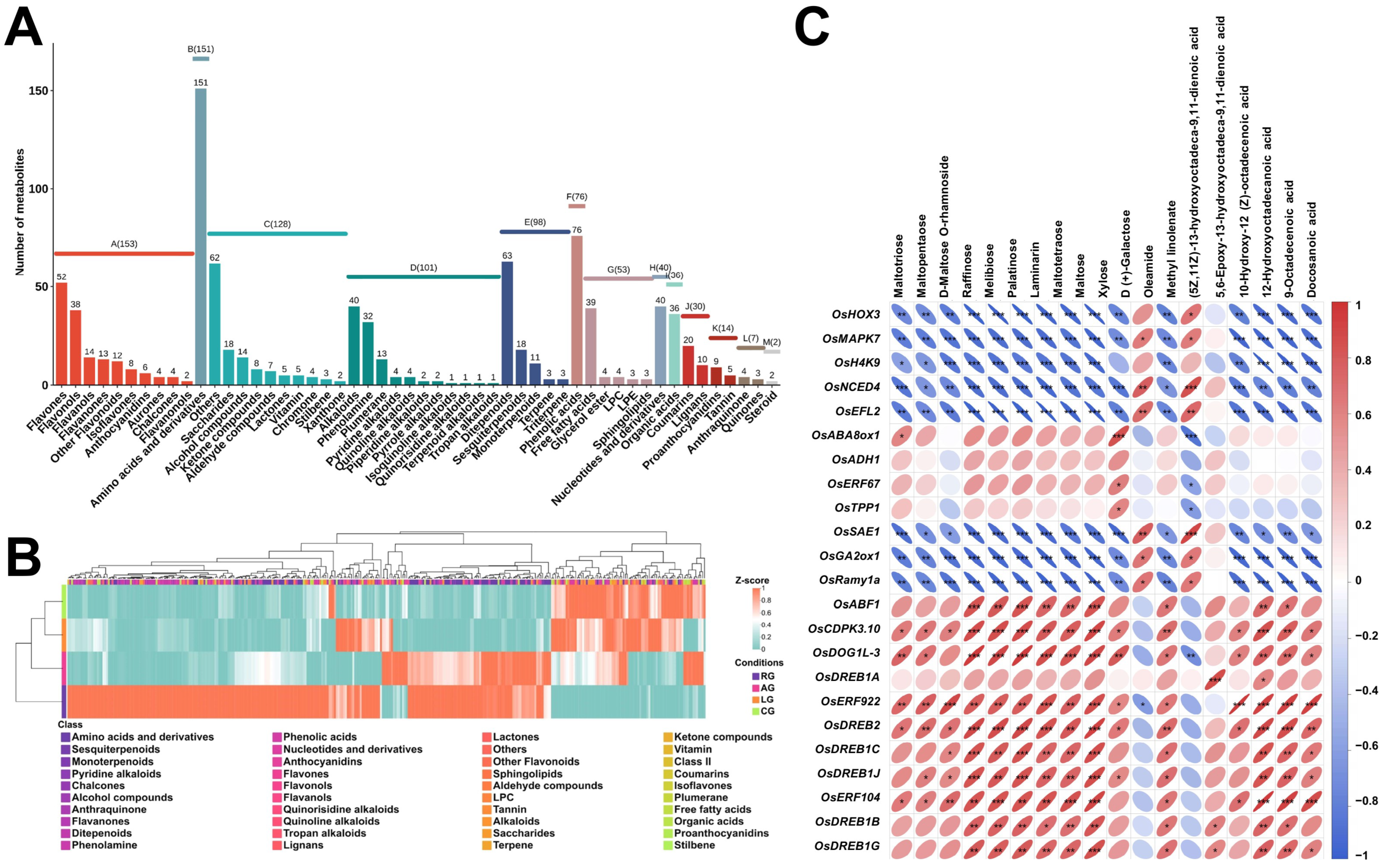
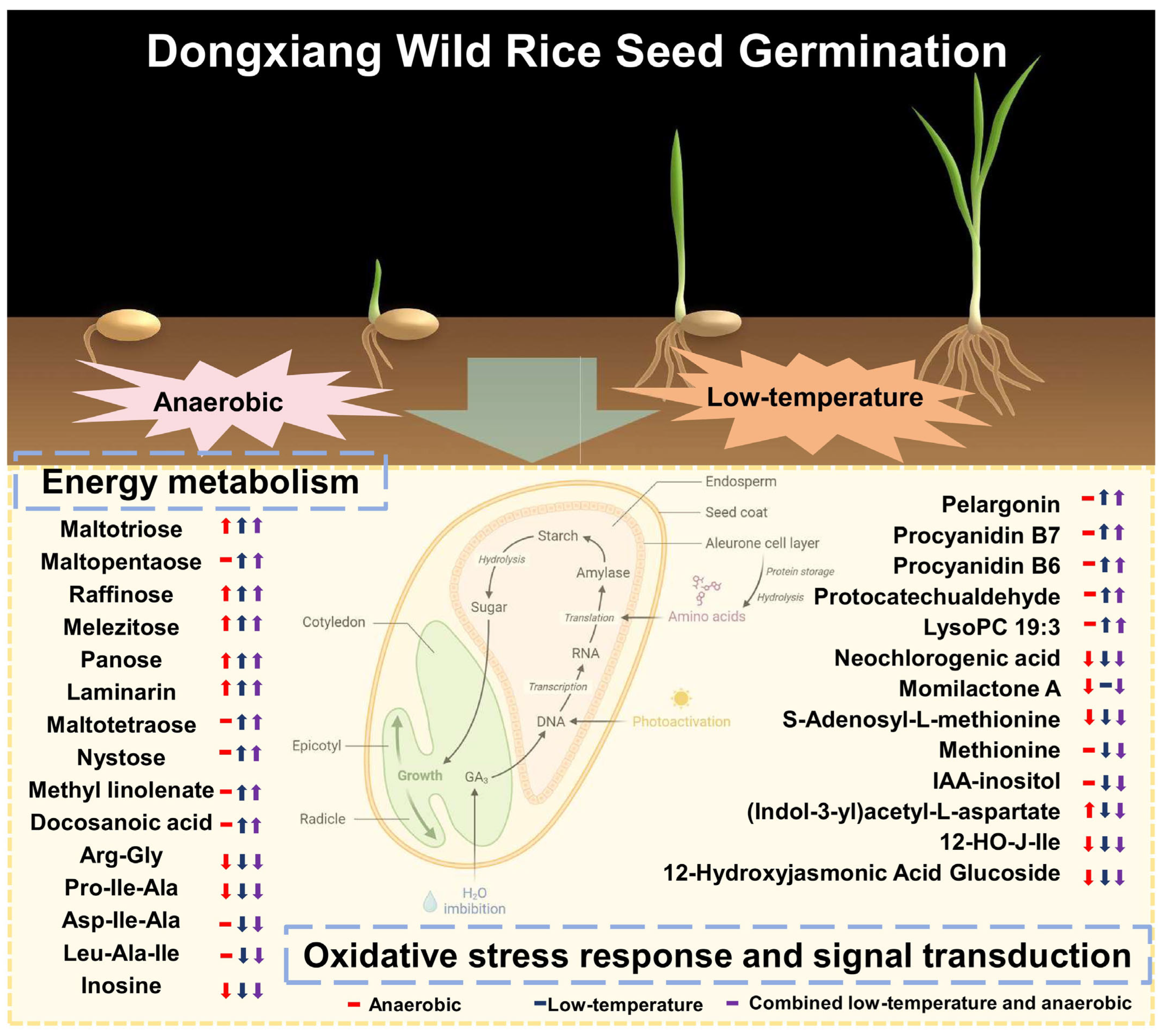
2.4. Differential Accumulation and Regulatory Patterns of Stress-Responsive Metabolites
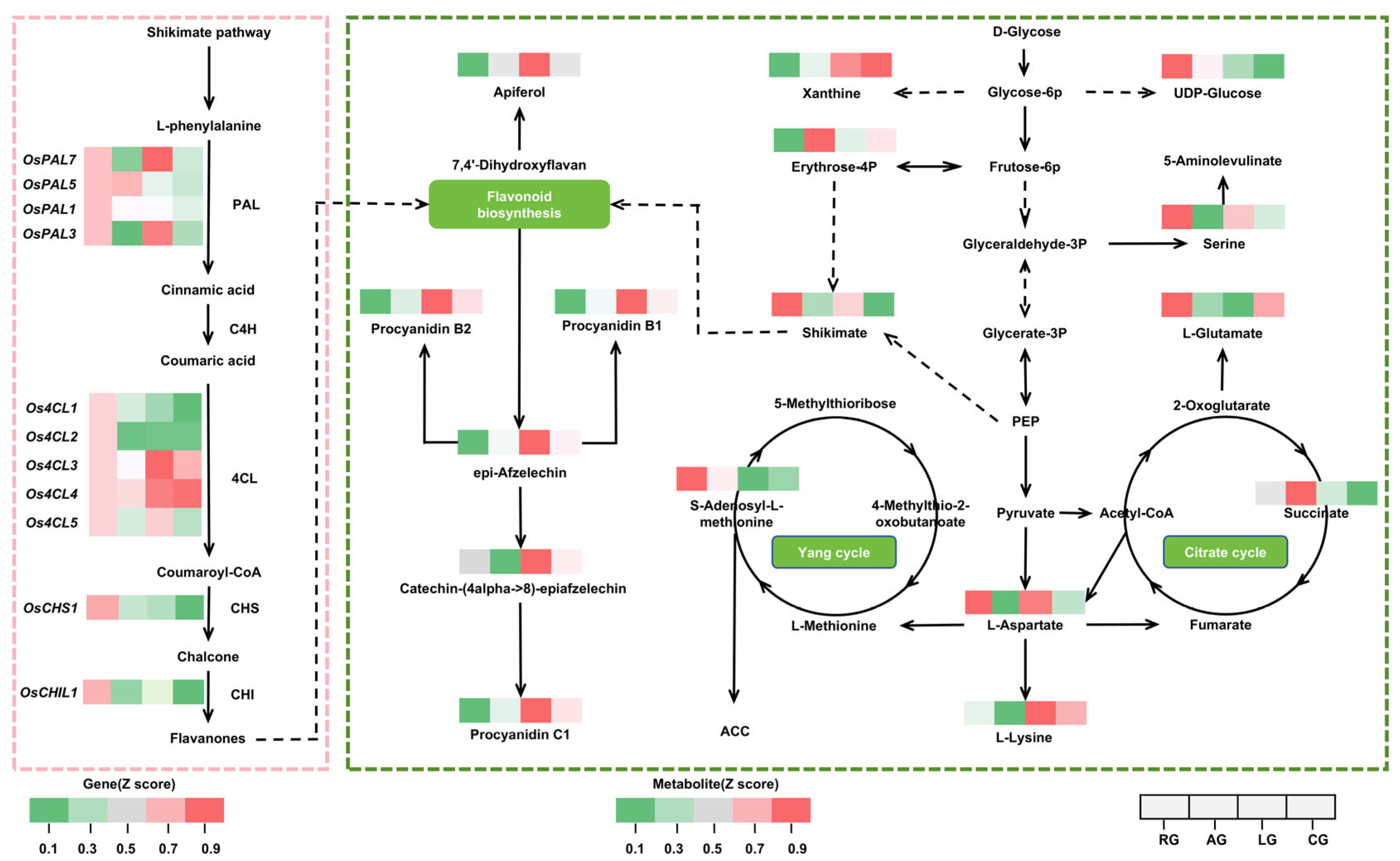
2.5. Coordinated Environmental Regulation of Secondary Metabolism
3. Discussion
3.1. A Hierarchical Genetic Framework: The Genetic Basis of DXWR’s Extreme Stress Tolerance
3.2. Metabolic Reprogramming: Synergistic Energy Optimization and Antioxidant Defense
3.3. Gene–Metabolite Networks and Hormonal Crosstalk: Fine-Tuning the Growth–Defense Balance
3.4. Implications for Breeding Climate-Adapted Direct-Seeded Rice
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Germination Treatments and Experimental Design
4.3. Germination Kinetics and Phenotypic Analysis
4.4. Sample Collection for Multi-Omics Analysis
4.5. Orthologous Gene Identification
4.6. RNA-Seq Analysis: Sampling, Library Preparation, Sequencing, and Bioinformatics
4.7. Metabolomic Profiling and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DXWR | Dongxiang Wild Rice |
| RG | Room-temperature aerobic germination |
| AG | Low-temperature aerobic germination |
| CG | Combined low-temperature and anaerobic germination |
| UDPGT | UDP-Glycosyltransferase |
| GH18 | Glycoside hydrolases 18 |
References
- Fujino, K.; Sekiguchi, H.; Sato, T.; Kiuchi, H.; Nonoue, Y.; Takeuchi, Y.; Ando, T.; Lin, S.Y.; Yano, M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 2004, 108, 794–799. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Yang, T.; Zeng, Z.; Huang, Z.; Liu, Q.; Wang, X.; Leach, J.; Leung, H.; Liu, B. Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol. Plant 2015, 154, 381–394. [Google Scholar] [CrossRef]
- Ismail, A.M.; Johnson, D.E.; Ella, E.S.; Vergara, G.V.; Baltazar, A.M. Adaptation to flooding during emergence and seedling growth in rice and weeds, and implications for crop establishment. AoB Plants 2012, 2012, pls019. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shahid, M.Q.; Bai, L.; Lu, Z.; Chen, Y.; Jiang, L.; Diao, M.; Liu, X.; Lu, Y. Evaluation of Genetic Diversity and Development of a Core Collection of Wild Rice (Oryza rufipogon Griff.) Populations in China. PLoS ONE 2015, 10, e0145990. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, T.; Mao, L.; Yan, S.; Chen, X.; Wu, Z.; Chen, R.; Luo, X.; Xie, J.; Gao, S. Genome-wide analysis of Dongxiang wild rice (Oryza rufipogon Griff.) to investigate lost/acquired genes during rice domestication. BMC Plant Biol. 2016, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 2007, 144, 218–231. [Google Scholar] [CrossRef]
- Ucker, D.S. Exploiting death: Apoptotic immunity in microbial pathogenesis. Cell Death Differ. 2016, 23, 990–996. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Pelayo, M.A.; Trijatmiko, K.R.; Gabunada, L.F.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.H.; Sreenivasulu, N.; Bailey-Serres, J.; et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124. [Google Scholar] [CrossRef]
- Hoffmann-Benning, S.; Kende, H. On the role of abscisic Acid and gibberellin in the regulation of growth in rice. Plant Physiol. 1992, 99, 1156–1161. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, G.; Cui, Z.; Kong, X.; Yu, X.; Gui, R.; Han, Y.; Li, Z.; Lang, H.; Hua, Y.; et al. Regain flood adaptation in rice through a 14-3-3 protein OsGF14h. Nat. Commun. 2022, 13, 5664. [Google Scholar] [CrossRef]
- He, Y.; Sun, S.; Zhao, J.; Huang, Z.; Peng, L.; Huang, C.; Tang, Z.; Huang, Q.; Wang, Z. UDP-glucosyltransferase OsUGT75A promotes submergence tolerance during rice seed germination. Nat. Commun. 2023, 14, 2296. [Google Scholar] [CrossRef]
- Miyashita, Y.; Good, A.G. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 92–102. [Google Scholar] [CrossRef]
- Chatterjee, Y.; Tomar, S.; Mishra, M.; Pareek, A.; Singla-Pareek, S.L. OsLdh7 Overexpression in Rice Confers Submergence Tolerance by Regulating Key Metabolic Pathways: Anaerobic Glycolysis, Ethanolic Fermentation and Amino Acid Metabolism. Plant Cell Environ. 2025, 48, 2804–2820. [Google Scholar] [CrossRef]
- Fujino, K.; Matsuda, Y. Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Mol. Biol. 2010, 72, 137–152. [Google Scholar] [CrossRef]
- Shim, K.C.; Kim, S.H.; Lee, H.S.; Adeva, C.; Jeon, Y.A.; Luong, N.H.; Kim, W.J.; Akhtamov, M.; Park, Y.J.; Ahn, S.N. Characterization of a New qLTG3-1 Allele for Low-temperature Germinability in Rice from the Wild Species Oryza rufipogon. Rice 2020, 13, 10. [Google Scholar] [CrossRef]
- Wang, W.; Huang, R.; Wu, G.; Sun, J.; Zhu, Y.; Wang, H. Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6. Plants 2023, 12, 871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, H.; Tian, X.; Lin, X.; Han, Y.; Han, Z.; Sha, H.; Liu, J.; Liu, J.; Zhang, J.; et al. A transposon insertion in the promoter of OsUBC12 enhances cold tolerance during japonica rice germination. Nat. Commun. 2024, 15, 2211. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, S.; Yuan, X.; Chen, J.; Tian, M.; Zhao, Z.; Guo, T.; Xiao, W. OsNAL11 and OsBURP12 Affect Rice Seed Germination at Low Temperature. Plant Cell Environ. 2025, 48, 6118–6139. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Teng, Z.; Liu, B.; Lv, J.; Chen, Y.; Qin, Z.; Peng, Y.; Meng, S.; He, Y.; Duan, M.; et al. Transcription factor OsMYB30 increases trehalose content to inhibit alpha-amylase and seed germination at low temperature. Plant Physiol. 2024, 194, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, J.; Su, L.; Sun, K.; Li, D.; Liu, Y.; Wang, H.; Chen, Z.; Guo, T. Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress. Plant Sci. 2019, 289, 110282. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, D.; Chen, H.; Deng, W.; Chen, D.; Chen, P.; Wang, J. Genome-Wide Identification of DUF26 Domain-Containing Genes in Dongxiang Wild Rice and Analysis of Their Expression Responses under Submergence. Curr. Issues Mol. Biol. 2022, 44, 3351–3363. [Google Scholar] [CrossRef]
- Wang, D.; Xiao, Y.; Chen, H.; Huang, C.; Chen, P.; Chen, D.; Deng, W.; Wang, J. Combination of Genomics, Transcriptomics Identifies Candidate Loci Related to Cold Tolerance in Dongxiang Wild Rice. Plants 2022, 11, 2329. [Google Scholar] [CrossRef]
- Huang, C.; Wang, J.; Wang, D.; Chang, J.; Chen, H.; Chen, D.; Deng, W.; Tian, C. Genome-Wide Identification and Analysis of OsSPXs Revealed Its Genetic Influence on Cold Tolerance of Dongxiang Wild Rice (DXWR). Int. J. Mol. Sci. 2023, 24, 8755. [Google Scholar] [CrossRef]
- Wang, J.; Huang, C.; Tang, L.; Chen, H.; Chen, P.; Chen, D.; Wang, D. Identification of Submergence Tolerance Loci in Dongxiang Wild Rice (DXWR) by Genetic Linkage and Transcriptome Analyses. Int. J. Mol. Sci. 2025, 26, 1829. [Google Scholar] [CrossRef]
- Wang, L.; Wang, R.; Cai, X.; Zheng, H.; Huang, Y.; Li, Y.; Cui, M.; Lin, M.; Tang, H. A loss-of-function mutation in OsTZF5 confers sensitivity to low temperature and effects the growth and development in rice. Plant Mol. Biol. 2024, 114, 116. [Google Scholar] [CrossRef]
- Bing, H.; Gu, J.; Xia, B.; Kong, X.; Luo, Y.; Wang, X.; Liu, C.; Zhao, J.; Xiang, W. Endophytic fungus Stagonosporopsis ajaci NEAU-BLH1 from Adonis amurensis enhances seed germination under low-temperature stress and increases grain yield in direct-seeded rice. Microbiol. Res. 2025, 295, 128111. [Google Scholar] [CrossRef]
- Zhang, P.; Yu, H.; Huang, Z.; Yang, P.; Li, H.; Huang, G.; Tang, L.; Zhong, Z.; Hu, G.; Yu, G.; et al. Combined Analysis of Transcriptome and Metabolome Reveals the Heat Stress Resistance of Dongxiang Wild Rice at Seedling Stage. Plants 2025, 14, 1192. [Google Scholar] [CrossRef]
- Deng, Q.W.; Luo, X.D.; Chen, Y.L.; Zhou, Y.; Zhang, F.T.; Hu, B.L.; Xie, J.K. Transcriptome analysis of phosphorus stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.). Biol. Res. 2018, 51, 7. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhu, X.; Zhang, J.; Shen, M.; Chen, K.; Fu, X.; Ma, L.; Liu, X.; Zhou, C.; Zhou, D.X.; et al. eQTLs play critical roles in regulating gene expression and identifying key regulators in rice. Plant Biotechnol. J. 2022, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Yao, J.; Chu, Q.; Guo, X.; Shao, W.; Shang, N.; Luo, K.; Li, X.; Chen, H.; Cheng, Q.; Mo, F.; et al. Spatiotemporal transcriptomic landscape of rice embryonic cells during seed germination. Dev. Cell 2024, 59, 2320–2332.e5. [Google Scholar] [CrossRef]
- Wang, N.; Cao, S.; Liu, Z.; Xiao, H.; Hu, J.; Xu, X.; Chen, P.; Ma, Z.; Ye, J.; Chai, L.; et al. Genomic conservation of crop wild relatives: A case study of citrus. PLoS Genet. 2023, 19, e1010811. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Liu, W.; Zhao, Y. Harness the wild: Progress and perspectives in wheat genetic improvement. J. Genet. Genom. 2025. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pang, H.; Wang, J.; Yao, X.; Song, Y.; Li, F.; Lou, D.; Ge, J.; Zhao, Z.; Qiao, W.; et al. Genomic signatures of domestication and adaptation during geographical expansions of rice cultivation. Plant Biotechnol. J. 2022, 20, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef]
- Zubrycka, A.; Dambire, C.; Dalle Carbonare, L.; Sharma, G.; Boeckx, T.; Swarup, K.; Sturrock, C.J.; Atkinson, B.S.; Swarup, R.; Corbineau, F.; et al. ERFVII action and modulation through oxygen-sensing in Arabidopsis thaliana. Nat. Commun. 2023, 14, 4665. [Google Scholar] [CrossRef]
- Maruyama, K.; Takeda, M.; Kidokoro, S.; Yamada, K.; Sakuma, Y.; Urano, K.; Fujita, M.; Yoshiwara, K.; Matsukura, S.; Morishita, Y.; et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009, 150, 1972–1980. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Song, S.; Ye, X.; Ding, Z.; Liu, J.; Wang, Z.; Li, J.; Hou, X.; Xu, B.; et al. MaDREB1F confers cold and drought stress resistance through common regulation of hormone synthesis and protectant metabolite contents in banana. Hortic. Res. 2023, 10, uhac275. [Google Scholar] [CrossRef]
- Lv, K.; Li, J.; Zhao, K.; Chen, S.; Nie, J.; Zhang, W.; Liu, G.; Wei, H. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 2020, 292, 110375. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Tanaka, J.; Takashima, T.; Abe, N.; Fukamizo, T.; Numata, T.; Ohnuma, T. Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins. Plant Sci. 2023, 326, 111524. [Google Scholar] [CrossRef]
- Cao, P.J.; Bartley, L.E.; Jung, K.H.; Ronald, P.C. Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol. Plant 2008, 1, 858–877. [Google Scholar] [CrossRef]
- Liao, B.; Liu, X.; Li, Y.; Ge, Y.; Liang, X.; Liao, Z.; Zhao, C.; Cao, J.; Wang, H.; Li, S.; et al. Functional Characterization of a Highly Efficient UDP-Glucosyltransferase CitUGT72AZ4 Involved in the Biosynthesis of Flavonoid Glycosides in Citrus. J. Agric. Food Chem. 2025, 73, 5450–5464. [Google Scholar] [CrossRef] [PubMed]
- Petrich, J.; Alvarez, C.E.; Gomez Cano, L.; Dewberry, R.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Functional characterization of a maize UDP-glucosyltransferase involved in the biosynthesis of flavonoid 7-O-glucosides and di-O-glucosides. Plant Physiol. Biochem. 2025, 221, 109583. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhao, S.; Ma, X.; Dong, G.; Liu, C.; Ding, Y.; Hou, B. A high temperature responsive UDP-glucosyltransferase gene OsUGT72F1 enhances heat tolerance in rice and Arabidopsis. Plant Cell Rep. 2025, 44, 48. [Google Scholar] [CrossRef]
- Blochl, A.; Peterbauer, T.; Richter, A. Inhibition of raffinose oligosaccharide breakdown delays germination of pea seeds. J. Plant Physiol. 2007, 164, 1093–1096. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. ZmAGA1 Hydrolyzes RFOs Late during the Lag Phase of Seed Germination, Shifting Sugar Metabolism toward Seed Germination Over Seed Aging Tolerance. J. Agric. Food Chem. 2021, 69, 11606–11615. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Wang, G.; Zhang, J.; Wang, G. Analysis of gene expression in early seed germination of rice: Landscape and genetic regulation. BMC Plant Biol. 2022, 22, 70. [Google Scholar] [CrossRef]
- Cohen, H.; Pajak, A.; Pandurangan, S.; Amir, R.; Marsolais, F. Higher endogenous methionine in transgenic Arabidopsis seeds affects the composition of storage proteins and lipids. Amino Acids 2016, 48, 1413–1422. [Google Scholar] [CrossRef]
- Angelovici, R.; Fait, A.; Zhu, X.; Szymanski, J.; Feldmesser, E.; Fernie, A.R.; Galili, G. Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol. 2009, 151, 2058–2072. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, L.; Wang, H.; Guo, J.; Li, Y.; Tan, Y.; Shu, Q.; Qian, Q.; Yu, H.; Chen, Y.; et al. The phosphatidylethanolamine-binding proteins OsMFT1 and OsMFT2 regulate seed dormancy in rice. Plant Cell 2024, 36, 3857–3874. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xu, H.; Wang, Y.; Wang, H.; Li, Z.; Liu, X.; Shu, Y.; Li, G.; Liu, W.; Ying, J.; et al. OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 12220. [Google Scholar] [CrossRef] [PubMed]
- Heitz, T.; Smirnova, E.; Marquis, V.; Poirier, L. Metabolic Control within the Jasmonate Biochemical Pathway. Plant Cell Physiol. 2019, 60, 2621–2628. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Dubugnon, L.; Liechti, R.; Farmer, E.E. Jasmonate biochemical pathway. Sci. Signal 2010, 3, cm3. [Google Scholar] [CrossRef] [PubMed]
- Brunoni, F.; Pencik, A.; Zukauskaite, A.; Ament, A.; Kopecna, M.; Collani, S.; Kopecny, D.; Novak, O. Amino acid conjugation of oxIAA is a secondary metabolic regulation involved in auxin homeostasis. New Phytol. 2023, 238, 2264–2270. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and S-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar] [CrossRef]
- Narsai, R.; Howell, K.A.; Carroll, A.; Ivanova, A.; Millar, A.H.; Whelan, J. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 2009, 151, 306–322. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Cassan, O.; Lebre, S.; Martin, A. Inferring and analyzing gene regulatory networks from multi-factorial expression data: A complete and interactive suite. BMC Genom. 2021, 22, 387. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Huang, C.; Chen, H.; Tang, L.; Wang, D. Integrated Multi-Omics Analysis Reveals the Survival Strategy of Dongxiang Wild Rice (DXWR, Oryza rufipogon Griff.) Under Low-Temperature and Anaerobic Stress. Plants 2025, 14, 3120. https://doi.org/10.3390/plants14203120
Wang J, Huang C, Chen H, Tang L, Wang D. Integrated Multi-Omics Analysis Reveals the Survival Strategy of Dongxiang Wild Rice (DXWR, Oryza rufipogon Griff.) Under Low-Temperature and Anaerobic Stress. Plants. 2025; 14(20):3120. https://doi.org/10.3390/plants14203120
Chicago/Turabian StyleWang, Jilin, Cheng Huang, Hongping Chen, Lijuan Tang, and Dianwen Wang. 2025. "Integrated Multi-Omics Analysis Reveals the Survival Strategy of Dongxiang Wild Rice (DXWR, Oryza rufipogon Griff.) Under Low-Temperature and Anaerobic Stress" Plants 14, no. 20: 3120. https://doi.org/10.3390/plants14203120
APA StyleWang, J., Huang, C., Chen, H., Tang, L., & Wang, D. (2025). Integrated Multi-Omics Analysis Reveals the Survival Strategy of Dongxiang Wild Rice (DXWR, Oryza rufipogon Griff.) Under Low-Temperature and Anaerobic Stress. Plants, 14(20), 3120. https://doi.org/10.3390/plants14203120



