Comparative Transcriptome Analysis of Walnuts (Juglans regia L.) in Response to Freezing Stress
Abstract
1. Introduction
2. Results
2.1. Overview of Transcriptome Data
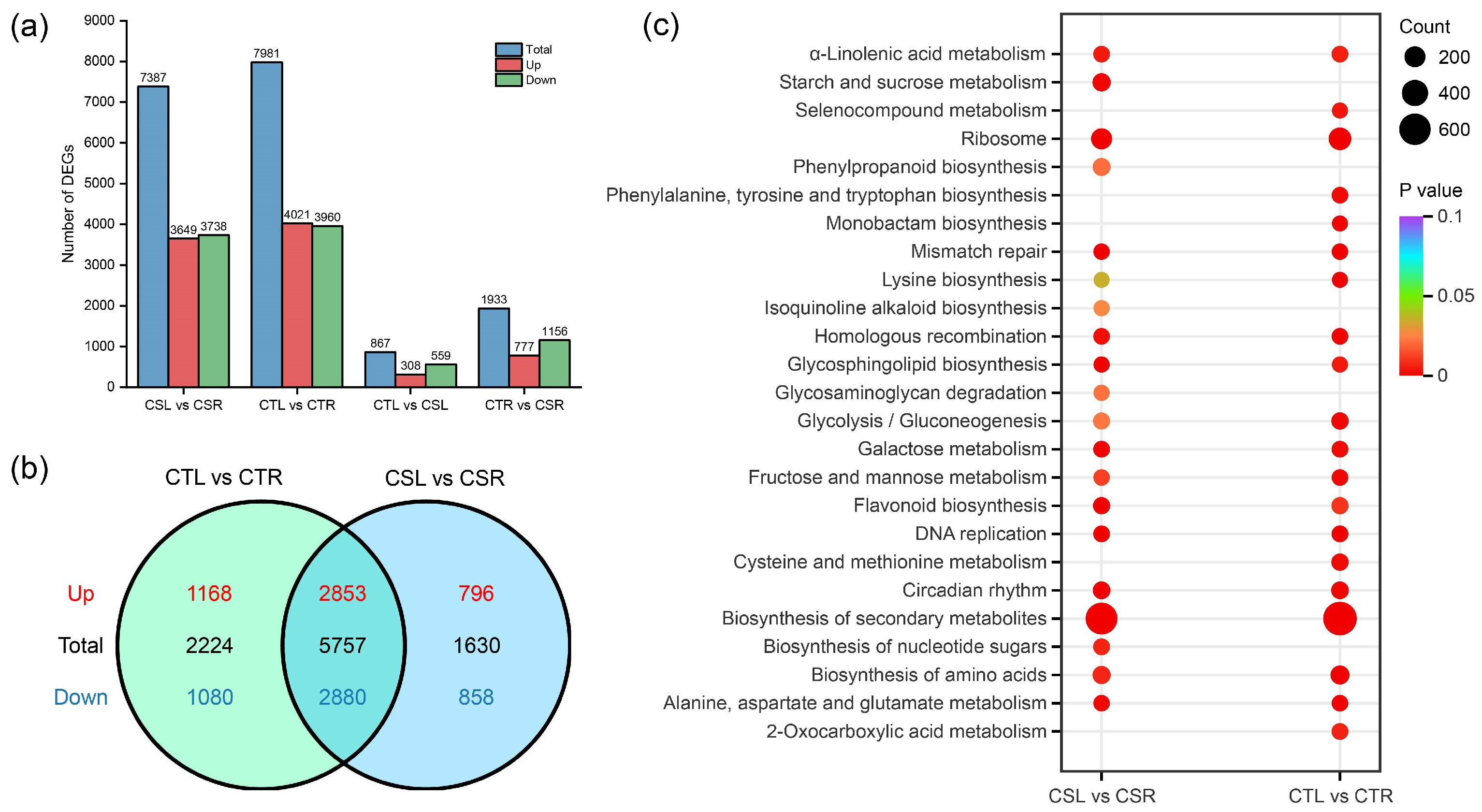
2.2. Analysis of Differentially Expressed Genes
2.3. Analysis of Genes Involved in Flavonoid Biosynthesis Under Freezing Stress
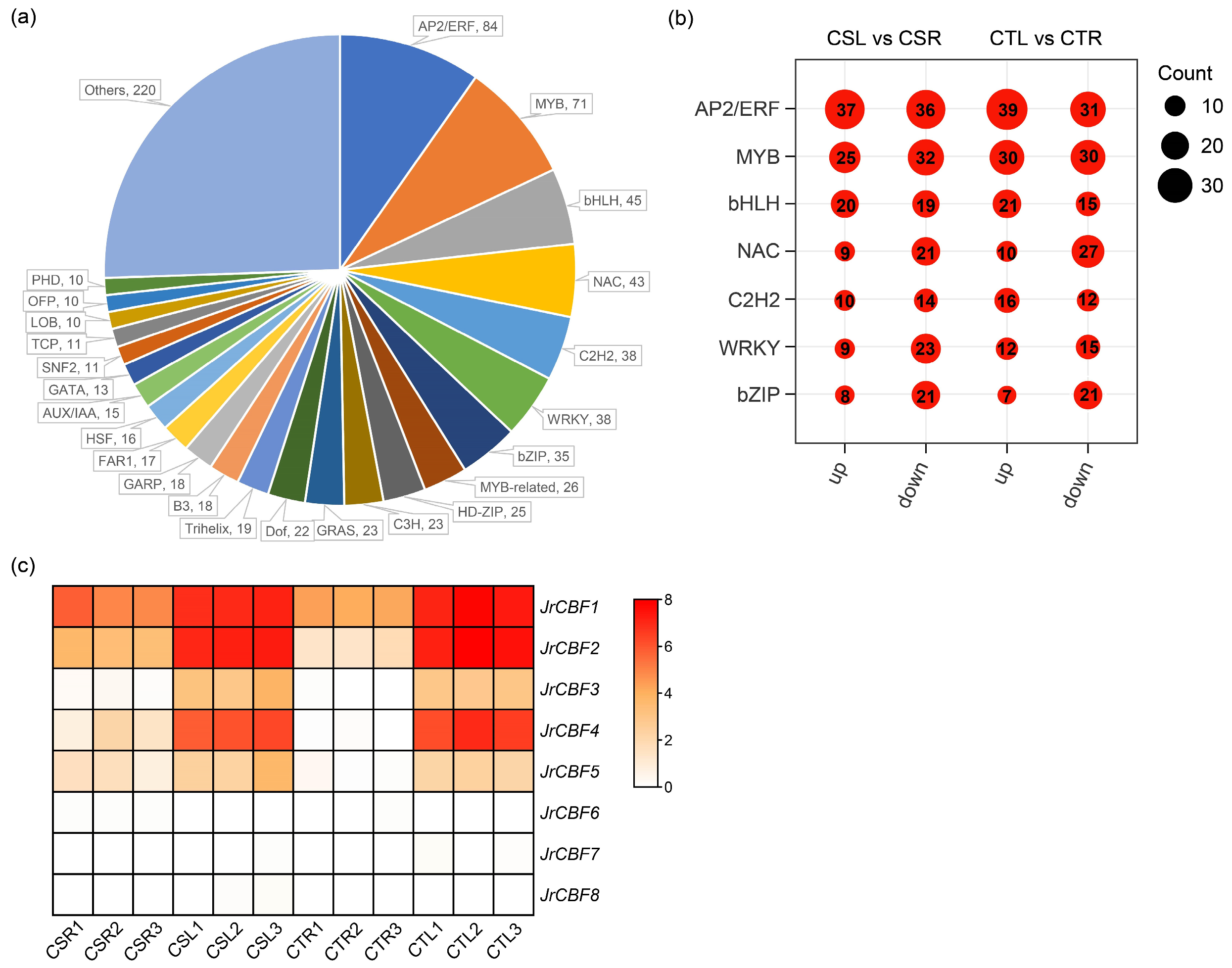
2.4. Analysis of Transcription Factors Under Freezing Stress
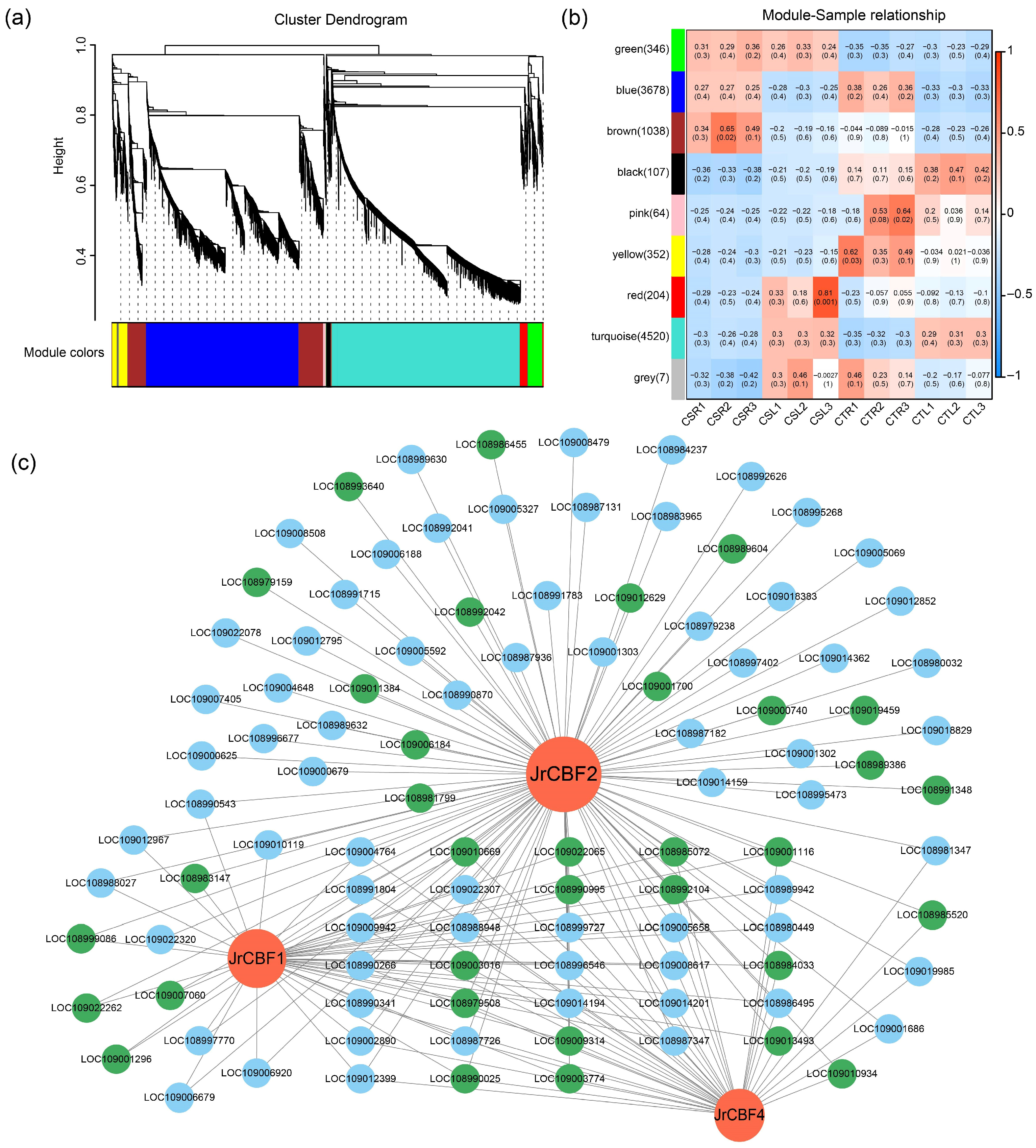
2.5. Weighted Gene Correlation Network Analysis of DEGs
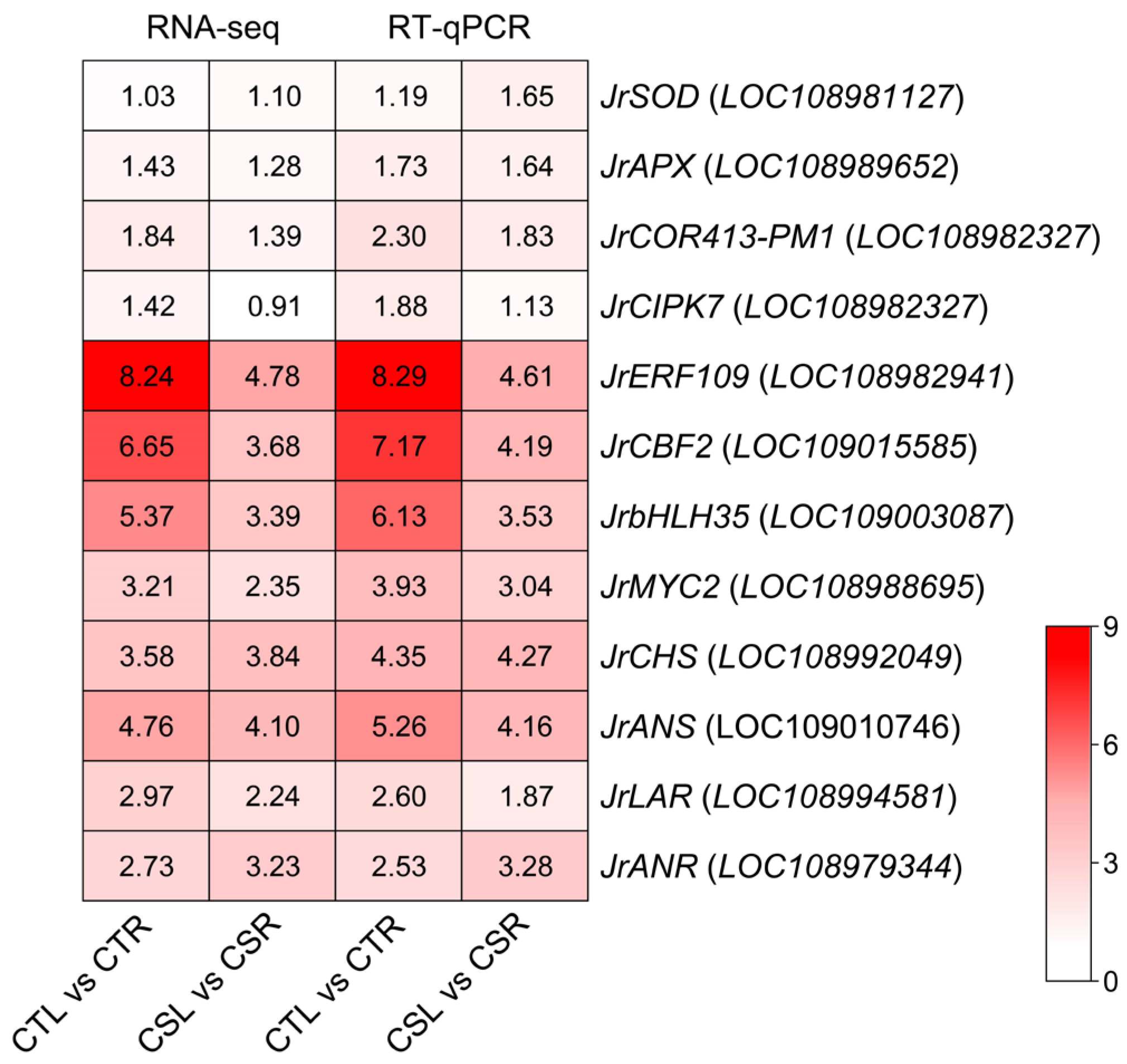
2.6. RT-qPCR Validation
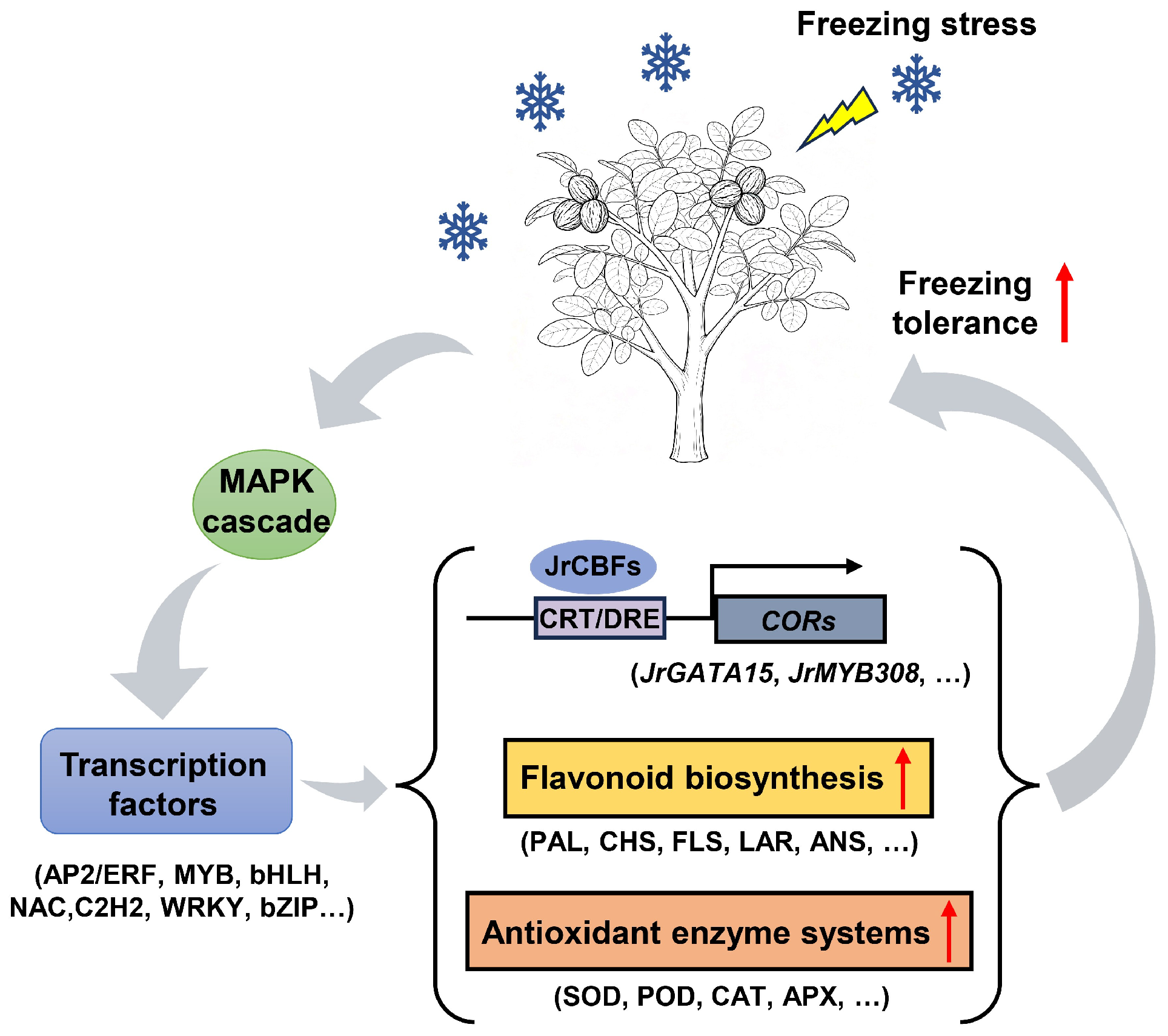
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Freezing Treatment and Sample Collection
4.3. Determination of Relative Electrical Conductivity
4.4. RNA Sequencing and Data Analysis
4.5. Genome-Wide Identification of CBF Genes in Walnut
4.6. Real-Time Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Panahi, B.; Tajaddod, S.; Mohammadzadeh Jallali, H.; Hejazi, M.A.; Zeinalabedini, M. Variability and association among some pomological and physiochemical traits in spring frost tolerant genotypes of Persian walnut (Juglans regia L.) and selection of genotypes with superior traits based on machine learning algorithms. Genet. Resour. Crop Evol. 2022, 69, 959–971. [Google Scholar] [CrossRef]
- Ramos, D.E. Walnut Production Manual; UCANR Publications: Oakland, CA, USA, 1998. [Google Scholar]
- Ebrahimi, A.; Zarei, A.; McKenna, J.R.; Bujdoso, G.; Woeste, K.E. Genetic diversity of Persian walnut (Juglans regia) in the cold-temperate zone of the United States and Europe. Sci. Hortic. 2017, 220, 36–41. [Google Scholar] [CrossRef]
- Theocharis, A.; Clement, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Furumoto, T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef]
- Khadivi, A.; Montazeran, A.; Yadegari, P. Superior spring frost resistant walnut (Juglans regia L.) genotypes identified among mature seedling origin trees. Sci. Hortic. 2019, 253, 147–153. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Lawson, S.S.; McKenna, J.R.; Jacobs, D.F. Morpho-Physiological and Genomic Evaluation of Juglans Species Reveals Regional Maladaptation to Cold Stress. Front. Plant Sci. 2020, 11, 229. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Sarwar, R.; Zhang, W.; Geng, R.; Zhu, K.-M.; Tan, X.-L. Research progress on the physiological response and molecular mechanism of cold response in plants. Front. Plant Sci. 2024, 15, 1334913. [Google Scholar] [CrossRef]
- Xin, Z.; Browse, J. Cold comfort farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 2000, 23, 893–902. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Wang, D.Z.; Jin, Y.N.; Ding, X.H.; Wang, W.J.; Zhai, S.S.; Bai, L.P.; Guo, Z.F. Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry 2017, 82, 1103–1117. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular Basis of Plant Cold Acclimation: Insights Gained from Studying the CBF Cold Response Pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef]
- Liu, N.; Du, H.; Xue, Y.; Liao, Y.; Zhang, W.; Ye, J.; Wang, Q.; Xu, F. Genome-wide identification and expression analysis of the walnut C-repeat binding factor gene family under low-temperature stress. Forests 2023, 14, 2274. [Google Scholar] [CrossRef]
- Fang, H.; Liu, X.; Dong, Y.; Feng, S.; Zhou, R.; Wang, C.; Ma, X.; Liu, J.; Yang, K.Q. Transcriptome and proteome analysis of walnut (Juglans regia L.) fruit in response to infection by Colletotrichum gloeosporioides. BMC Plant Biol. 2021, 21, 249. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, R.; Gao, S.; Pan, Z.; Guo, Z.; Yu, S.; Wang, Y.; Jin, Q.; Chen, X.; Zhang, L. Transcriptome analysis and phenotyping of walnut seedling roots under nitrogen stresses. Sci. Rep. 2022, 12, 12066. [Google Scholar] [CrossRef]
- An, X.-H.; Wang, N.; Wang, H.; Li, Y.; Si, X.-Y.; Zhao, S.; Tian, Y. Physiological and transcriptomic analyses of response of walnuts (Juglans regia) to Pantoea agglomerans infection. Front. Plant Sci. 2023, 14, 1294643. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, T. Combined transcriptomic and metabolomic analysis of the mechanism by which Bacillus velezensis induces resistance to anthracnose in walnut. Front. Microbiol. 2024, 15, 1420922. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Nie, R.; Li, A.; Wu, C.; Ji, X.; Tang, J.; Zhang, J. Integrated physio-biochemical and transcriptomic analysis reveals mechanism underlying salt tolerance in walnut. Plant Growth Regul. 2024, 104, 727–743. [Google Scholar] [CrossRef]
- Zhao, H.; Luo, X.; Guo, C.; Zhang, Z.; Ma, K.; Niu, J.; Quan, S. Transcriptome and microRNA analysis of Juglans regia in response to low-temperature stress. Int. J. Mol. Sci. 2025, 26, 1401. [Google Scholar] [CrossRef]
- Han, L.; Luo, X.; Zhao, Y.; Li, N.; Xu, Y.; Ma, K. A haplotype-resolved genome provides insight into allele-specific expression in wild walnut (Juglans regia L.). Sci. Data 2024, 11, 278. [Google Scholar] [CrossRef]
- Ma, Y.; Su, S.; Li, Y.; Chong, P. Cold tolerance of different walnut varieties. J. Fruit Sci. 2018, 35, 987–996. (In Chinese) [Google Scholar]
- Juurakko, C.L.; di Cenzo, G.C.; Walker, V.K. Cold acclimation and prospects for cold-resilient crops. Plant Stress 2021, 2, 100028. [Google Scholar] [CrossRef]
- Li, J.; Yu, Q.; Liu, C.; Zhang, N.; Xu, W. Flavonoids as key players in cold tolerance: Molecular insights and applications in horticultural crops. Hortic. Res. 2025, 12, uhae366. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Sang, Y.; Zhang, Y.; An, X.; Wang, H.; Zhang, R. Metabolite profile and metabolic network analysis of walnuts (Juglans regia L.) in response to chilling stress. Metabolites 2025, 15, 394. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef]
- Guo, J.; Ren, Y.; Tang, Z.; Shi, W.; Zhou, M. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. PeerJ 2019, 7, e8190. [Google Scholar] [CrossRef]
- An, J.-P.; Yao, J.-F.; Wang, X.-N.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J. Plant Physiol. 2017, 218, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Shua, Z.; Zhao, D.; Liu, B.; Luo, H.; Chen, Y.; Meng, D.; Song, Z.; Yang, Q.; Wang, Z.; et al. Genome assembly of pomegranate highlights structural variations driving population differentiation and key loci underpinning cold adaption. Hortic. Res. 2025, 12, uhaf022. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Ma, K.; Zhao, Y.; Mei, C.; Mamat, A.; Wang, J.; Qin, L.; He, T. The cold-stress responsive gene DREB1A involved in low-temperature tolerance in Xinjiang wild walnut. PeerJ 2022, 10, e4021. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ma, J.; Liu, H.; Zhao, P. Genome-wide identification of the CBF gene family and ICE transcription factors in walnuts and expression profiles under cold conditions. Int. J. Mol. Sci. 2024, 25, 25. [Google Scholar] [CrossRef]
- Marrano, A.; Britton, M.; Zaini, P.A.; Zimin, A.V.; Workman, R.E.; Puiu, D.; Bianco, L.; Di Pierro, E.A.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 2020, 9, giaa050. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, T.; Li, Z.; Huang, K.; Kim, N.-E.; Ma, Z.; Kwon, S.-W.; Jiang, W.; Du, X. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45-1 at the seedling stage in rice. Rice 2021, 14, 42. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef]
- Huang, X.; Shi, H.; Hu, Z.; Liu, A.; Amombo, E.; Chen, L.; Fu, J. ABA Is Involved in Regulation of Cold Stress Response in Bermudagrass. Front. Plant Sci. 2017, 8, 1613. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Meth. 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, L.; Niu, J.; Quan, S. Identification of appropriate reference genes for RT-qPCR analysis in Juglans regia L. PLoS ONE 2018, 13, e0209424. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Wang, J.; Zhang, Q.; Xu, T.; Ji, Z.; Hao, H.; Wang, J.; Shi, G.; Li, J. Comparative Transcriptome Analysis of Walnuts (Juglans regia L.) in Response to Freezing Stress. Plants 2025, 14, 3089. https://doi.org/10.3390/plants14193089
Chen L, Wang J, Zhang Q, Xu T, Ji Z, Hao H, Wang J, Shi G, Li J. Comparative Transcriptome Analysis of Walnuts (Juglans regia L.) in Response to Freezing Stress. Plants. 2025; 14(19):3089. https://doi.org/10.3390/plants14193089
Chicago/Turabian StyleChen, Lin, Juntao Wang, Qi Zhang, Taoyu Xu, Zhongrui Ji, Huazheng Hao, Jing Wang, Gensheng Shi, and Jian Li. 2025. "Comparative Transcriptome Analysis of Walnuts (Juglans regia L.) in Response to Freezing Stress" Plants 14, no. 19: 3089. https://doi.org/10.3390/plants14193089
APA StyleChen, L., Wang, J., Zhang, Q., Xu, T., Ji, Z., Hao, H., Wang, J., Shi, G., & Li, J. (2025). Comparative Transcriptome Analysis of Walnuts (Juglans regia L.) in Response to Freezing Stress. Plants, 14(19), 3089. https://doi.org/10.3390/plants14193089





