Geranyl Diphosphate Synthases GDS 1 and GDS7 Facilitate Natural Rubber Biosynthesis in Taraxacum kok-saghyz Roots
Abstract
1. Introduction
2. Results
2.1. Identification of TkGDS and Phylogenetic Analysis
2.2. GDS Protein Motifs, Domains, Gene Structures Analysis and Syntenic Profiling
2.3. Secondary and Tertiary Structural Analysis of TkGDS Proteins and Interaction Network
2.4. Cis-Acting Elements Analysis of the Promoter of TkGDS
2.5. Subcellular Localization of TkGDS
2.6. Potential Role of GDS1 and GDS7 in the Regulatory Network of T. kok-saghyz
2.7. Functional Characterization of GDS1 and GDS7 Overexpression Lines in T. kok-saghyz
3. Discussion
4. Materials and Methods
4.1. Cultivation Condition and Selection of the Materials
4.2. Identification and Phylogenetic Analysis of GDS in T. kok-saghyz
4.3. Motif, Domain, and Gene Structural Analysis of GDS
4.4. Structural Prediction of TkGDS Proteins
4.5. Protein Interaction Network Analysis
4.6. Cis-Acting Regulatory Elements Analysis
4.7. Transient Transformation Assay
4.8. Expression Pattern Analysis of TkGDS Genes
4.9. Construction of Recombinant Plasmids for Overexpression
4.10. Transformation and Molecular Identification
4.11. Analysis of TkGDS Genes in Overexpression Materials
4.12. Quantification of Natural Rubber Content in T. kok-saghyz Roots
4.13. Morphology of T. kok-saghyz Photosynthesis
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MVA | Mevalonate |
| MeJA | Methyl jasmonate |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| GFP | Green fluorescent protein |
| CDB | Cut-dip-budding Cut-dip-budding |
| OE | Overexpression |
| PN | Net photosynthetic rate |
| TR | Transpiration rate |
| GS | Stomatal conductance |
| CI | Intercellular CO2 concentration |
| WUE | Water use efficiency |
| PAR | Photosynthetically active radiation |
References
- Van Beilen, J.B.; Poirier, Y. Guayule and Russian dandelion as alternative sources of natural rubber. Crit. Rev. Biotechnol. 2007, 27, 217–231. [Google Scholar] [CrossRef]
- Senevirathna, A.M.; Stirling, C.M.; Rodrigo, V.H. Growth, photosynthetic performance and shade adaptation of rubber (Hevea brasiliensis) grown in natural shade. Tree Physiol. 2003, 23, 705–712. [Google Scholar] [CrossRef]
- Zhai, D.; Thaler, P.; Worthy, F.; Xu, J. Rubber latex yield is affected by interactions between antecedent temperature, rubber phenology, and powdery mildew disease. Int. J. Biometeorol. 2023, 67, 1569–1579. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Cadavid, D.A.; Valles-Ramirez, S.; Cornish, K.; Michel, F.C. Simultaneous quantification of rubber, inulin, and resins in Taraxacum kok-saghyz (TK) roots by sequential solvent extraction. Ind. Crops Prod. 2018, 122, 647–656. [Google Scholar] [CrossRef]
- Wang, S.M.; Zhang, Y.; Zhu, W.; Duan, B.; Chuan, X.X.; Zhou, M. The current status on pests and diseases of natural rubber tree under monitoring and early-warning system. Trop. Agric. Sci. Technol. 2018, 41, 10–13+17. [Google Scholar]
- Krotkov, G. A review of literature on Taraxacum koksaghyz Rod. Bot. Rev. 1945, 11, 417–461. [Google Scholar] [CrossRef]
- Xie, Q.; Ding, G.; Zhu, L.; Yu, L.; Yuan, B.; Gao, X.; Wang, D.; Sun, Y.; Liu, Y.; Li, H.; et al. Proteomic landscape of the mature roots in a rubber-producing grass Taraxacum kok-saghyz. Int. J. Mol. Sci. 2019, 20, 2596. [Google Scholar] [CrossRef]
- Salehi, M.; Cornish, K.; Bahmankar, M.; Naghavi, M.R. Natural rubber-producing sources, systems, and perspectives for breeding and biotechnology studies of Taraxacum kok-saghyz. Ind. Crops Prod. 2021, 170, 113667. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, C.; Yao, M.; Fang, F.; Wang, B.; Jin, F.; Chen, F.; Ji, M.; Liu, H.; Yu, Y.; et al. Auxin response factor 3 positively affects natural rubber biosynthesis by targeting farnesyl diphosphate synthase 1 in Taraxacum kok-saghyz. Ind. Crops Prod. 2025, 233, 118426. [Google Scholar] [CrossRef]
- Hodgson-Kratky, K.J.; Wolyn, D.J. Inheritance of flowering habit in Russian dandelion. J. Am. Soc. Hortic. Sci. 2015, 140, 614–619. [Google Scholar] [CrossRef]
- Wang, Y.F.; Yang, Z.C.; Yuan, B.X.; He, L.; Han, Y.Y.; Wang, J.Y.; Wang, X.C. Genome-wide identification of oxidosqualene cyclase genes regulating natural rubber in Taraxacum kok-saghyz. Planta. 2024, 260, 88. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Sa, R.N.; Sun, S.C.; Zhang, J.W.; Li, H.M.; Zhang, X.Q.; Zhang, L.Q.; Liu, S.Z. Dandelion rubber plant: A natural rubber in urgent need of intensive research. Rubber Ind. 2011, 58, 632–637. [Google Scholar]
- Cherian, S.; Ryu, S.B.; Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019, 17, 2041–2061. [Google Scholar] [CrossRef]
- Wang, G.; Dixon, R.A. Heterodimeric geranyl(geranyl) diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 2009, 106, 9914–9919. [Google Scholar]
- Ogura, K.; Koyama, T. Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 1998, 98, 1263–1276. [Google Scholar] [CrossRef]
- Kellogg, B.A.; Poulter, C.D. Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1997, 1, 570–578. [Google Scholar] [CrossRef]
- Wang, X.C.; Wang, D.; Sun, Y.; Yang, Q.; Chang, L.L.; Wang, L.M.; Meng, X.R.; Huang, Q.X.; Jin, X.; Tong, Z. Comprehensive proteomics analysis of laticifer latex reveals new insights into ethylene stimulation of natural rubber production. Sci. Rep. 2015, 5, 13778. [Google Scholar] [CrossRef] [PubMed]
- Córdoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2018, 60, 2933–2943. [Google Scholar]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Structure and dynamics of the isoprenoid pathway network. Mol. Plant 2012, 5, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.M.; Yu, J.; Tian, W.M. Cloning and expression analysis of GDS genes from the laticifer cells of Hevea brasiliensis. J. Southern Agric. 2018, 49, 2117–2128. [Google Scholar]
- Orlova, I.; Nagegowda, D.A.; Kish, C.M.; Gutensohn, M.; Maeda, H.; Varbanova, M.; Fridman, E.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; et al. The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of Nicotiana benthamiana geranylgeranyl diphosphate synthase in planta. Plant Cell 2009, 21, 4002–4017. [Google Scholar] [CrossRef]
- Bouvier, F.; Suire, C.; d’Harlingue, A.; Backhaus, R.A.; Camara, B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 2000, 24, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Chem. Biol. 2018, 4, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, P.S.; Zerbe, P. Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 2019, 10, 1166. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, H.T.; Feng, M.Y.; Liang, C.Y.; Li, W.L. Prokaryotic expression and RNA interference vector construction for geranyl diphosphate synthase of Mentha haplocalyx Briq. Bio Bull. 2013, 9, 84–88. [Google Scholar]
- Zhang, H.Y.; Chen, S.Y.; Lu, C.H.; Deng, W.J.; He, M.L.; Yan, H.J. Cloning and bioinformatic analysis of GDS genes of Pogostemon cablin. Lishizhen Med. Mater. Medica 2020, 31, 1234–1237. [Google Scholar]
- Lin, T.; Xu, X.; Du, H.; Fan, X.; Chen, Q.; Hai, C.; Zhou, Z.; Su, X.; Kou, L.; Gao, Q.; et al. Extensive sequence divergence between the reference genomes of Taraxacum kok-saghyz and Taraxacum mongolicum. Sci. China Life Sci. 2022, 65, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Takahashi, S. Molecular mechanisms of natural rubber biosynthesis. Annu. Rev. Biochem. 2020, 89, 821–851. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.; Paponov, I.; Palme, K. Auxin in action: Signaling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Yan, L.; Fang, Z.; Zhang, N.; Yang, L.; Zhang, Y.; Zhuang, M.; Lv, H.; Ji, J.; Wang, Y. Genome-wide identification, characterization, and expression analysis of the geranylgeranyl pyrophosphate synthase (GGPPS) gene family reveals its importance in chloroplasts of Brassica oleracea L. Agriculture 2023, 13, 1615. [Google Scholar] [CrossRef]
- Sakuma, K.; Kobayashi, N.; Sugiki, T.; Nagashima, T.; Fujiwara, T.; Suzuki, K.; Kobayashi, A.; Murata, T.; Kosugi, T.; Tatsumi, K.R.; et al. Design of complicated all α protein structures. Nat. Struct. Mol. Biol. 2024, 31, 275–282. [Google Scholar] [CrossRef]
- Roychoudhry, S.; Kepinski, S. Auxin in root development. Cold Spring Harb. Perspect. Biol. 2022, 14, a039933. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qin, Y.Y.; Wei, N.N.; Xue, H.Y.; Dai, W.S. Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation in citrus seeds and its application in gene functional analysis. Front. Plant Sci. 2023, 14, 1293374. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Y.; Liang, J.; Hu, X.; He, Y.; Miao, T.; Ouyang, Z.; Yang, Z.; Amin, A.K.; Ling, C.; et al. Agrobacterium rhizogenes-mediated marker-free transformation and gene editing system revealed that AeCBL3 mediates the formation of calcium oxalate crystal in kiwifruit. Mol. Hortic. 2024, 4, 1. [Google Scholar] [CrossRef]
- Lee, K.R.; Kim, K.H.; Kim, J.B.; Hong, S.B.; Jeon, I.; Kim, H.U.; Lee, M.H.; Kim, J.K. High accumulation of γ-linolenic acid and Stearidonic acid in transgenic Perilla (Perilla frutescens var. frutescens) seeds. BMC Plant Biol. 2019, 19, 120. [Google Scholar] [CrossRef]
- Mitchell, M.C.; Pritchard, J.; Okada, S.; Zhang, J.; Venables, I.; Vanhercke, T.; Ral, J.P. Increasing growth and yield by altering carbon metabolism in a transgenic leaf oil crop. Plant Biotechnol. J. 2020, 18, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Collins-Silva, J.; Nural, A.T.; Skaggs, A.; Scott, D.; Hathwaik, U.; Woolsey, R.; Schegg, K.; McMahan, C.; Whalen, M.; Cornish, K.; et al. Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry 2012, 79, 46–56. [Google Scholar] [CrossRef]
- Gao, K.; Zou, Y.L.; Wei, Z.P.; Wang, Z.Y.; Wu, J.; Fan, B.Y. Cloning and bioinformatic analysis of geranyl diphosphate gene from Paeonia lactiflora Pall. North. Hortic. 2023, 10, 73–80. [Google Scholar]
- Koichiro, T.; Glen, S.; Sudhi, K. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, 427–432. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, 200–203. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol. Plant-Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [PubMed]
- Kan, Y.; Simons, E.; Citovsky, V. Confocal microscopy analysis of protein sorting to plasmodesmata in Nicotiana benthamiana. J. Vis. Exp. 2024, 213, 67378. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.J. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2022, 4, 100345. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Li, Y.; Liu, X.; Yang, X.; Huang, Z.; Xing, J.; Liang, C.; Li, J.; Tan, Y.; Zhang, S.; et al. Rubber biosynthesis drives the biogenesis and development of rubber particles, the rubber-producing organelles. Plant Biotechnol. J. 2025, 23, 2303–2316. [Google Scholar] [CrossRef] [PubMed]
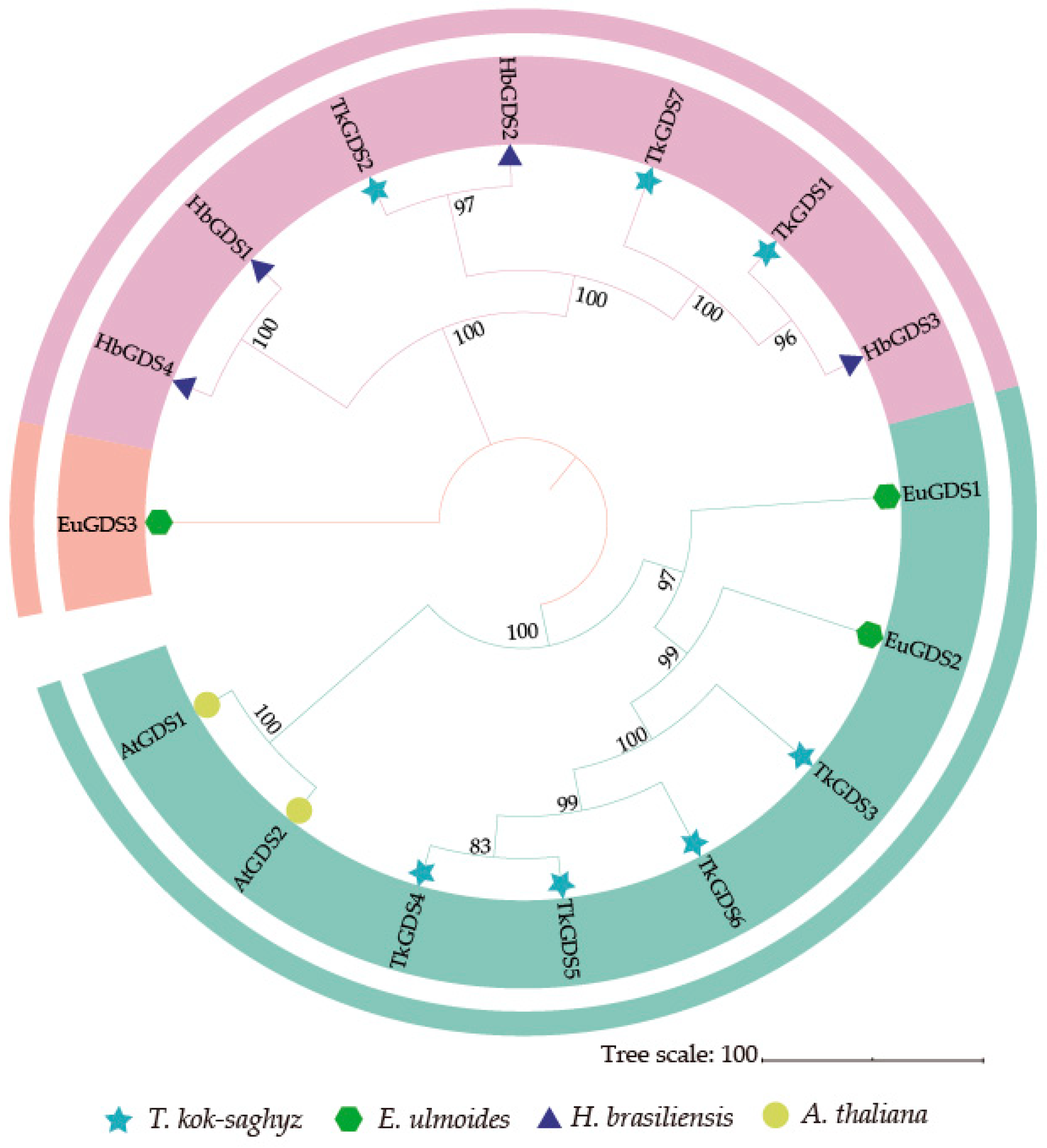


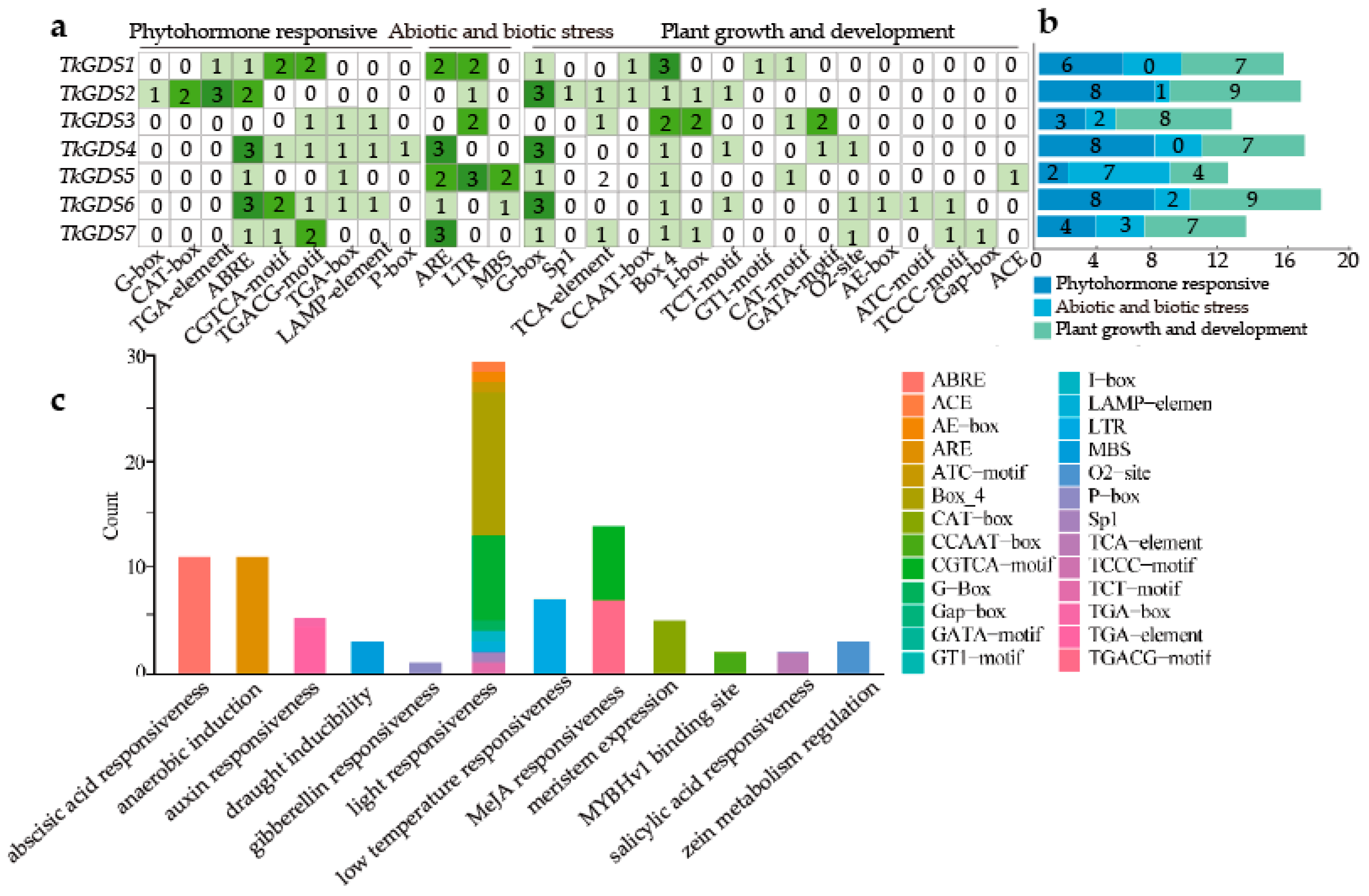


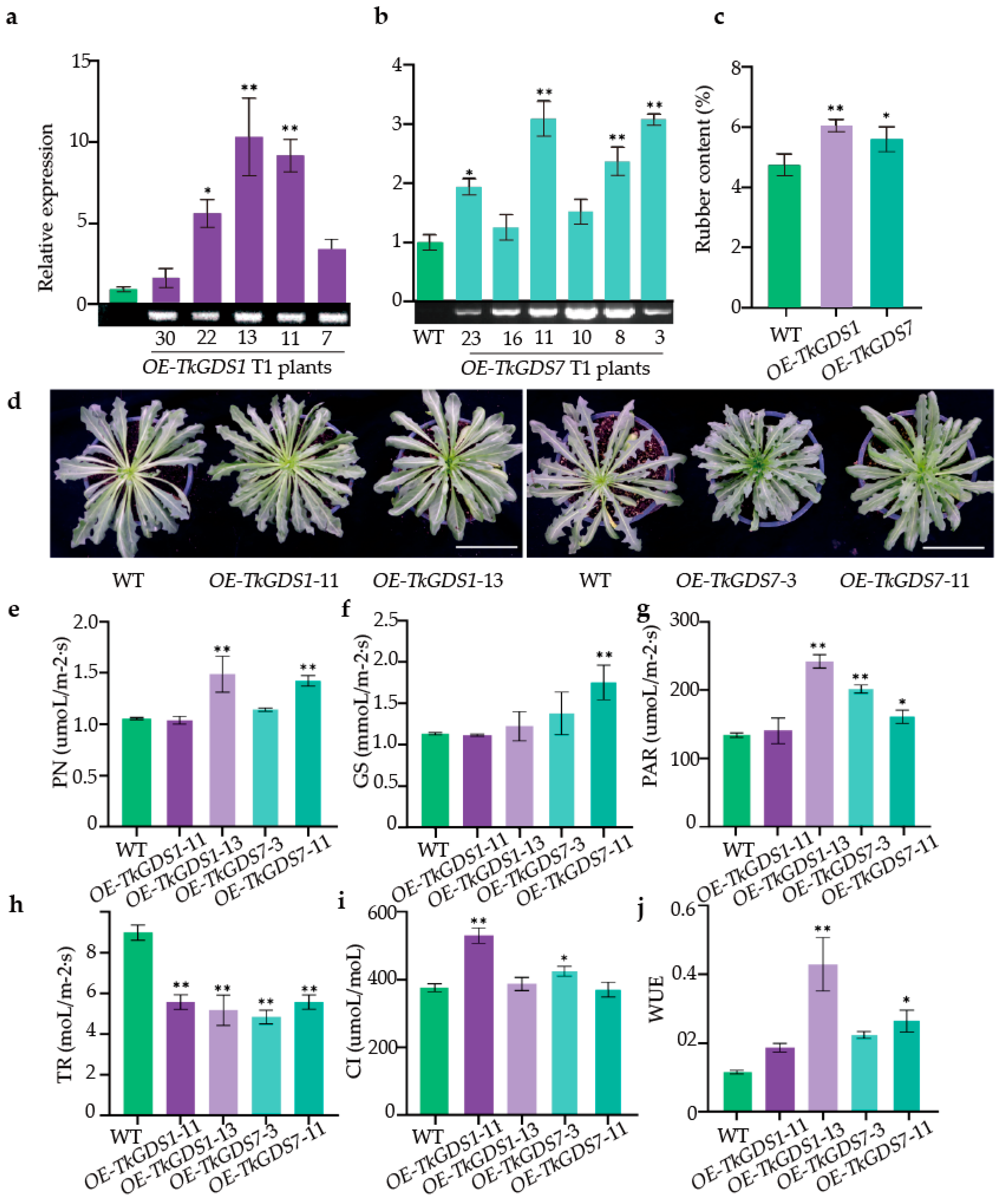
| Gene | Gene ID | ORF/bp | PI | MW/ kDa | Size/ aa | Subcellular Localization * |
|---|---|---|---|---|---|---|
| TkGDS1 | TkA06G134330 | 812 | 5.14 | 29.30 | 268 | Chloroplast |
| TkGDS2 | TkA06G097970 | 799 | 6.01 | 28.53 | 264 | Chloroplast |
| TkGDS3 | TkA07G129020 | 975 | 5.54 | 35.03 | 321 | Chloroplast |
| TkGDS4 | TkA07G129470 | 1269 | 6.33 | 46.38 | 419 | Chloroplast |
| TkGDS5 | TkA07G129240 | 1045 | 5.67 | 37.88 | 345 | Chloroplast & Mitochondrial |
| TkGDS6 | TkA07G129530 | 1272 | 6.08 | 46.30 | 419 | Chloroplast |
| TkGDS7 | TkA07G403060 | 1102 | 5.16 | 38.82 | 364 | Chloroplast |
| Proteins | α-Helix (%) | β-Turn (%) | Extended Strand (%) | Random Coil (%) | Structure |
|---|---|---|---|---|---|
| TkGDS1 | 63.26 | 6.44 | 4.92 | 25.38 |  |
| TkGDS2 | 61.57 | 7.09 | 5.97 | 25.37 |  |
| TkGDS3 | 63.64 | 5.67 | 5.39 | 25.30 |  |
| TkGDS4 | 66.67 | 5.22 | 4.35 | 23.77 |  |
| TkGDS5 | 53.94 | 4.06 | 5.73 | 36.28 |  |
| TkGDS6 | 62.31 | 6.54 | 5.61 | 25.55 |  |
| TkGDS7 | 59.07 | 5.49 | 7.42 | 28.02 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Yuan, B.; Ao, G.; Wu, X.; Fang, F.; Long, S.; Hui, S. Geranyl Diphosphate Synthases GDS 1 and GDS7 Facilitate Natural Rubber Biosynthesis in Taraxacum kok-saghyz Roots. Plants 2025, 14, 2980. https://doi.org/10.3390/plants14192980
Wang B, Yuan B, Ao G, Wu X, Fang F, Long S, Hui S. Geranyl Diphosphate Synthases GDS 1 and GDS7 Facilitate Natural Rubber Biosynthesis in Taraxacum kok-saghyz Roots. Plants. 2025; 14(19):2980. https://doi.org/10.3390/plants14192980
Chicago/Turabian StyleWang, Baoqiang, Boxuan Yuan, Guoen Ao, Xiaoyou Wu, Fengyan Fang, Shiqi Long, and Shugang Hui. 2025. "Geranyl Diphosphate Synthases GDS 1 and GDS7 Facilitate Natural Rubber Biosynthesis in Taraxacum kok-saghyz Roots" Plants 14, no. 19: 2980. https://doi.org/10.3390/plants14192980
APA StyleWang, B., Yuan, B., Ao, G., Wu, X., Fang, F., Long, S., & Hui, S. (2025). Geranyl Diphosphate Synthases GDS 1 and GDS7 Facilitate Natural Rubber Biosynthesis in Taraxacum kok-saghyz Roots. Plants, 14(19), 2980. https://doi.org/10.3390/plants14192980






