A Comprehensive Functional Analysis of OsPEAMT1 and OsPEAMT2 Genes in Rice (Oryza sativa L. ssp. japonica)
Abstract
1. Introduction
2. Results
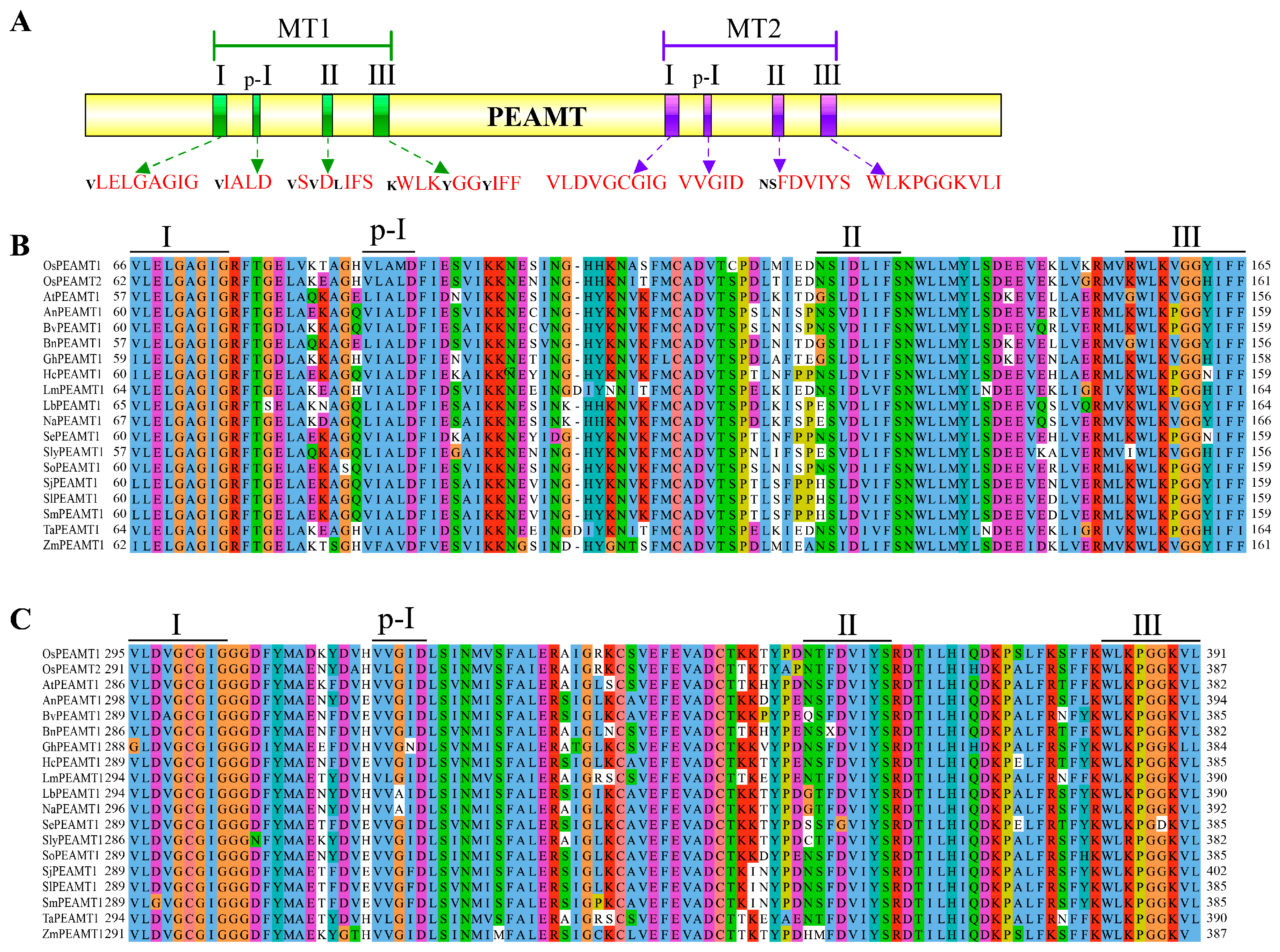
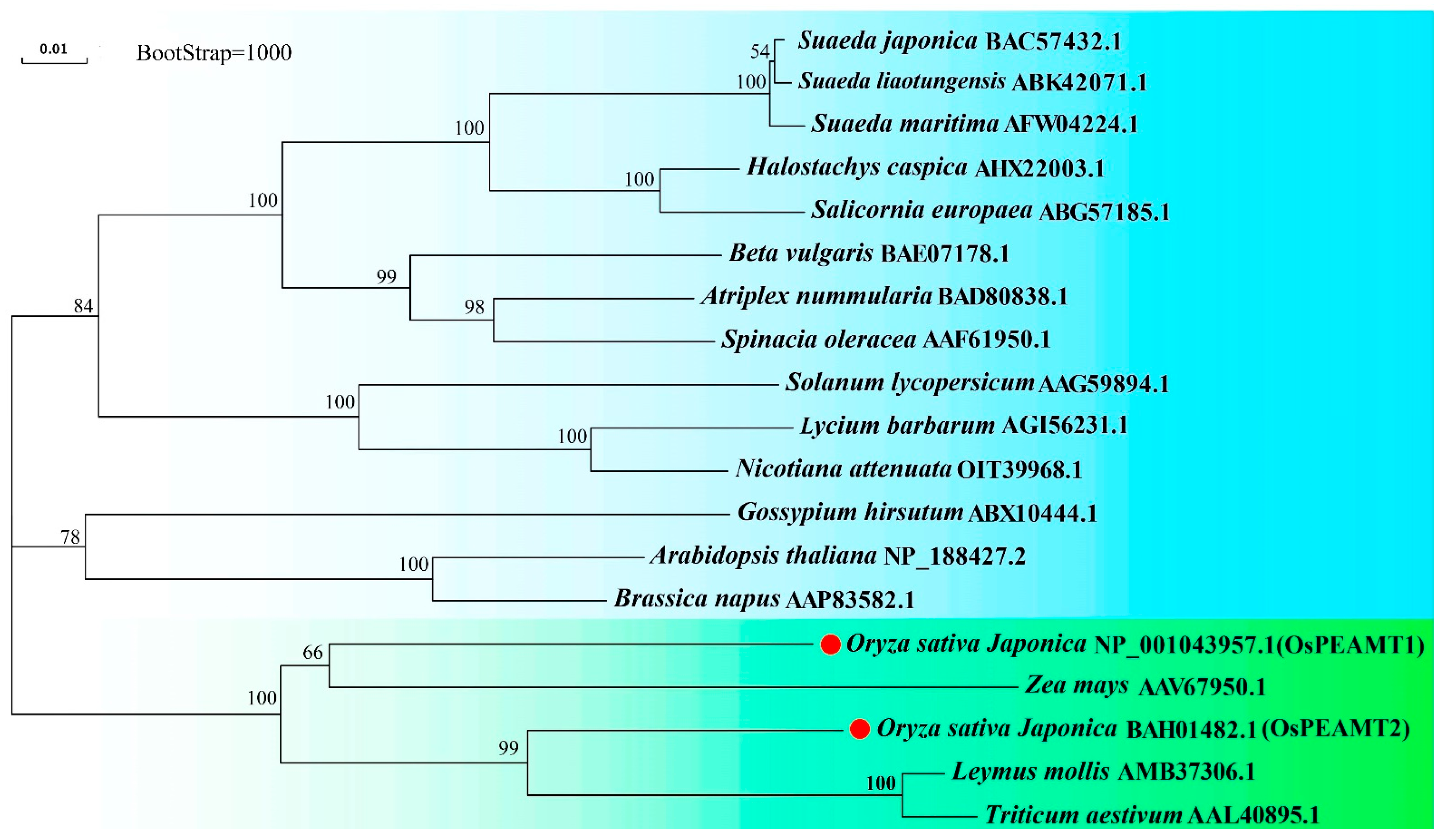
2.1. Identification of OsPEAMT1 and OsPEAMT2
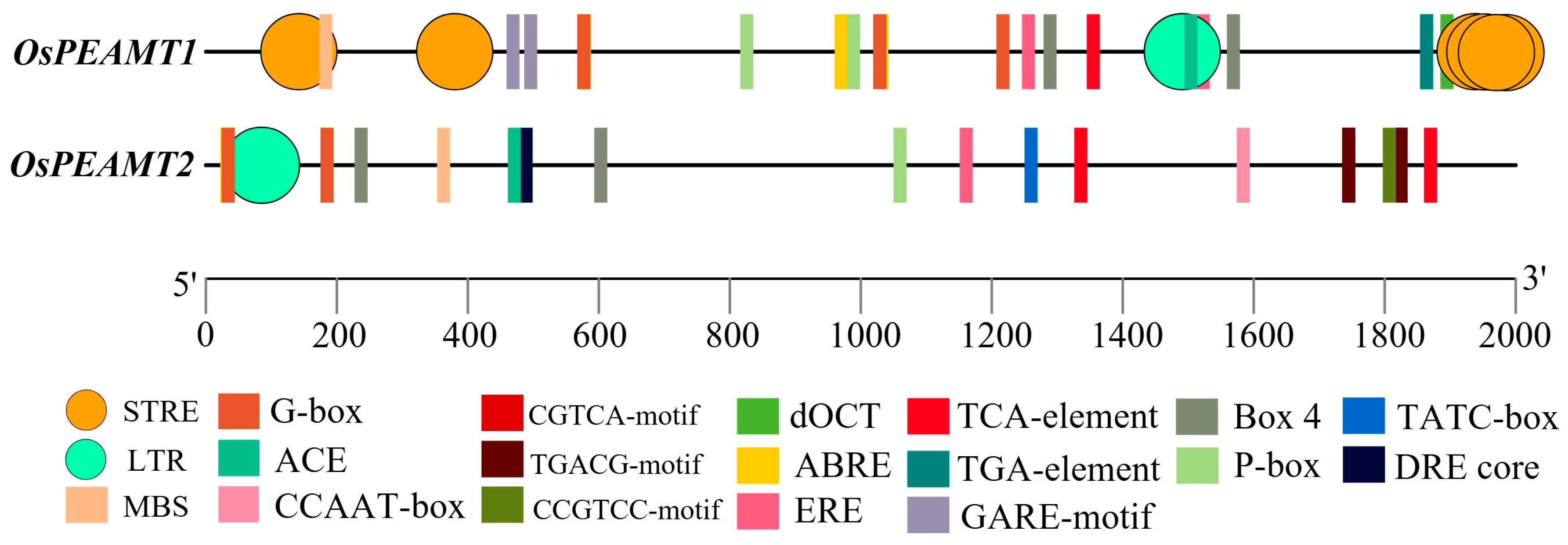
2.2. Identification of OsPEAMT1 and OsPEAMT2 Promoters
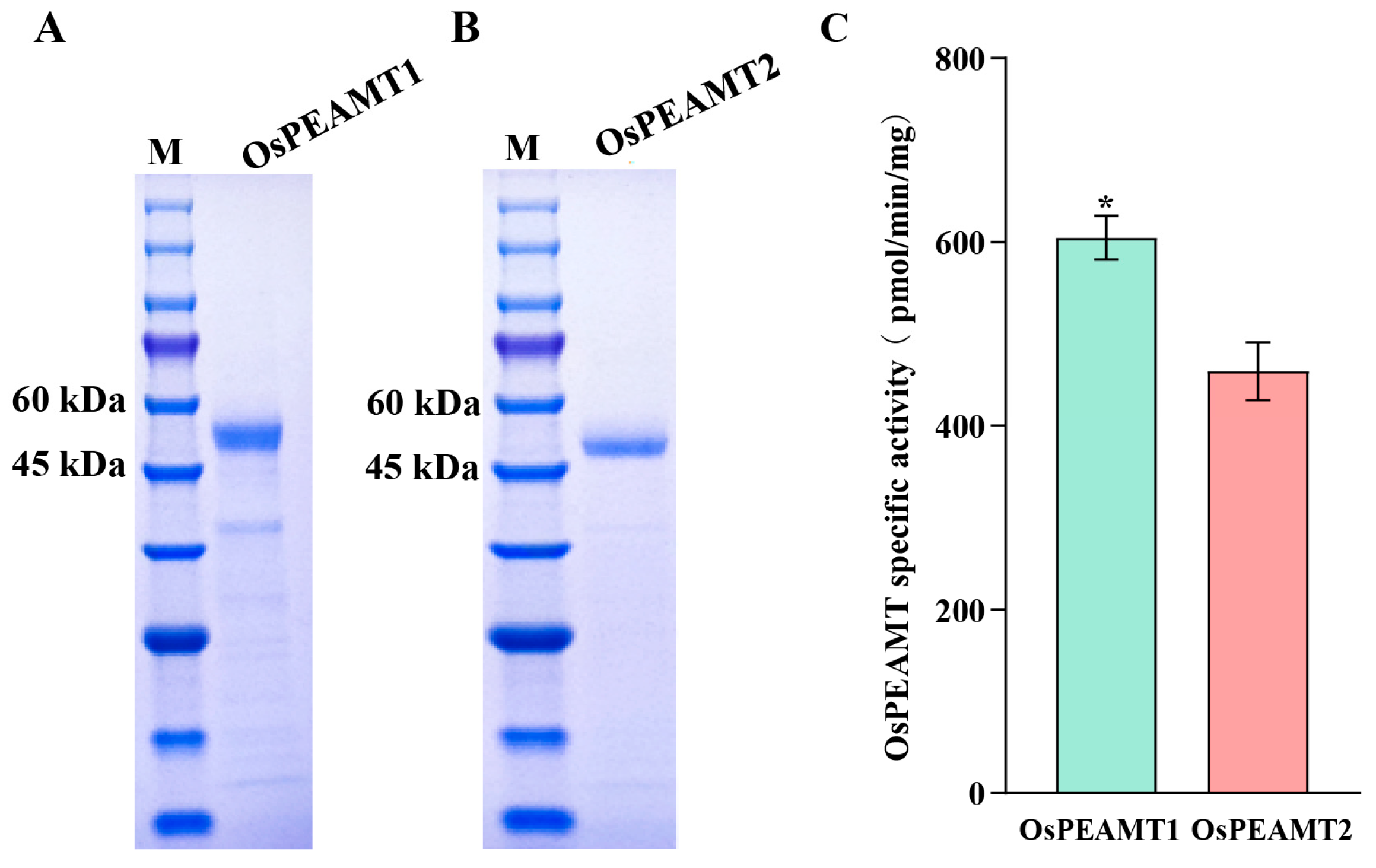
2.3. Detection of OsPEAMT1 and OsPEAMT2 Activity
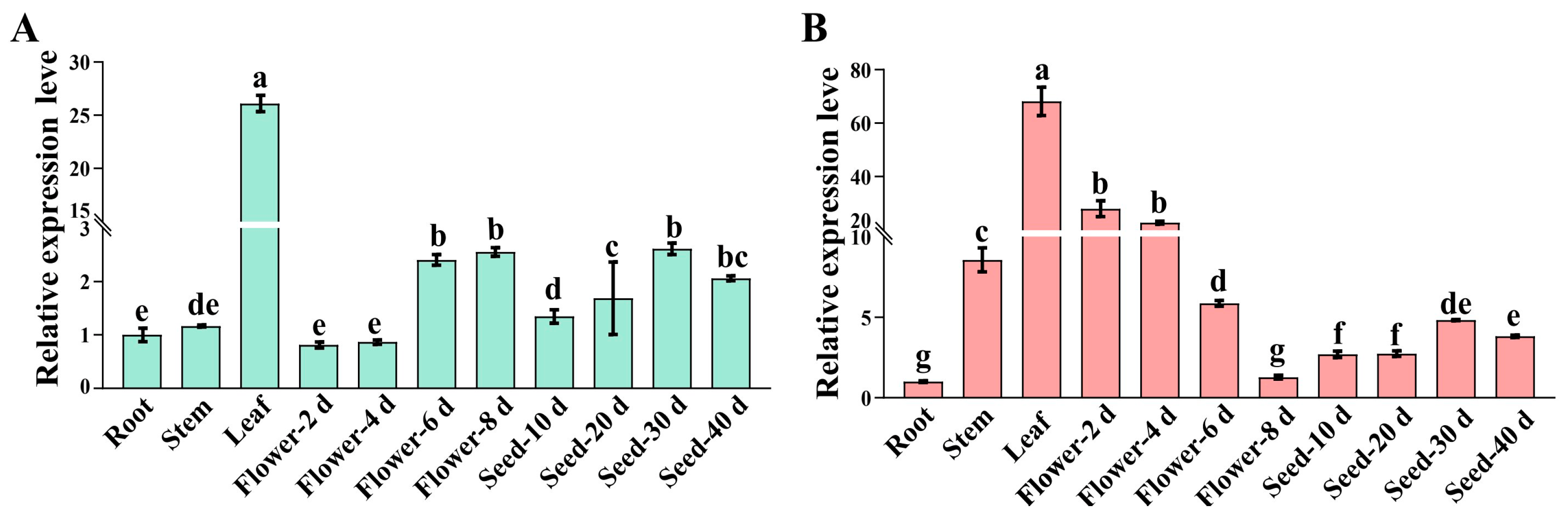
2.4. Expression Analysis of OsPEAMT1 and OsPEAMT2 in Different Tissues, Flowers, and Seeds at Various Developmental Stages
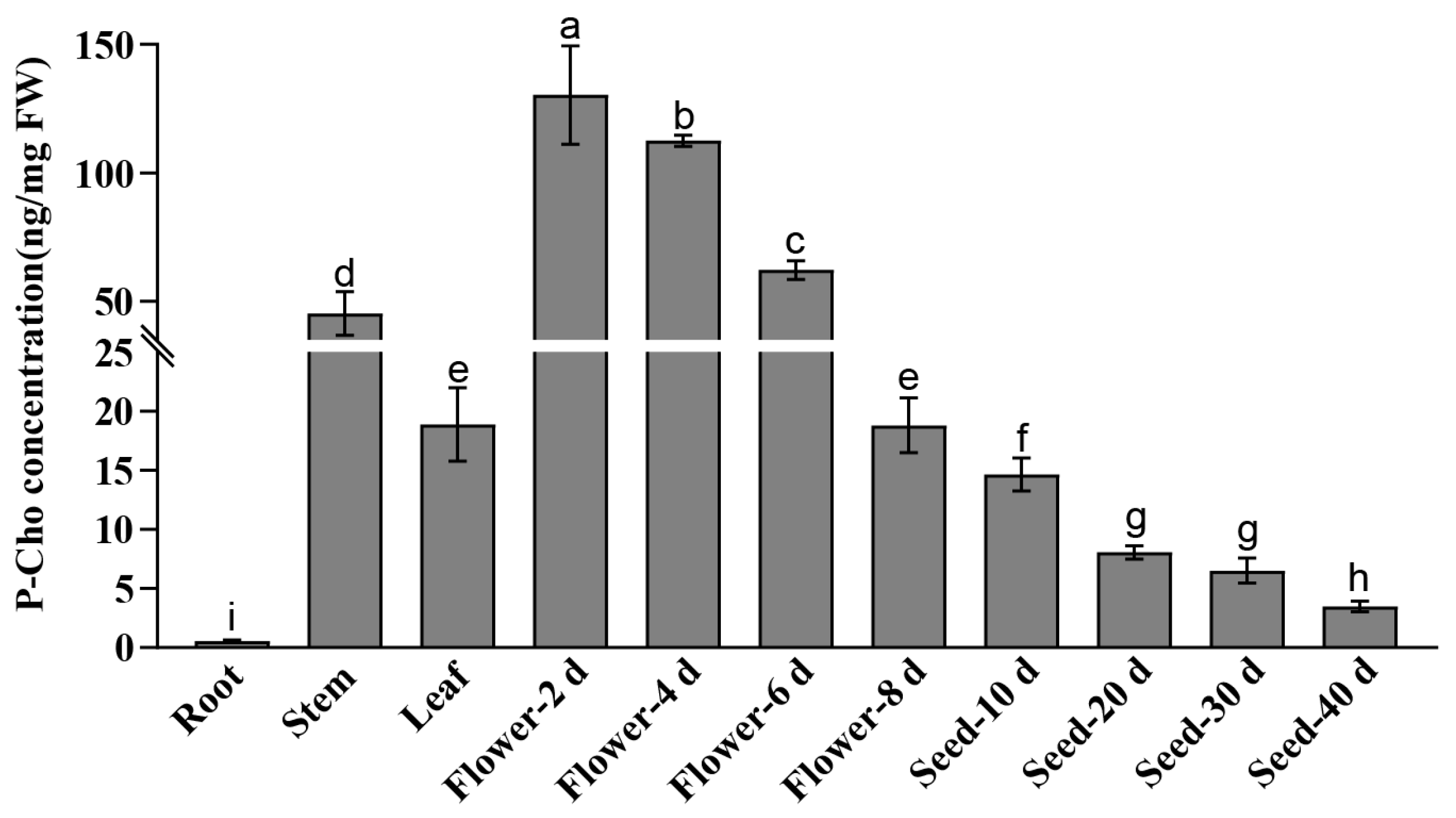
2.5. Determination of P-Cho Content in Different Tissues, Flowers, and Seeds at Various Developmental Stages
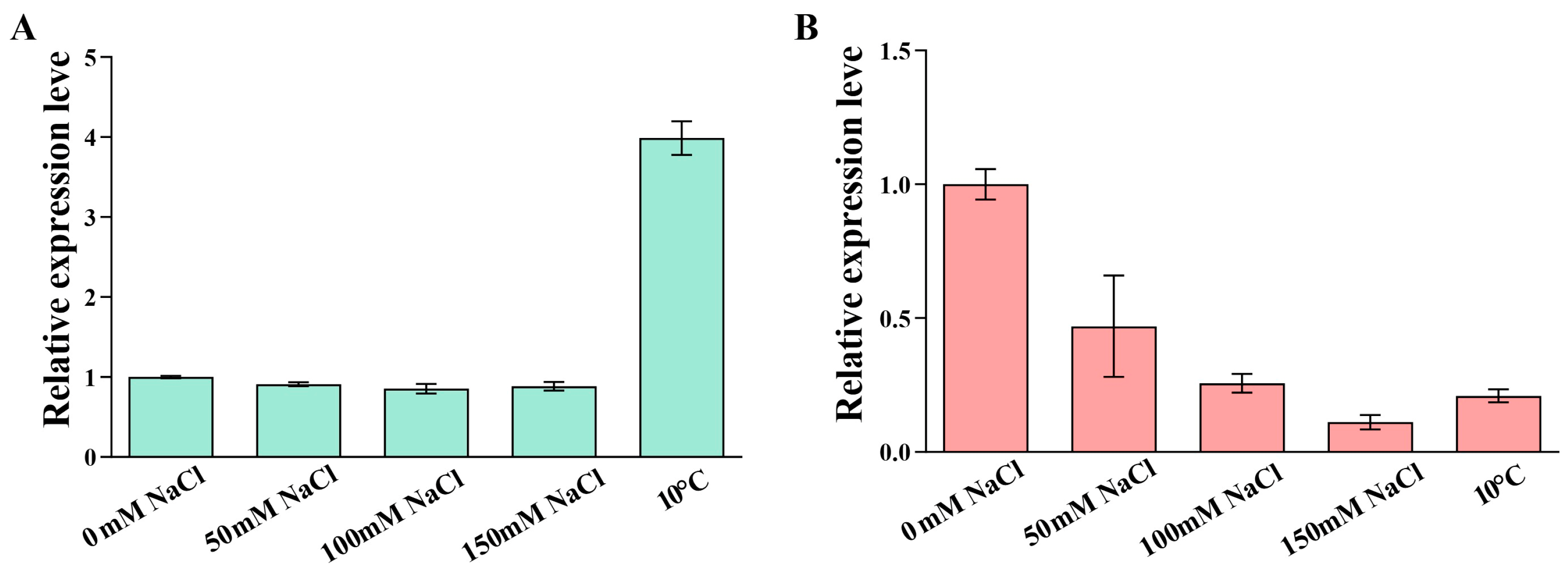
2.6. Expression Analysis of OsPEAMT1 and OsPEAMT2 Under Salt or Low-Temperature Stress
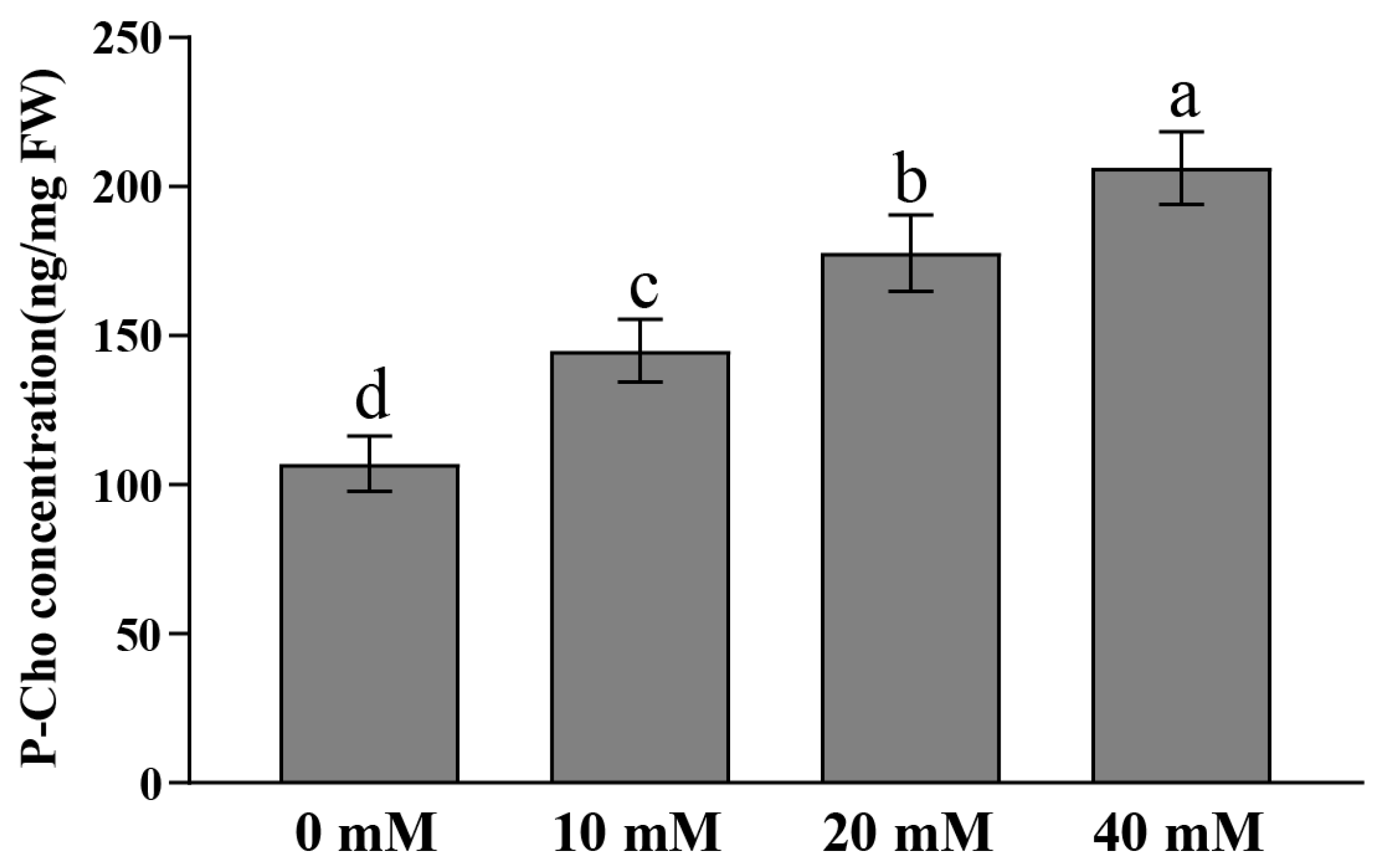
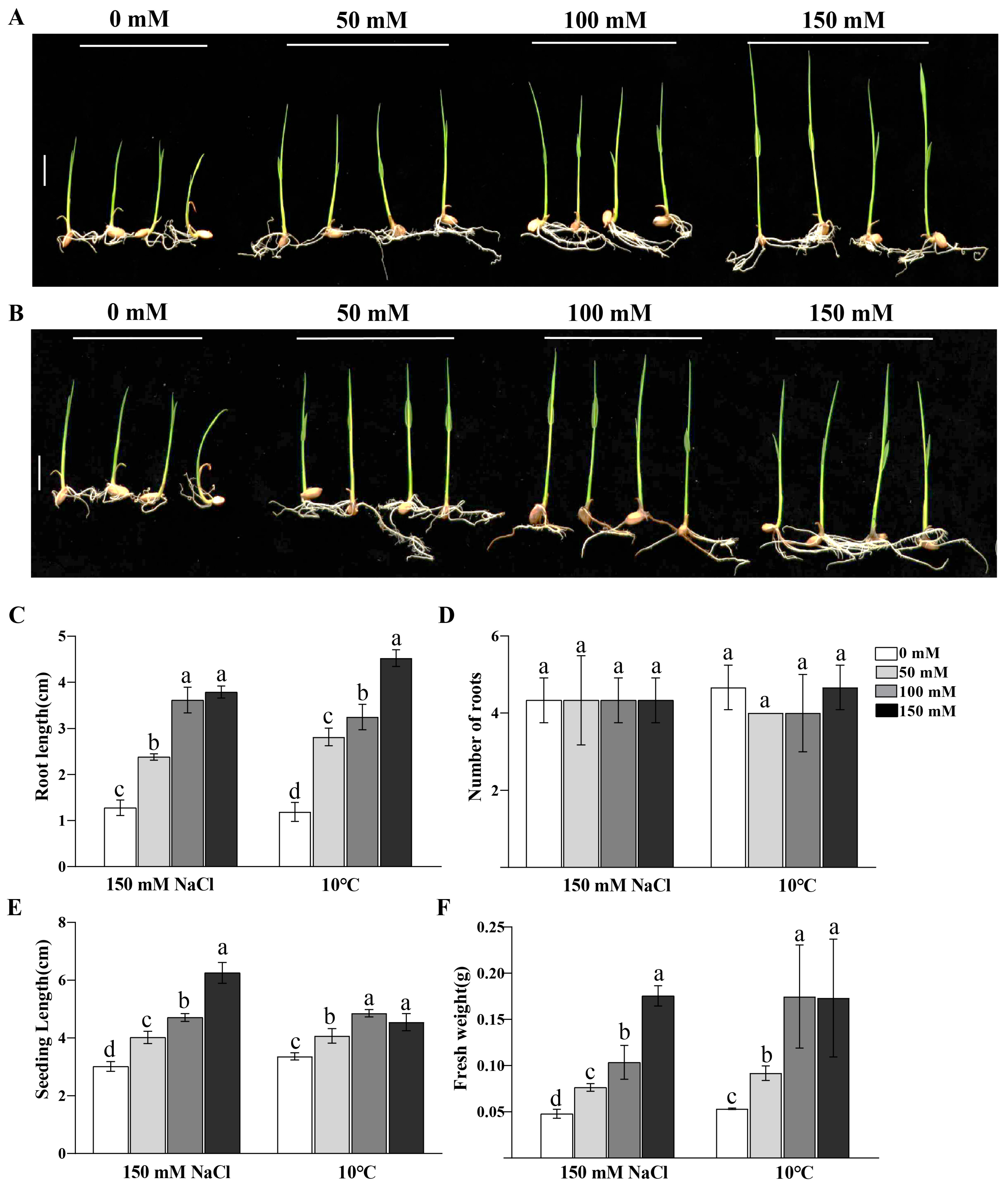
2.7. Response Analysis of Exogenous P-Cho to Salt and Low-Temperature Stress in Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of OsPEAMT1 and OsPEAMT2 Promoters
4.3. Sequence and Phylogenetic Analysis of OsPEAMT1 and OsPEAMT2 Proteins
4.4. Expression and Purification of OsPEAMT1 and OsPEAMT2 Proteins
4.5. Enzyme Assays of OsPEAMT1 and OsPEAMT2
4.6. Expression Analysis of OsPEAMT1 and OsPEAMT2
4.7. Quantification of P-Cho
4.8. Analysis of Response of Exogenous P-Cho to Salt and Low-Temperature Stress in Rice
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BADH | Betaine aldehyde dehydrogenase |
| CDP | Cytidine diphosphate |
| CMO | Choline monooxygenase |
| CTAB | Cetyltrimethylammonium bromide |
| CTP | Cytidine-5′ triphosphate |
| DAGs | Diacylglycerols |
| GB | Glycine betaine |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| LTR | Low-temperature response element |
| MBS | MYB-binding site |
| MT | Methyltransferase |
| NCBI | National Center for Biotechnology Information |
| PA | Phosphatidic acid |
| PC | Phosphatidylcholine |
| P-Cho | Phosphocholine |
| PDMEA | Phosphodimethylethanolamine |
| P-EA | Phosphoethanolamine |
| PEAMT | Phosphoethanolamine N-methyltransferase |
| PMMEA | Phosphomonomethylethanolamine |
| RT-qPCR | Real-time fluorescence quantitative PCR |
| STREs | Stress response elements |
References
- Lin, Y.; Liu, Y.; Nakamura, Y. The choline/ethanolamine kinase family in Arabidopsis: Essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 2015, 27, 1497–1511. [Google Scholar] [CrossRef]
- Cruz-Ramiérez, A.; Loépez-Bucio, J.; Ramiérez-Pimentel, G.; Zurita-Silva, A.; Saénchez-Calderon, L.; Ramiérez-Chaévez, E.; Gonzaélez-Ortega, E.; Herrera-Estrella, L. The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 2004, 16, 2020–2034. [Google Scholar] [CrossRef]
- Potocký, M.; Eliás, M.; Profotová, B.; Novotná, Z.; Valentová, O.; Zársky, V. Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 2003, 217, 122–130. [Google Scholar] [CrossRef]
- Chen, W.; Salari, H.; Taylor, M.C.; Jost, R.; Berkowitz, O.; Barrow, R.; Qiu, D.; Branco, R.; Masle, J. NMT1 and NMT3 N-methyltransferase activity is critical to lipid homeostasis, morphogenesis, and reproduction. Plant Physiol. 2018, 177, 1605–1628. [Google Scholar] [CrossRef]
- Tian, F.; Wang, W.; Liang, C.; Wang, X.; Wang, G.; Wang, W. Overaccumulation of glycine betaine makes the function of the thylakoid membrane better in wheat under salt stress. Crop J. 2017, 5, 73–82. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, T.; Zuo, J.; Gao, L.; Fan, L. Amelioration of postharvest chilling injury in sweet pepper by glycine betaine. Postharvest Biol. Technol. 2016, 112, 114–120. [Google Scholar] [CrossRef]
- Bolognese, C.P.; McGraw, P. The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine: Phospho-ethanolamine N-methyltransferase. Plant Physiol. 2000, 124, 1800–1813. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, M.L.; Ziemak, M.J.; Henry, S.A.; Weretilnyk, E.A.; Hanson, A.D. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 2000, 275, 14095–14101. [Google Scholar] [CrossRef]
- Charron, J.F.; Breton, G.; Danyluk, J.; Muzac, I.; Ibrahim, R.K.; Sarhan, F. Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol. 2002, 129, 363–373. [Google Scholar] [CrossRef]
- Wu, S.; Yu, Z.; Wang, F.; Li, W.; Ye, C.; Li, J.; Tang, J.; Ding, J.; Zhao, J.; Wang, B. Cloning, characterization, and transformation of the phosphoethanolamine N-methyltransferase gene (ZmPEAMT1) in maize (Zea mays L.). Mol. Biotechnol. 2007, 36, 102–112. [Google Scholar] [CrossRef]
- BeGora, M.D.; Macleod, M.J.R.; McCarry, B.E.; Summers, P.S. Identification of phosphomethylethanolamine N-methyltransferase from Arabidopsis and its role in choline and phospholipid metabolism. J. Biol. Chem. 2010, 285, 29147–29155. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Jez, J.M. Conformational changes in the di-domain structure of Arabidopsis phosphoethanolamine methyltransferase leads to active-site formation. J. Biol. Chem. 2017, 292, 21690–21702. [Google Scholar] [CrossRef]
- Mou, Z.; Wang, X.; Fu, Z.; Dai, Y.; Han, C.; Ouyang, J.; Bao, F.; Hu, Y.; Li, J. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 2002, 14, 2031–2043. [Google Scholar] [CrossRef] [PubMed]
- Alatorre-Cobos, F.; Cruz-Ramírez, A.; Hayden, C.A.; Pérez-Torres, C.-A.; Chauvin, A.-L.; Ibarra-Laclette, E.; Alva-Cortés, E.; Jorgensen, R.A.; Herrera-Estrella, L. Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J. Exp. Biol. 2012, 63, 5203–5221. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, X.; Tan, Y.; Huang, J.; Zheng, Z.; Tao, L. Phosphoethanolamine N-methyltransferase 1 contributes to maintenance of root apical meristem by affecting ROS and auxin-regulated cell differentiation in Arabidopsis. New Phytol. 2019, 224, 258–273. [Google Scholar] [CrossRef]
- He, Q.Y.; Jin, J.F.; Lou, H.Q.; Dang, F.F.; Xu, J.M.; Zheng, S.J.; Yang, J.L. Abscisic acid-dependent PMT1 expression regulates salt tolerance by alleviating abscisic acid-mediated reactive oxygen species production in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1803–1820. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, J.; Di, H.; Huo, Y. Clone and expression characterization of CsPEAMT gene in Cleistogenes songorica. Acta Bot. Boreal.-Occident. Sin. 2015, 34, 2367–2373. [Google Scholar]
- Wang, A.H.; Yang, L.; Yao, X.Z.; Wen, X.P. Overexpression of the pitaya phosphoethanolamine N-methyltransferase gene (HpPEAMT) enhanced simulated drought stress in tobacco. Plant Cell Tiss. Organ 2021, 146, 29–40. [Google Scholar] [CrossRef]
- Ma, P.; Li, J.; Sun, G.; Zhu, J. Comparative transcriptome analysis reveals the adaptive mechanisms of halophyte Suaeda dendroides encountering high saline environment. Front. Plant Sci. 2024, 15, 1283912. [Google Scholar] [CrossRef]
- Ye, C.; Wu, S.; Yang, H.; Ma, C.; Yang, G.; Wang, B. Cloning, sequencing and salt induced expression of PEAMT and BADH in oilseed rape (Brassica napus). DNA Seq. 2005, 16, 364–371. [Google Scholar] [CrossRef]
- Zhang, H.; Murzello, C.; Sun, Y.; Kim, M.-S.; Xie, X.; Jeter, R.M.; Zak, J.C.; Dowd, S.E.; Paré, P.W. Choline and osmotic-stress tolerance induced in Arabidopsis by the soil microbe Bacillus subtilis (GB03). Mol. Plant Microbe Interact. 2010, 23, 1097–1104. [Google Scholar] [CrossRef]
- Adak, M.K.; Chakraborty, N.; Roy, S. Plant-Microbe Interaction Under Environmental Extremities with Special Reference to Rhizospheric Engineering. In Plant-Microbe Interaction Under Xenobiotic Exposure; Springer Nature: Singapore, 2025; pp. 305–322. [Google Scholar]
- Jost, R.; Berkowitz, O.; Shaw, J.; Masle, J. Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic aci. J. Biol. Chem. 2009, 284, 31962–31971. [Google Scholar]
- Niu, G.; Gou, W.; Han, X.; Qin, C.; Zhang, L.-X.; Abomohra, A.E.-F.; Ashraf, M. Cloning and functional analysis of phosphoethanolamine methyltransferase promoter from maize (Zea mays L.). Int. J. Mol. Sci. 2018, 19, 191. [Google Scholar] [CrossRef]
- Du, K.; Zhao, W.; Mao, Y.; Lv, Z.; Khattak, W.A.; Ali, S.; Wang, Y. Maize ear growth is stimulated at the fourth day after pollination by cell wall remodeling and changes in lipid and hormone signaling. J. Sci. Food Agric. 2022, 102, 5429–5439. [Google Scholar] [CrossRef]
- Hashim, N.; Ali, M.M.; Mahadi, M.R.; Abdullah, A.F.; Wayayok, A.; Kassim, M.S.M.; Jamaluddin, A. Smart farming for sustainable rice production: An insight into application, challenge, and future prospect. Rice Sci. 2024, 31, 47–61. [Google Scholar] [CrossRef]
- Guo, T.; Yu, H.; Qiu, J.; Li, J.; Han, B.; Lin, H. Advances in rice genetics and breeding by molecular design in China. Sci. Sin. Vitae 2019, 49, 1185–1212. [Google Scholar]
- Liu, M.; Wu, X.; Li, M.; Li, S.; Xiong, T.; Li, C.; Tang, Y. Multi-omics analysis reveals the physiological and molecular response to cold stress in different spring wheat cultivars at the booting stage. Front. Plant Sci. 2025, 16, 1594676. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Y.; Tang, W.; Liu, J.; Lu, B.-R.; Liu, Y. The accumulation of glycine betaine is dependent on choline monooxygenase (OsCMO), not on phosphoethanolamine N-methyltransferase (OsPEAMT1), in rice (Oryza sativa L. ssp. japonica). Plant Mol. Biol. Rep. 2014, 32, 916–922. [Google Scholar]
- Hirashima, T.; Toyoshima, M.; Moriyama, T.; Sato, N. Evolution of the phosphatidylcholine biosynthesis pathways in green algae: Combinatorial diversity of methyltransferases. J. Mol. Evol. 2018, 86, 68–76. [Google Scholar] [CrossRef]
- Chen, W.; Taylor, M.C.; Barrow, R.A.; Croyal, M.; Masle, J. Loss of phosphoethanolamine N-methyltransferases abolishes phosphatidylcholine synthesis and is lethal. Plant Physiol. 2019, 179, 124–142. [Google Scholar] [PubMed]
- Luo, D.; Niu, X.; Yu., J.; Yan, J.; Gou, X.; Lu, B.-R.; Liu, Y. Rice choline monooxygenase (OsCMO) protein functions in enhancing glycine betaine biosynthesis in transgenic tobacco but does not accumulate in rice (Oryza sativa L. ssp. japonica). Plant Cell Rep. 2012, 31, 1625–1635. [Google Scholar] [CrossRef]
- Ji, X.; Wu, X.; Chen, W.; Yuan, Q.; Shen, Y.; Chi, Y. Cloning and functional identification of phosphoethanolamine methyltransferase in soybean (Glycine max). Front. Plant Sci. 2021, 12, 612158. [Google Scholar] [CrossRef]
- McNeil, S.D.; Nuccio, M.L.; Ziemak, M.J.; Hanson, A.D. Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 2001, 98, 10001–10005. [Google Scholar] [CrossRef]
- Liu, J.H.; Peng, T.; Dai, W. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, D.; Liu, X.; Zai, Z.; Gu, H.; Zheng, D.; Wang, L.; Yue, Y.; Yang, X. Systematic identification and characterization of Group IV WRKYs in Osmanthus fragrans and induction of salt-sensitivity by OfWRKY157. Plant Stress. 2025, 17, 100956. [Google Scholar] [CrossRef]
- Jiang, L.; Xiao, W.; Chen, H.; Qi, Y.; Kuang, X.; Shi, J.; Liu, Z.; Gao, J.; Lin, Q.; Yu, F.; et al. The OsGAPC1-OsSGL module negatively regulates salt tolerance by mediating abscisic acid biosynthesis in rice. New Phytol. 2024, 244, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, S.; Shinozaki, K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001, 26, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T. Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 2001, 6, 227–233. [Google Scholar] [CrossRef]
- Sun, M.; Liu, X.; Zhang, B.; Yu, W.; Xiao, Y.; Peng, F. Lipid metabolomic and transcriptomic analyses reveal that phosphatidylcholine enhanced the resistance of peach seedlings to salt stress through phosphatidic acid. J. Agric. Food Chem. 2023, 71, 8846–8858. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Z.; Xie, Y.B.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Mussakhmetov, A.; Utepbergenov, D.; Khassenov, B. High affinity of recombinant DJ-1 (PARK7) protein to Ni-NTA. Eurasian J. Appl. Biotechnol. 2022, 2, 33–37. [Google Scholar] [CrossRef]
- Hirashima, T.; Toyoshima, M.; Moriyama, T.; Nakamura, Y.; Sato, N. Characterization of phosphoethanolamine-N-methyltransferases in green algae. Biochem. Biophys. Res. Commun. 2017, 488, 141–146. [Google Scholar] [CrossRef]
- Kanani, P.; Shukla, Y.M.; Modi, A.R.; Subhash, N.; Kumar, S. Standardization of an efficient protocol for isolation of RNA from Cuminum cyminum. J. King Saud. Univ. Sci. 2019, 31, 1202–1207. [Google Scholar] [CrossRef]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Zhang, Y.; Ma, S.; Wen, X.; Zhao, N.; Feng, X.; Zong, D.; Li, J. A Comprehensive Functional Analysis of OsPEAMT1 and OsPEAMT2 Genes in Rice (Oryza sativa L. ssp. japonica). Plants 2025, 14, 2935. https://doi.org/10.3390/plants14182935
Yu J, Zhang Y, Ma S, Wen X, Zhao N, Feng X, Zong D, Li J. A Comprehensive Functional Analysis of OsPEAMT1 and OsPEAMT2 Genes in Rice (Oryza sativa L. ssp. japonica). Plants. 2025; 14(18):2935. https://doi.org/10.3390/plants14182935
Chicago/Turabian StyleYu, Jinde, Yuying Zhang, Shaojie Ma, Xia Wen, Ning Zhao, Xiaofei Feng, Dan Zong, and Jing Li. 2025. "A Comprehensive Functional Analysis of OsPEAMT1 and OsPEAMT2 Genes in Rice (Oryza sativa L. ssp. japonica)" Plants 14, no. 18: 2935. https://doi.org/10.3390/plants14182935
APA StyleYu, J., Zhang, Y., Ma, S., Wen, X., Zhao, N., Feng, X., Zong, D., & Li, J. (2025). A Comprehensive Functional Analysis of OsPEAMT1 and OsPEAMT2 Genes in Rice (Oryza sativa L. ssp. japonica). Plants, 14(18), 2935. https://doi.org/10.3390/plants14182935






