Molecular Diversification of the Genus Clinopodium (Lamiaceae) from the Balkans with an Emphasis on the Transferred Groups Calamintha, Acinos, and the Sect. Pseudomelissa
Abstract
1. Introduction
2. Results
2.1. DNA Regions and Alignments
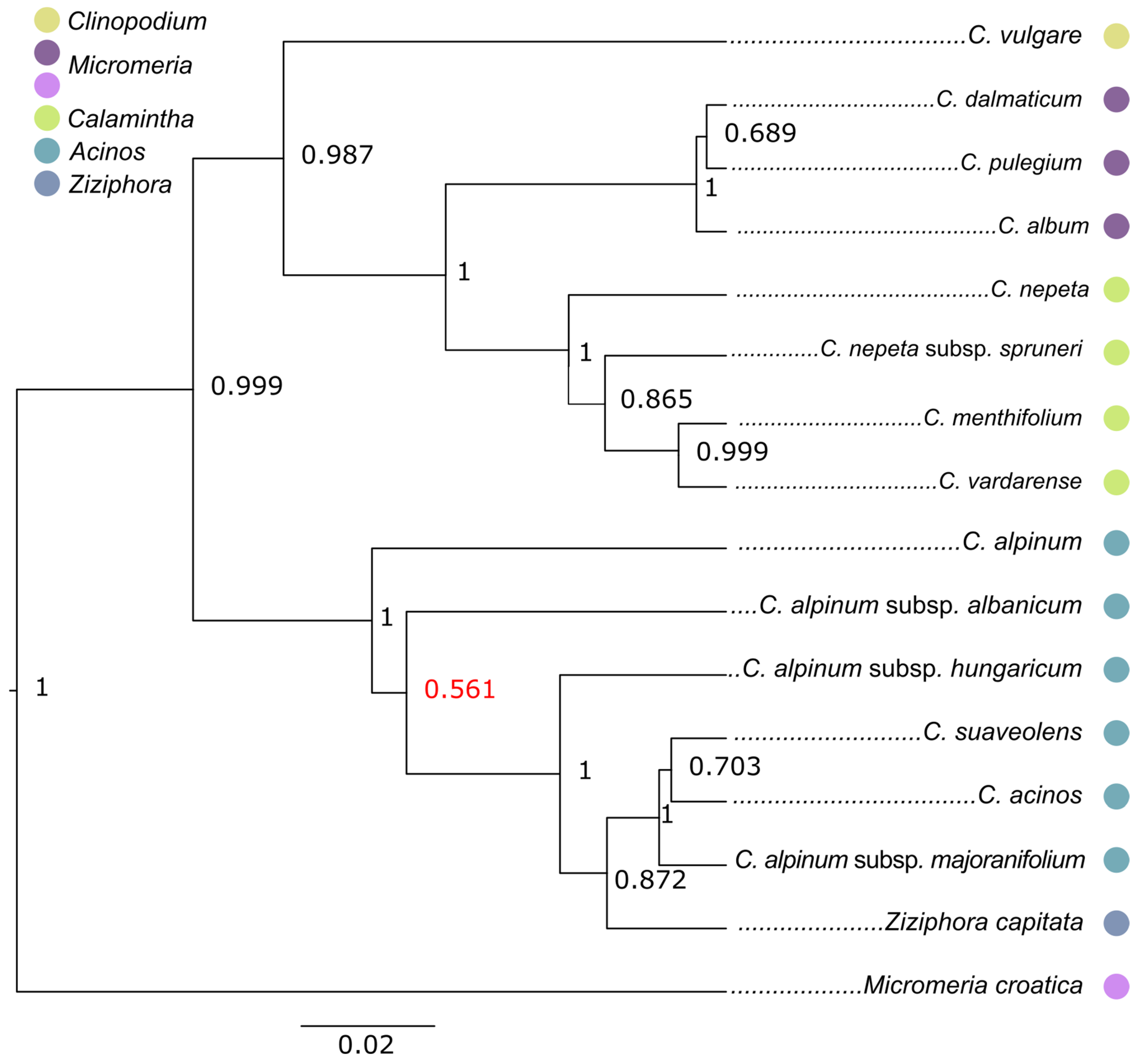
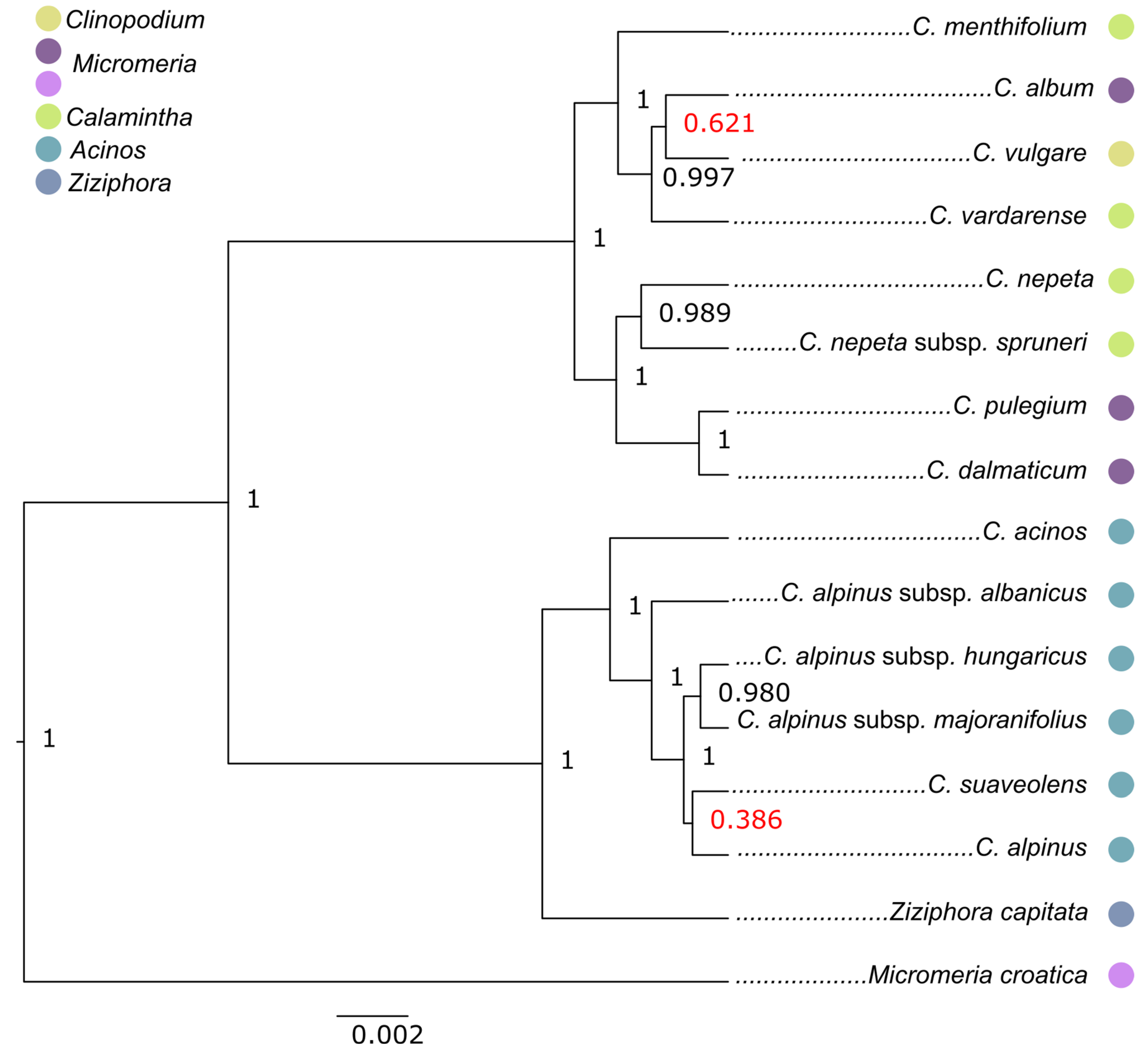
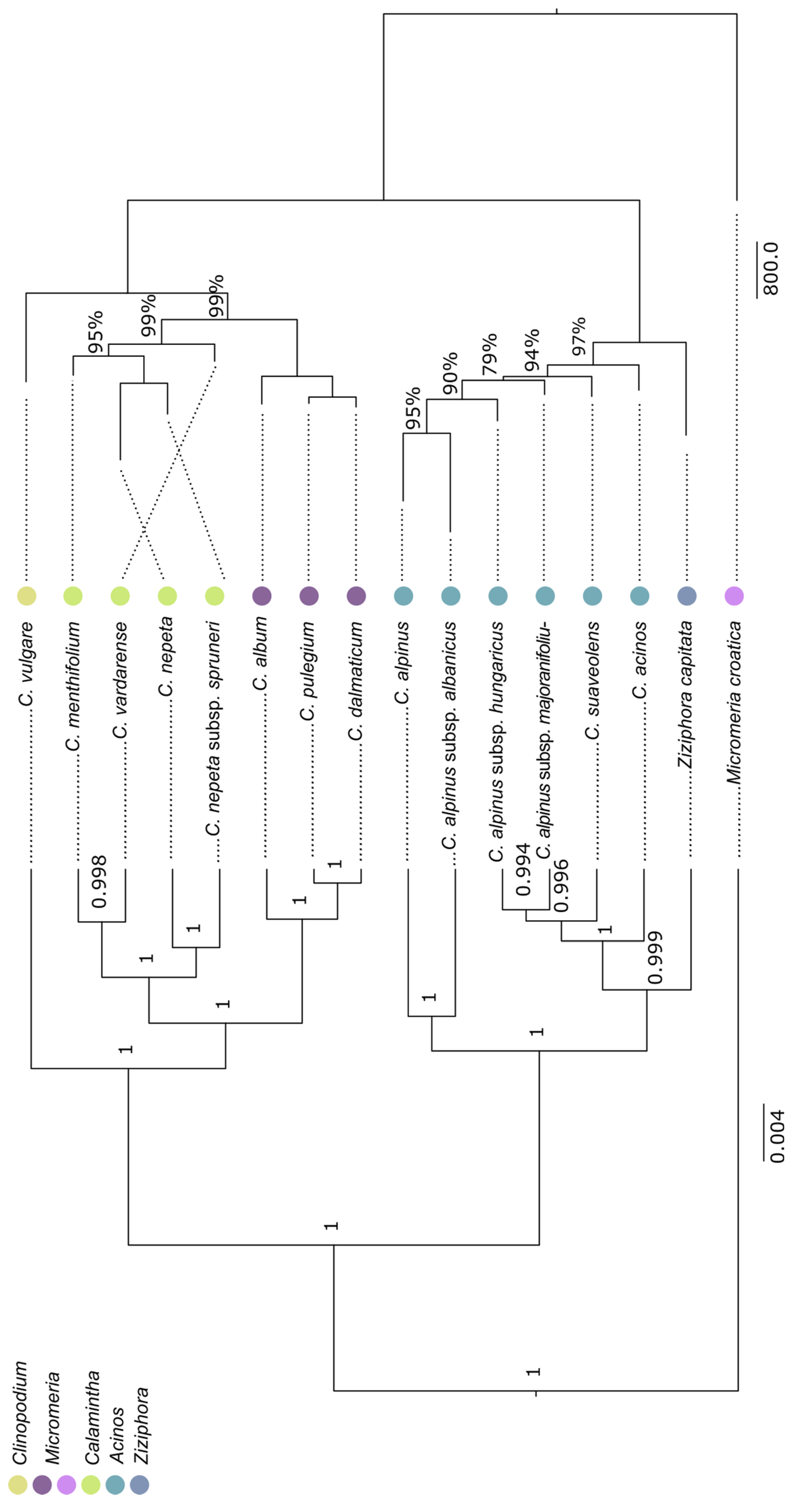
2.2. Phylogenetic Reconstructions
3. Discussion
3.1. Acinos/Ziziphora Clade
3.2. Clinopodium–Pseudomelissa–Calamintha Clade
3.3. General Discussion on Taxonomic Position of Clinopodium Taxa from the Balkans
4. Materials and Methods
4.1. Taxon Sampling
4.2. Choice of Molecular Markers
4.3. DNA Extraction, Amplification, and Sequencing
4.4. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chrysargyris, A. It Runs in the Family: The Importance of the Lamiaceae Family Species. Agronomy 2024, 14, 1274. [Google Scholar] [CrossRef]
- Alimpić, A.; Pljevljakušić, D.; Šavikin, K.; Knežević, A.; Ćurčić, M.; Veličković, D.; Stević, T.; Petrović, G.; Matevski, V.; Vukojević, J.; et al. Composition and Biological Effects of Salvia Ringens (Lamiaceae) Essential Oil and Extracts. Ind. Crops Prod. 2015, 76, 702–709. [Google Scholar] [CrossRef]
- Ebadollahi, A.; Ziaee, M.; Palla, F. Essential Oils Extracted from Different Species of the Lamiaceae Plant Family as Prospective Bioagents against Several Detrimental Pests. Molecules 2020, 25, 1556. [Google Scholar] [CrossRef]
- Wang, Z.; Poudyal, S.; Kopp, K.; Zhang, Y. Response of Ornamental Plants to Salinity: Impact on Species-Specific Growth, Visual Quality, Photosynthetic Parameters, and Ion Uptake. Front. Plant Sci. 2025, 16, 1611767. [Google Scholar] [CrossRef]
- Bentham, G. Labiatarum Genera et Species: Or, a Description of the Genera and Species of Plants of the Order Labiatae; with Their General History, Characters, Affinities, and Geographical Distribution. Available online: https://bibdigital.rjb.csic.es/records/item/12582-redirection (accessed on 8 December 2023).
- Boissier, E. Flora Orientalis; Biodiversity Heritage Library: Washington, DC, USA, 1879. [Google Scholar]
- Visiani, R.D. Flora Dalmatica—Sive Enumeratio Stirpium Vascularium Quas Hactenus In Dalmatia Lectas Et Sibi Digessit; Hofmeister: Leipzig, Germany, 1847. [Google Scholar]
- Šilić, Č. Monografija Rodova Satureja L., Calamintha Miller, Micromeria Bentham, Acinos Miller i Clinopodium L. u Flori Jugoslavije; Zemaljski muzej BiH: Sarajevo, Bosnia and Herzegovina, 1979. [Google Scholar]
- Marin, P. Orasice i Trihome u Familiji Lamiaceae; Biološki fakultet: Beograd, Serbia, 1996; pp. 159–168. [Google Scholar]
- Engler, A.; Krause, K.; Pilger, R.; Prantl, K. Die Natürlichen Pflanzenfamilien Nebst Ihren Gattungen Und Wichtigeren Arten, Insbesondere Den Nutzpflanzen, Unter Mitwirkung Zahlreicher Hervorragender Fachgelehrten Begründet; W. Engelmann: Leipzig, Germany, 1887. [Google Scholar]
- Wagstaff, S.J.; Olmstead, R.G.; Cantino, P.D. Parsimony Analysis of cpDNA Restriction Site Variation in Subfamily Nepetoideae (Labiatae). Am. J. Bot. 1995, 82, 886–892. [Google Scholar] [CrossRef]
- Bräuchler, C.; Meimberg, H.; Abele, T.; Heubl, G. Polyphyly of the Genus Micromeria (Lamiaceae)—Evidence from cpDNA Sequence Data. Taxon 2005, 54, 639–650. [Google Scholar] [CrossRef]
- Doroszenko, A.M. Taxonomic Studies on the Satureja Complex (Labiatae). Ph.D. Thesis, KB thesis scanning project 2015. The University of Edinburgh, Edinburgh, UK, 1986. [Google Scholar]
- Cantino, P.D.; Wagstaff, S.J. A Reexamination of North American Satureja s. l. (Lamiaceae) in Light of Molecular Evidence. Brittonia 1998, 50, 63. [Google Scholar] [CrossRef]
- Harley, R.M.; Paucar, A.G. List of Species of Tropical American Clinopodium (Labiatae), with New Combinations. Kew Bull. 2000, 55, 917. [Google Scholar] [CrossRef]
- Li, B.; Cantino, P.D.; Olmstead, R.G.; Bramley, G.L.C.; Xiang, C.-L.; Ma, Z.-H.; Tan, Y.-H.; Zhang, D.-X. A Large-Scale Chloroplast Phylogeny of the Lamiaceae Sheds New Light on Its Subfamilial Classification. Sci. Rep. 2016, 6, 34343. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.-P.; Salmaki, Y.; Drew, B.T.; Wilson, T.C.; Scheen, A.-C.; Celep, F.; Bräuchler, C.; Bendiksby, M.; Wang, Q.; et al. An Updated Tribal Classification of Lamiaceae Based on Plastome Phylogenomics. BMC Biol. 2021, 19, 2. [Google Scholar] [CrossRef]
- Bräuchler, C.; Meimberg, H.; Heubl, G. New Names in Old World Clinopodium—The Transfer of the Species of Micromeria Sect. Pseudomelissa to Clinopodium. Taxon 2006, 55, 977–981. [Google Scholar] [CrossRef]
- Ryding, O. Revision of the Clinopodium Abyssinicum Group (Labiatae). Bot. J. Linn. Soc. 2006, 150, 391–408. [Google Scholar] [CrossRef][Green Version]
- Ryding, O. Revision of the Clinopodium Simense Group (Labiatae). Kew Bull. 2006, 61, 419–432. [Google Scholar][Green Version]
- Bräuchler, C.; Meimberg, H.; Heubl, G. Molecular Phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae)—Taxonomy, Biogeography and Conflicts. Mol. Phylogenetics Evol. 2010, 55, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Janković, S.; Alimpić Aradski, A.; Dodoš, T.; Novaković, J.; Ivanović, S.; Vujisić, L.; Marin, P.D.; Rajčević, N. Clinopodium, L. Taxa from the Balkans—Are There Unique Leaf Micromorphological and Phytochemical Patterns? Plants 2024, 13, 251. [Google Scholar] [CrossRef]
- Španiel, S.; Rešetnik, I. Plant Phylogeography of the Balkan Peninsula: Spatiotemporal Patterns and Processes. Plant Syst. Evol. 2022, 308, 38. [Google Scholar] [CrossRef]
- Dimopoulos, P.; Raus, T.; Bergmeier, E.; Constantinidis, T.; Iatrou, G.; Kokkini, S.; Strid, A.; Tzanoudakis, D. Vascular Plants of Greece: An Annotated Checklist; Botanic Garden and Botanical Museum Berlin-Dahlem & Hellenic Botanical Society: Berlin, Germany, 2013; ISBN 978-3-921800-88-1. [Google Scholar]
- Rose, J.P.; Xiang, C.-L.; Sytsma, K.J.; Drew, B.T. A Timeframe for Mint Evolution: Towards a Better Understanding of Trait Evolution and Historical Biogeography in Lamiaceae. Bot. J. Linn. Soc. 2022, 200, 15–38. [Google Scholar] [CrossRef]
- Trusty, J.L.; Olmstead, R.G.; Bogler, D.J.; Santos-Guerra, A.; Francisco-Ortega, J. Using Molecular Data to Test a Biogeographic Connection of the Macaronesian Genus Bystropogon (Lamiaceae) to the New World: A Case of Conflicting Phylogenies. Syst. Bot. 2004, 29, 702–715. [Google Scholar] [CrossRef]
- Melnikov, D.G. Taxonomic and Nomenclatural Notes on Clinopodium L. and Ziziphora L. (Lamiaceae). Novit. Syst. Plant. Vasc. 2016, 47, 103–107. [Google Scholar] [CrossRef]
- Jandaghi, M.; Vaezi, J.; Shakeri, A. Subspecies Delineation of Ziziphora Clinopodioides Complex (Lamiaceae) Based on Morphological, Sequence-Related Amplified Polymorphism and Essential Oils Analyses. Phytotaxa 2025, 684, 55–77. [Google Scholar] [CrossRef]
- Kremer, D.; Stabentheiner, E.; Bogunić, F.; Ballian, D.; Eleftheriadou, E.; Stešević, D.; Matevski, V.; Ranđelović, V.; Ivanova, D.; Ruščić, M.; et al. Micromorphological Traits of Balcanic Micromeria and Closely Related Clinopodium Species (Lamiaceae). Plants 2021, 10, 1666. [Google Scholar] [CrossRef]
- Duminil, J.; Di Michele, M. Plant Species Delimitation: A Comparison of Morphological and Molecular Markers. Plant Biosyst.-Int. J. Deal. All Asp. Plant Biol. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Candolle, A.P. de Prodromus systematis naturalis regni vegetabilis: Sive enumeratio contracta (...); Éditeur non identifié: Anvers, Belgium, 1848. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 9, 11–15. [Google Scholar]
- Dodos, T.; Aleksic, J.; Rajcevic, N.; Marin, P.D. A Robust and Cost-Effective Method for DNA Isolation from Satureja Species (Lamiaceae). Arch. Biol. Sci. 2014, 66, 285–297. [Google Scholar] [CrossRef]
- Rajčević, N.; Dodoš, T.; Novaković, J.; Kuzmanović, N.; Janaćković, P.; Marin, P. Are Environmental Factors Responsible for Essential Oil Chemotype Distribution of Balkan Juniperus Communis Var. Saxatilis Populations? Plant Biosyst.-Int. J. Deal. All. Asp. Plant Biol. 2022, 157, 102–111. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of Whole Chloroplast Genome Sequences to Choose Noncoding Regions for Phylogenetic Studies in Angiosperms: The Tortoise and the Hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA Barcodes to Identify Flowering Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Zhou, S. Complete Chloroplast Genome of Sedum Sarmentosum and Chloroplast Genome Evolution in Saxifragales. PLoS ONE 2013, 8, e77965. [Google Scholar] [CrossRef] [PubMed]
- Douzery, E.J.P.; Pridgeon, A.M.; Kores, P.; Linder, H.P.; Kurzweil, H.; Chase, M.W. Molecular Phylogenetics of Diseae (Orchidaceae): A Contribution from Nuclear Ribosomal ITS Sequences. Am. J. Bot. 1999, 86, 887–899. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Markos, S. Phylogenetic Utility of the External Transcribed Spacer (ETS) of 18S–26S rDNA: Congruence of ETS and ITS Trees of Calycadenia(Compositae). Mol. Phylogenetics Evol. 1998, 10, 449–463. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A Graphical Interface and Toolkit for Phylogenetic Analyses Using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Tavaré, S. Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Yang, Z. Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef]
| Locus | Length 1 | No. of Indels 2 | Indel Length in bp 3 | Substitutions | Microsatellite Loci | |
|---|---|---|---|---|---|---|
| Transitions | Transversions | |||||
| trnSUGA-psbK | 703 | 21 | 90 | 11 | 19 | 13 |
| trnLUAG-rpl32 | 903 | 16 | 91 | 27 | 34 | 34 |
| rps16-trnQUUG | 1002 | 22 | 206 | 21 | 31 | 41 |
| rps16-trnKUUU | 710 | 15 | 67 | 19 | 23 | 18 |
| rps15-ycf1 | 611 | 12 | 144 | 24 | 12 | 19 |
| psbA-trnHGUG | 424 | 10 | 61 | 13 | 21 | 13 |
| petN-psbM | 1057 | 19 | 47 | 21 | 17 | 25 |
| ITS | 702 | 15 | 21 | 77 | 47 | 17 |
| ETS | 408 | 16 | 24 | 138 | 68 | 8 |
| Taxon | Locality | Long. | Lat. | Alt [m a.s.l.] | BEOU |
|---|---|---|---|---|---|
| Former Acinos taxa | |||||
| Clinopodium acinos (L.) Kuntze | Serbia, Mt. Tara | 43.865 | 19.406 | 872 | 17,987 |
| Clinopodium alpinum (L.) Kuntze subsp. alpinum | Serbia, Topli Do | 43.334 | 22.664 | 690 | 17,990 |
| Clinopodium alpinum subsp. albanicum (Kümmerle & Jáv.) Govaerts | Serbia, Mt. Rogozna | 43.045 | 20.521 | 882 | 17,992 |
| Clinopodium alpinum subsp. hungaricum (Simonk.) Govaerts | Serbia, Mt. Fruska gora | 45.156 | 19.778 | 387 | 17,993 |
| Clinopodium alpinum subsp. majoranifolium (Mill.) Govaerts | Montenegro, Kotor | 42.423 | 18.791 | 534 | 18,037 |
| Clinopodium suaveolens (Sm.) Kuntze | Serbia, Mt. Rtanj | 43.767 | 21.926 | 791 | 18,001 |
| Former Calamintha taxa | |||||
| Clinopodium menthifolium (Host.) Merino | Serbia, Mt. Tara | 43.969 | 19.344 | 1010 | 18,039 |
| Clinopodium vardarense (Šilić) Govaerts | N. Macedonia, Stenje | 40.934 | 20.930 | 818 | 18,042 |
| Clinopodium nepeta subsp. spruneri (Boiss.) Bartolucci & F.Conti | Croatia, Vrana lake | 43.848 | 15.636 | 9 | 18,041 |
| Clinopodium nepeta (L.) Kuntze | Serbia, Belgrade | 44.815 | 20.472 | 143 | 18,040 |
| Former Pseudomelissa taxa | |||||
| Clinopodium album (Waldst. & Kit.) Bräuchler & Govaerts | Serbia, Mt. Tara | 43.866 | 19.407 | 872 | 18,004 |
| Clinopodium pulegium (Rochel) Bräuchler | Serbia, Svrljig gorge | 43.542 | 22.177 | 259 | 17,999 |
| Clinopodium dalmaticum (Benth.) Bräuchler & Heubl | Montenegro, Njeguši | 42.408 | 18.787 | 912 | 18,038 |
| Clinopodium L. | |||||
| Clinopodium vulgare L. | Serbia, Zlot | 44.029 | 21.961 | 303 | 18,043 |
| Outgroup taxa | |||||
| Ziziphora capitata L. | Serbia, Svrljig gorge | 43.542 | 22.177 | 259 | 18,045 |
| Micromeria croatica (Pers.) Schott | Serbia, Mt. Tara | 43.864 | 19.409 | 844 | 18,044 |
| Locus | Primer Sequence (Seq 5′-3′) | Ta [°C] | Ext. [s] | Reference | |
|---|---|---|---|---|---|
| rps16-trnKUUU | F R | TTAAAAGCCGAGTACTCTACC AAAGTGGGTTTTTATGATCC | 53 | 60 | [35] |
| rpl32-trnLUAG | F R | CAGTTCCAAAAAAACGTACTTC CTGCTTCCTAAGAGCAGCGT | 53 | 60 | [35] |
| rps15-ycf1 | F R | CAATTYCAAATGTGAAGTAAGTCTCC CTTGTATGRATCGTTATTGKTTTG | 58 | 60 | [35] |
| psbA-trnHGUG | F R | GTTATGCATGAACGTAATGCTC CGCGCATGGTGGATTCACAATCC | 53 | 60 | [36] |
| rps16-trnKUUU | F R | GTTTCAAACGAAGTTTTACCAT TCGAATCCTTCCGTCCC | 51 | 75 | [37] |
| petN-psbM | F R | ATGGATATAGTAAGTCTCGCTTG ATGGAAGTAAATATTCTTGCAT | 51 | 90 | [37] |
| psbK-trnSUGA | F R | TTTGGCAGGCTGCTGTAAGTT ACTAAAGCGTCGGATTGCT | 56.5 | 60 | * |
| ITS | F R | GGAAGTAAAAGTCGTAACAAGG TCCTCCGCTTATTGATATGC | 63 | 45 | [38] |
| ITS | F R | GTCCACTGAACCTTATCATTTAG TCCTCCGCTTATTGATATGC | 55 | 60 | [21] |
| ETS | F R | GTGAGTGGTGKTTGGCGYGT GCAGGATCAACCAGGTAGCA | 55 | 60 | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janković, S.; Dodoš, T.; Marin, P.D.; Obradović Novaković, J.; Rajčević, N. Molecular Diversification of the Genus Clinopodium (Lamiaceae) from the Balkans with an Emphasis on the Transferred Groups Calamintha, Acinos, and the Sect. Pseudomelissa. Plants 2025, 14, 2940. https://doi.org/10.3390/plants14182940
Janković S, Dodoš T, Marin PD, Obradović Novaković J, Rajčević N. Molecular Diversification of the Genus Clinopodium (Lamiaceae) from the Balkans with an Emphasis on the Transferred Groups Calamintha, Acinos, and the Sect. Pseudomelissa. Plants. 2025; 14(18):2940. https://doi.org/10.3390/plants14182940
Chicago/Turabian StyleJanković, Smiljana, Tanja Dodoš, Petar D. Marin, Jelica Obradović Novaković, and Nemanja Rajčević. 2025. "Molecular Diversification of the Genus Clinopodium (Lamiaceae) from the Balkans with an Emphasis on the Transferred Groups Calamintha, Acinos, and the Sect. Pseudomelissa" Plants 14, no. 18: 2940. https://doi.org/10.3390/plants14182940
APA StyleJanković, S., Dodoš, T., Marin, P. D., Obradović Novaković, J., & Rajčević, N. (2025). Molecular Diversification of the Genus Clinopodium (Lamiaceae) from the Balkans with an Emphasis on the Transferred Groups Calamintha, Acinos, and the Sect. Pseudomelissa. Plants, 14(18), 2940. https://doi.org/10.3390/plants14182940









