Genome-Wide Identification of Arachis hypogaea LEC1s, FUS3s, and WRIs and Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a and AhWRI1d Increased Oil Content in Arabidopsis Seeds
Abstract
1. Introduction
2. Results
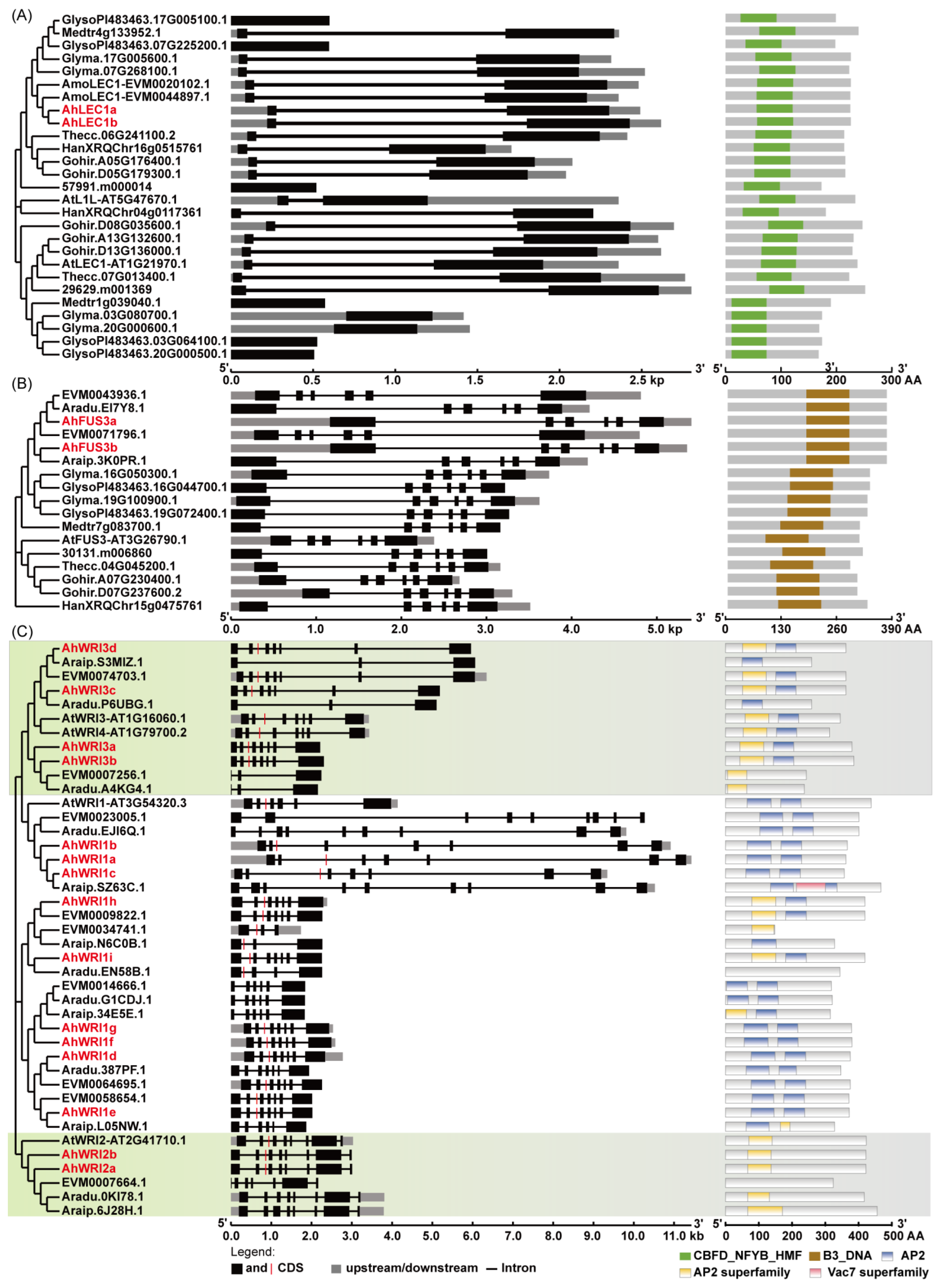
2.1. Cloning and Identification of AhLEC1s, AhFUS3s, and AhWRIs in A. hypogaea
2.2. Protein Alignment and Identity Analysis
2.3. Chromosomal Distribution and Collinearity Analysis
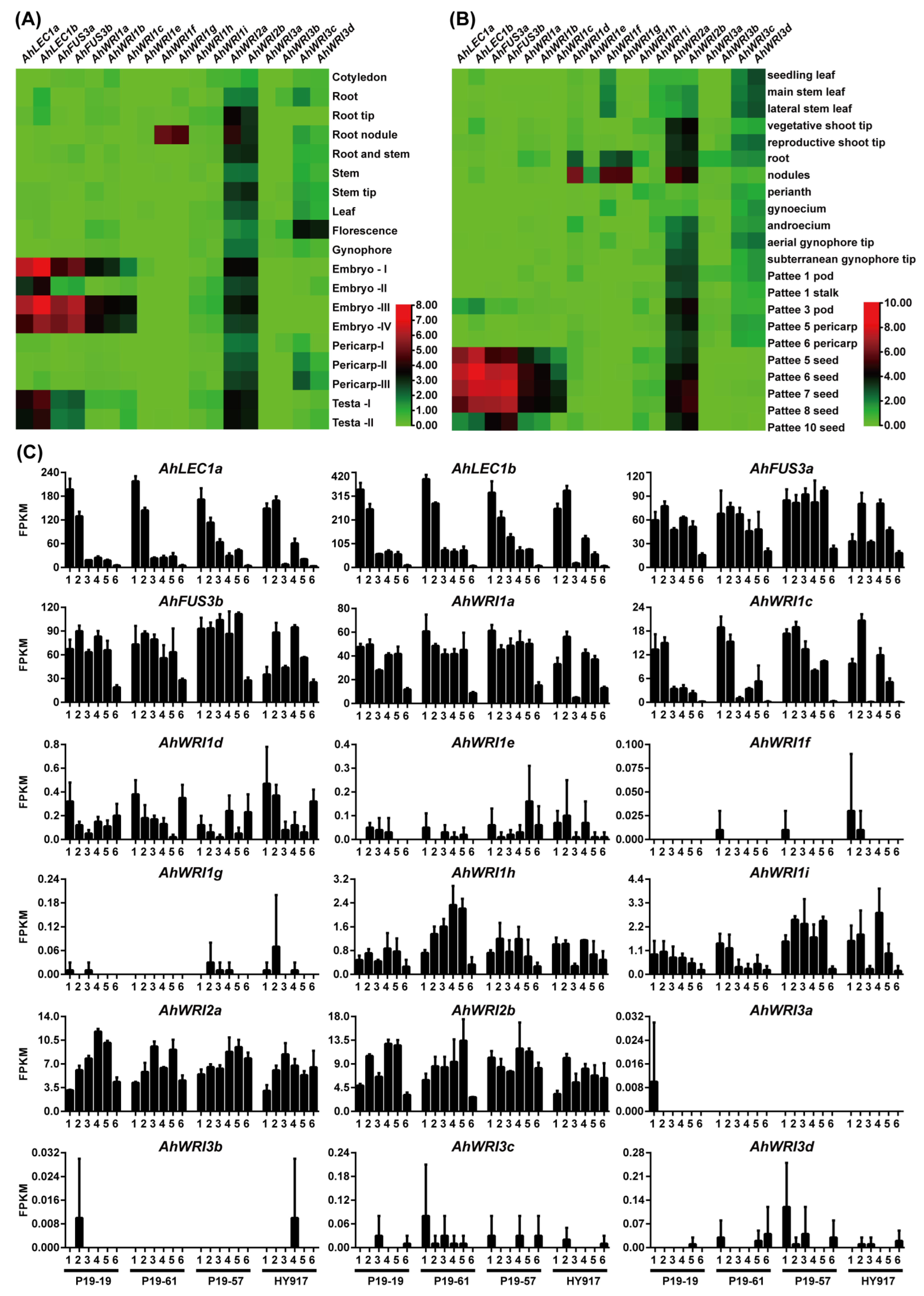
2.4. Transcriptional Profiles of AhLEC1s, AhFUS3s, and AhWRIs
2.5. Putative Cis-Elements in the Promoters of AhLEC1s, AhFUS3s, and AhWRIs and miRNA Regulating AhLEC1s, AhFUS3s, and AhWRIs
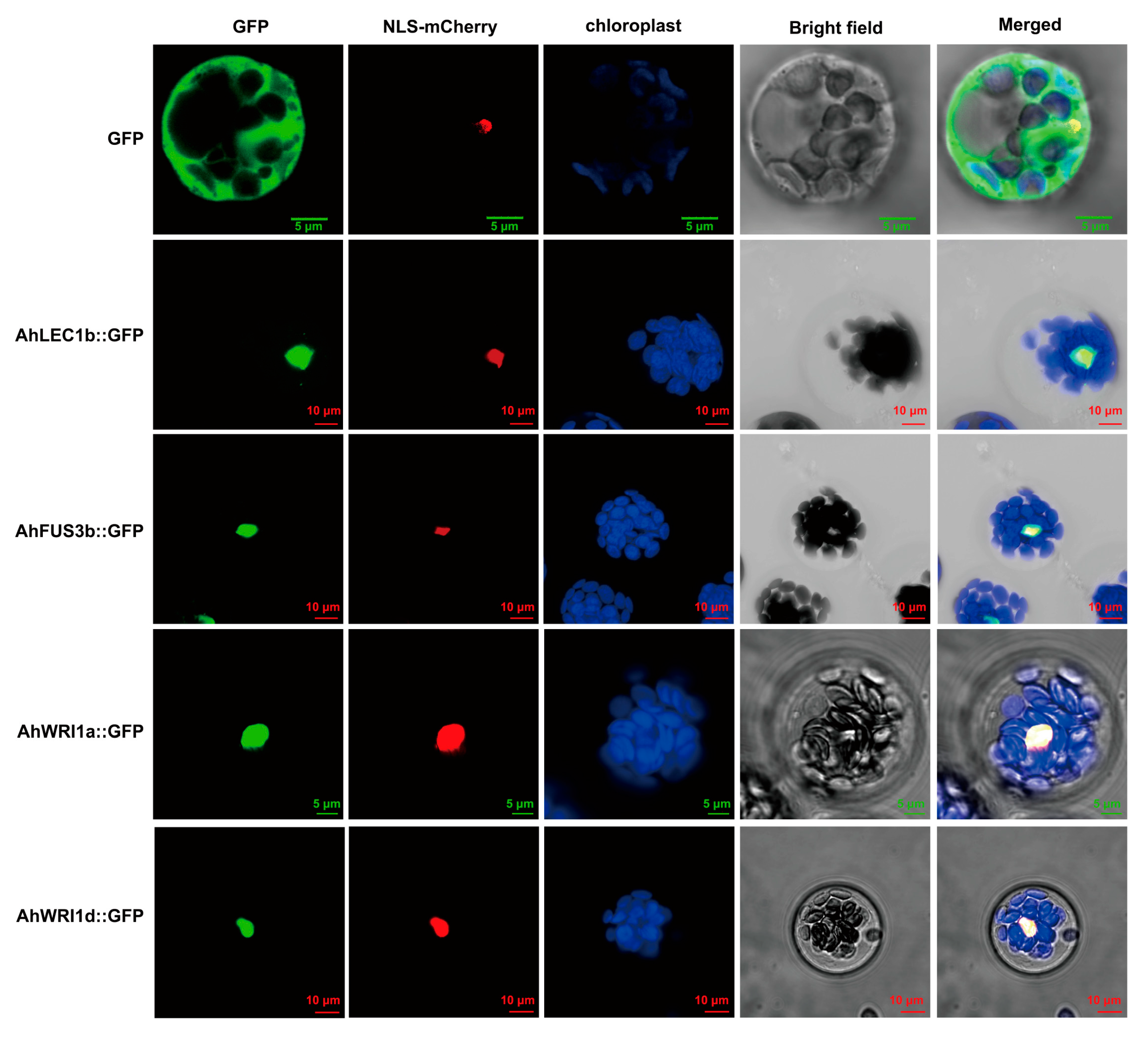
2.6. Subcellular Localization of AhLEC1s, AhFUS3s, and AhWRIs
2.7. Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a, and AhWRI1d Increased Fatty Acids Accumulation and Thousand-Seed Weight, but Decreased Germination Rate, Plant Height, and Silique Length
3. Discussion
3.1. Gene Duplication and Functional Diversification of AhLEC1s in A. hypogaea
3.2. Gene Duplication and Functional Diversification of AhFUS3s in A. hypogaea
3.3. Gene Duplication and Functional Diversification of AhWRIs in A. hypogaea
3.4. Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a, and AhWRI1d Alter Key Major Agronomic Traits
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. BLASTing and Cloning of LEC1, FUS3, and WRI Family Members in A. hypogaea
4.3. Construction of Plant Expression Vectors and Genetic Transformation
4.4. Analysis of Phylogenetic Tree, Conserved Domain, Physicochemical Properties, Sequence Alignments, Gene Structures, Identity, and PEST Motifs
4.5. Chromosomal Location and Gene Synteny Analysis
4.6. Analysis of Expression Patterns
4.7. Analyses of Cis-Acting Elements and miRNAs Targeting AhLEC1s, AhFUS3s, and AhWRIs
4.8. Analysis of the Content and Composition of Fatty Acids in Seeds
4.9. Measurement of Thousand-Seed Weight, Plant Height, and Silique Length, and Warm Germination Test
4.10. Quantitative RT–PCR (qRT–PCR) Analysis of Gene Expression
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP2/EREBP | APETALA2-ethylene-responsive element-binding protein |
| CDD | the Conserved Domain Database |
| CDS | coding sequence |
| FA | fatty acid |
| FPKM | Fragments Per Kilobase of transcript per Million mapped reads |
| FUS3 | FUSCA3 |
| GRAVY | the grand average of hydropathicity |
| GSDS | Gene Structure Display Serve |
| JTT | Jones Taylor Thornton |
| LEC1 | LEAFY COTYLEDON1 |
| L1L | LEAFY COTYLEDON1-LIKE |
| NLS | nuclear localization signal |
| MW | molecular weight |
| PGR | Peanut Genome Resource |
| pI | isoelectric point |
| PKL | PICKLE |
| TAIR | The Arabidopsis Information Resource |
| TPM | Transcripts Per Million |
| VAL | VIVIPAROUS ABI3-LIKE protein |
| WRI1 | WRINKLED1 |
| WT | wild-type |
References
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef] [PubMed]
- Covas, M.I.; de la Torre, R.; Fito, M. Virgin olive oil: A key food for cardiovascular risk protection. Br. J. Nutr. 2015, 113, S19–S28. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kong, Q.; Lim, A.R.Q.; Lu, S.; Zhao, H.; Guo, L.; Yuan, L.; Ma, W. Transcriptional regulation of oil biosynthesis in seed plants: Current understanding, applications, and perspectives. Plant Commun. 2022, 3, 100328. [Google Scholar] [CrossRef] [PubMed]
- Tyczewska, A.; Woźniak, E.; Gracz, J.; Kuczyński, J.; Twardowski, T. Towards food security: Current state and future prospects of agrobiotechnology. Trends Biotechnol. 2018, 36, 1219–1229. [Google Scholar] [CrossRef]
- Li, D.; Jin, C.; Duan, S.; Zhu, Y.; Qi, S.; Liu, K.; Gao, C.; Ma, H.; Zhang, M.; Liao, Y.; et al. MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 2017, 173, 1211–1225. [Google Scholar] [CrossRef]
- Santos-Mendoza, M.; Dubreucq, B.; Baud, S.; Parcy, F.; Caboche, M.; Lepiniec, L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008, 54, 608–620. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.-J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, X.; Jia, Q.; Ohlrogge, J. FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 2016, 88, 95–107. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef]
- Lotan, T.; Ohto, M.-a.; Yee, K.M.; West, M.A.L.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 2003, 100, 2152–2156. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Zhu, Y.; Xie, L.; Chen, G.Q.; Lee, M.Y.; Loque, D.; Scheller, H.V. A transgene design for enhancing oil content in Arabidopsis and Camelina seeds. Biotechnol. Biofuels 2018, 11, 46. [Google Scholar] [CrossRef]
- Tan, H.; Yang, X.; Zhang, F.; Zheng, X.; Qu, C.; Mu, J.; Fu, F.; Li, J.; Guan, R.; Zhang, H.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef]
- Manan, S.; Alabbosh, K.F.; Al-Andal, A.; Ahmad, W.; Khan, K.A.; Zhao, J. Soybean LEAFY COTYLEDON 1: A key target for genetic enhancement of oil biosynthesis. Agronomy 2023, 13, 2810. [Google Scholar] [CrossRef]
- Tang, G.; Xu, P.; Jiang, C.; Li, G.; Shan, L.; Wan, S. Peanut LEAFY COTYLEDON1-type genes participate in regulating the embryo development and the accumulation of storage lipids. Plant Cell Rep. 2024, 43, 124. [Google Scholar] [CrossRef]
- Yang, Z.; Bai, T.; E, Z.; Niu, B.; Chen, C. OsNF-YB7 inactivates OsGLK1 to inhibit chlorophyll biosynthesis in rice embryo. eLife 2024, 13, RP96553. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhang, P.; Wu, Y.; Lian, G.; Yang, Z.; Liu, W.; Buerte, B.; Zhou, C.; Zhang, W.; Li, D.; et al. Rice LEAFY COTYLEDON1 hinders embryo greening during the seed development. Front. Plant Sci. 2022, 13, 887980. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Zhang, Z.; Zhang, J.; Zhou, Y.; Chen, C. The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm-specific sucrose synthase protein kinase, and functions in seed development. Plant J. 2021, 106, 1233–1246. [Google Scholar] [CrossRef]
- Luerßen, H.; Kirik, V.; Herrmann, P.; Miséra, S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998, 15, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.; Gazzarrini, S. Overlapping and distinct roles of AKIN10 and FUSCA3 in ABA and sugar signaling during seed germination. Plant Signal. Behav. 2012, 7, 1238–1242. [Google Scholar] [CrossRef]
- Ahmad, B.; Zhang, S.; Yao, J.; Chai, S.; Yadav, V.; Athar, H.-u.-R.; Rahman, M.U.; Wang, L.; Wang, X. Ectopic expression of VvFUS3, B3-domain transcription factor, in tomato influences seed development via affecting endoreduplication and hormones. Hortic. Plant J. 2022, 8, 351–360. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Li, X.; Jing, H.; Shao, Y.; Ma, R.; Hou, Q.; Chen, M. Linum usitatissimum FUSCA3–1 regulates plant architecture and seed storage reserve accumulation in Arabidopsis thaliana. Environ. Exp. Bot. 2022, 202, 105035. [Google Scholar] [CrossRef]
- Wang, H.; Guo, J.; Lambert, K.N.; Lin, Y. Developmental control of Arabidopsis seed oil biosynthesis. Planta 2007, 226, 773–783. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Usui, H.; Hobo, T.; Takeda, S.; Hattori, T. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 2010, 51, 2031–2046. [Google Scholar] [CrossRef]
- Manan, S.; Li, P.; Alfarraj, S.; Ansari, M.J.; Bilal, M.; Ullah, M.W.; Zhao, J. FUS3: Orchestrating soybean plant development and boosting stress tolerance through metabolic pathway regulation. Plant Physiol. Biochem. 2024, 213, 108803. [Google Scholar] [CrossRef]
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073. [Google Scholar] [CrossRef]
- Kagaya, Y.; Okuda, R.; Ban, A.; Toyoshima, R.; Tsutsumida, K.; Usui, H.; Yamamoto, A.; Hattori, T. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 2005, 46, 300–311. [Google Scholar] [CrossRef]
- Wang, F.; Perry, S.E. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013, 161, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Mitsui, N.; Tsukagoshi, H.; Nishii, T.; Morikami, A.; Nakamura, K. ACTIVATOR of Spomin::LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 2005, 46, 547–556. [Google Scholar] [CrossRef]
- To, A.; Joubès, J.; Barthole, G.; Lécureuil, A.; Scagnelli, A.; Jasinski, S.; Lepiniec, L.; Baud, S. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 2012, 24, 5007–5023. [Google Scholar] [CrossRef]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef]
- Kuczynski, C.; McCorkle, S.; Keereetaweep, J.; Shanklin, J.; Schwender, J. An expanded role for the transcription factor WRINKLED1 in the biosynthesis of triacylglycerols during seed development. Front Plant Sci 2022, 13, 955589. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoet, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, L.; Ma, W. WRINKLED1, a “Master Regulator” in Transcriptional Control of Plant Oil Biosynthesis. Plants 2019, 8, 238. [Google Scholar] [CrossRef]
- Fei, W.; Yang, S.; Hu, J.; Yang, F.; Qu, G.; Peng, D.; Zhou, B. Research advances of WRINKLED1 (WRI1) in plants. Funct. Plant Biol. 2020, 47, 185–194. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, G.; Li, P.; Yang, J.; Guo, L.; Benning, C.; Wang, X.; Zhao, J. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 2020, 18, 155–171. [Google Scholar] [CrossRef]
- Li, W.; Wang, L.; Qi, Y.; Xie, Y.; Zhao, W.; Dang, Z.; Zhang, J. Overexpression of WRINKLED1 improves the weight and oil content in seeds of flax (Linum usitatissimum L.). Front. Plant Sci. 2022, 13, 1003758. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Han, B.; Haque, M.E.; Li, Y.-L.; Wang, Y.; Wu, D.; Wu, S.-B.; Liu, A.-Z. The molecular mechanism of WRINKLED1 transcription factor regulating oil accumulation in developing seeds of castor bean. Plant Divers. 2023, 45, 469–478. [Google Scholar] [CrossRef]
- de Paula, A.F.; Dinato, N.B.; Vigna, B.B.Z.; Favero, A.P. Recombinants from the crosses between amphidiploid and cultivated peanut (Arachis hypogaea) for pest-resistance breeding programs. PLoS ONE 2017, 12, e0175940. [Google Scholar] [CrossRef] [PubMed]
- Fávero, A.P.; Simpson, C.E.; Valls, J.F.M.; Vello, N.A. Study of the evolution of cultivated peanut through crossability studies among Arachis ipaënsis, A. duranensis, and A. hypogaea. Crop Sci. 2006, 46, 1546–1552. [Google Scholar] [CrossRef]
- Seijo, G.; Lavia, G.I.; Fernández, A.; Krapovickas, A.; Ducasse, D.A.; Bertioli, D.J.; Moscone, E.A. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 2007, 94, 1963–1971. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Newman, C.S.; Andres, R.J.; Youngblood, R.C.; Campbell, J.D.; Simpson, S.A.; Cannon, S.B.; Scheffler, B.E.; Oakley, A.T.; Hulse-Kemp, A.M.; Dunne, J.C. Initiation of genomics-assisted breeding in Virginia-type peanuts through the generation of a de novo reference genome and informative markers. Front. Plant Sci. 2023, 13, 1073542. [Google Scholar] [CrossRef]
- Grabowski, P.P.; Dang, P.; Jenkins, J.J.; Sreedasyam, A.; Webber, J.; Lamb, M.; Zhang, Q.; Sanz-Saez, A.; Feng, Y.; Bunting, V.; et al. Relics of interspecific hybridization retained in the genome of a drought-adapted peanut cultivar. G3-Genes Genomes Genet. 2024, 14, jkae208. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Yin, D.; Ji, C.; Ma, X.; Li, H.; Zhang, W.; Li, S.; Liu, F.; Zhao, K.; Li, F.; Li, K.; et al. Genome of an allotetraploid wild peanut Arachis monticola: A de novo assembly. Gigascience 2018, 7, giy066. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Sinha, S.; Kim, I.S.; Sohn, K.Y.; de Crombrugghe, B.; Maity, S.N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell Biol. 1996, 16, 328–337. [Google Scholar] [CrossRef][Green Version]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.Á.; González, N.; Díaz, I.; Parcy, F.; Carbonero, P.; Vicente-Carbajosa, J. FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J. 2008, 53, 882–894. [Google Scholar] [CrossRef]
- Tsai, A.Y.-L.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef]
- Chan, A.; Carianopol, C.; Tsai, A.Y.-L.; Varatharajah, K.; Chiu, R.S.; Gazzarrini, S. SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J. Exp. Bot. 2017, 68, 4219–4231. [Google Scholar] [CrossRef]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef]
- Lu, Q.S.; Dela Paz, J.; Pathmanathan, A.; Chiu, R.S.; Tsai, A.Y.-L.; Gazzarrini, S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 2010, 64, 100–113. [Google Scholar] [CrossRef]
- Ji, X.-J.; Mao, X.; Hao, Q.-T.; Liu, B.-L.; Xue, J.-A.; Li, R.-Z. Splice variants of the castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int. J. Mol. Sci. 2018, 19, 146. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Wang, Y.-X.; Hu, X.; Zhang, Y.-L.; Wang, G.-L.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. An apple AP2/EREBP-type transcription factor, MdWRI4, enhances plant resistance to abiotic stress by increasing cuticular wax load. Environ. Exp. Bot. 2020, 180, 104206. [Google Scholar] [CrossRef]
- Cheng, C.; Hu, S.; Han, Y.; Xia, D.; Huang, B.-L.; Wu, W.; Hussain, J.; Zhang, X.; Huang, B. Yellow nutsedge WRI4-like gene improves drought tolerance in Arabidopsis thaliana by promoting cuticular wax biosynthesis. BMC Plant Biol. 2020, 20, 498. [Google Scholar] [CrossRef]
- Zang, X.; Pei, W.; Wu, M.; Geng, Y.; Wang, N.; Liu, G.; Ma, J.; Li, D.; Cui, Y.; Li, X.; et al. Genome-scale analysis of the WRI-like family in Gossypium and functional characterization of GhWRI1a controlling triacylglycerol content. Front. Plant Sci. 2018, 9, 1516. [Google Scholar] [CrossRef]
- Behera, J.R.; Rahman, M.M.; Bhatia, S.; Shockey, J.; Kilaru, A. Functional and predictive structural characterization of WRINKLED2, a unique oil biosynthesis regulator in avocado. Front. Plant Sci. 2021, 12, 648494. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of Camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Du, C.; Song, H.; Aziz, U.; Wang, L.; Zhao, C.; Zhang, M. Genome-wide analysis reveals the evolution and structural features of WRINKLED1 in plants. Mol. Genet. Genom. 2019, 294, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sanjaya; Sedbrook, J.; Benning, C. Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Mantyla, J.J.; Yang, Y.; Ohlrogge, J.B.; Benning, C. 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 2016, 88, 228–235. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lu, J.; Liu, S.; Liu, X.; Lin, Y.; Li, L. Identification of rapidly induced genes in the response of peanut (Arachis hypogaea) to water deficit and abscisic acid. BMC Biotechnol. 2014, 14, 58. [Google Scholar] [CrossRef]
- Pattee, H.E.; Johns, E.B.; Singleton, J.A.; Sanders, T.H. Composition Changes of Peanut Fruit Parts During Maturation. Peanut Sci. 1974, 1, 57–62. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Z.; Wang, X.; Wang, Y.; Hua, J. Molecular characterization and expression analysis of GhWRI1 in upland cotton. J. Plant Biol. 2018, 61, 186–197. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, Q.; Wang, W.; Yang, J.; Zhang, X.; Yu, N.; Zhou, Y.; Wang, E. Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. Plant 2018, 11, 1344–1359. [Google Scholar] [CrossRef]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef]
- Sun, F.; Liu, X.; Wei, Q.; Liu, J.; Yang, T.; Jia, L.; Wang, Y.; Yang, G.; He, G. Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front. Plant Sci. 2017, 8, 1133. [Google Scholar] [CrossRef]
- Yue, D.; Hao, X.; Han, B.; Xu, J.; Sun, W.; Guo, X.; Zhang, X.; Yang, X. GhL1L1 regulates the contents of unsaturated fatty acids by activating the expression of GhFAD2 genes in cotton. Gene 2024, 893, 147899. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Singh, S.K.; Mantyla, J.J.; Pattanaik, S.; Guo, L.; Yuan, L.; Benning, C.; Ma, W. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiol. 2020, 184, 658–665. [Google Scholar] [CrossRef]
- Fambrini, M.; Durante, C.; Cionini, G.; Geri, C.; Giorgetti, L.; Michelotti, V.; Salvini, M.; Pugliesi, C. Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus annuus and its relationship with zygotic and somatic embryogenesis. Dev. Genes Evol. 2006, 216, 253. [Google Scholar] [CrossRef]
- Alemanno, L.; Devic, M.; Niemenak, N.; Sanier, C.; Guilleminot, J.; Rio, M.; Verdeil, J.-L.; Montoro, P. Characterization of leafy cotyledon1-like during embryogenesis in Theobroma cacao L. Planta 2008, 227, 853–866. Planta 2008, 227, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Mantovani, R.; Hooft van Huijsduijnen, R.; Andre, I.; Benoist, C.; Mathis, D. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 1992, 20, 1087–1091. [Google Scholar] [CrossRef]
- Baud, S.; Kelemen, Z.; Thévenin, J.; Boulard, C.; Blanchet, S.; To, A.; Payre, M.; Berger, N.; Effroy-Cuzzi, D.; Franco-Zorrilla, J.M.; et al. Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol. 2016, 171, 1099–1112. [Google Scholar] [CrossRef]
- Xu, J.; Yang, X.; Li, B.; Chen, L.; Min, L.; Zhang, X. GhL1L1 affects cell fate specification by regulating GhPIN1-mediated auxin distribution. Plant Biotechnol. J. 2019, 17, 63–74. [Google Scholar] [CrossRef]
- Tang, G.; Xu, P.; Ma, W.; Wang, F.; Liu, Z.; Wan, S.; Shan, L. Seed-Specific Expression of AtLEC1 Increased Oil Content and Altered Fatty Acid Composition in Seeds of Peanut (Arachis hypogaea L.). Front. Plant Sci. 2018, 9, 260. [Google Scholar] [CrossRef]
- Casson, S.A.; Lindsey, K. The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 2006, 142, 526–541. [Google Scholar] [CrossRef]
- Tang, G.; Xu, P.; Liu, W.; Liu, Z.; Shan, L. Cloning and characterization of 5′ flanking regulatory sequences of AhLEC1B gene from Arachis hypogaea L. PLoS ONE 2015, 10, e0139213. PLoS ONE 2015, 10, e0139213. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Xu, P.; Li, P.; Zhu, J.; Chen, G.; Shan, L.; Wan, S. Cloning and functional characterization of seed-specific LEC1A promoter from peanut (Arachis hypogaea L.). PLoS ONE 2021, 16, e0242949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Tang, L.P.; Peng, J.; Zhai, L.M.; Ma, Q.L.; Zhang, X.S.; Su, Y.H. A WRI1-dependent module is essential for the accumulation of auxin and lipid in somatic embryogenesis of Arabidopsis thaliana. New Phytol. 2024, 242, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 2019, 61, 564–580. [Google Scholar] [CrossRef]
- Song, J.; Xie, X.; Chen, C.; Shu, J.; Thapa, R.K.; Nguyen, V.; Bian, S.; Kohalmi, S.E.; Marsolais, F.; Zou, J.; et al. LEAFY COTYLEDON1 expression in the endosperm enables embryo maturation in Arabidopsis. Nat. Commun. 2021, 12, 3963. [Google Scholar] [CrossRef]
- Junker, A.; Mönke, G.; Rutten, T.; Keilwagen, J.; Seifert, M.; Thi, T.M.N.; Renou, J.-P.; Balzergue, S.; Viehöver, P.; Hähnel, U.; et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012, 71, 427–442. [Google Scholar] [CrossRef]
- Zhang, S.; Wong, L.; Meng, L.; Lemaux, P.G. Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.). Planta 2002, 215, 191–194. [Google Scholar] [CrossRef]
- Liu, S.-J.; Zhang, H.; Jin, X.-T.; Niu, M.-X.; Feng, C.-H.; Liu, X.; Liu, C.; Wang, H.-L.; Yin, W.; Xia, X. PeFUS3 drives lateral root growth via auxin and ABA signalling under drought stress in populus. Plant Cell Environ. 2025, 48, 664–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Chen, Q.; Liu, S. FUSCA3, a multi-role regulator in the process of plant growth and development. Plant Cell Tiss. Organ Cult. 2022, 150, 1–13. [Google Scholar] [CrossRef]
- Duong, S.; Vonapartis, E.; Li, C.-Y.; Patel, S.; Gazzarrini, S. The E3 ligase ABI3-INTERACTING PROTEIN2 negatively regulates FUSCA3 and plays a role in cotyledon development in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 1555–1567. [Google Scholar] [CrossRef][Green Version]
- Baumbusch, L.O.; Hughes, D.W.; Galau, G.A.; Jakobsen, K.S. LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J. Exp. Bot. 2004, 55, 77–87. [Google Scholar] [CrossRef][Green Version]
- Tang, L.P.; Zhou, C.; Wang, S.S.; Yuan, J.; Zhang, X.S.; Su, Y.H. FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017, 213, 1740–1754. [Google Scholar] [CrossRef]
- To, A.; Valon, C.; Savino, G.; Guilleminot, J.; Devic, M.; Giraudat, J.; Parcy, F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 2006, 18, 1642–1651. [Google Scholar] [CrossRef]
- Roscoe, T.J.; Vaissayre, V.; Paszkiewicz, G.; Clavijo, F.; Kelemen, Z.; Michaud, C.; Lepiniec, L.; Dubreucq, B.; Zhou, D.-X.; Devic, M. Regulation of FUSCA3 Expression during seed development in Arabidopsis. Plant Cell Physiol. 2019, 60, 476–487. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, M.; Chen, T.; Xuan, L.; Li, Z.; Du, X.; Zhou, L.; Zhang, G.; Jiang, L. TRANSPARENT TESTA2 regulates embryonic fatty acid biosynthesis by targeting FUSCA3 during the early developmental stage of Arabidopsis seeds. Plant J. 2014, 77, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xuan, L.; Wang, Z.; Zhou, L.; Li, Z.; Du, X.; Ali, E.; Zhang, G.; Jiang, L. TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 2014, 165, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Han, X.; Peng, Y.; Yin, S.; Zhao, H.; Zong, Z.; Tan, Z.; Zhang, Y.; Ma, W.; Guo, L. Transcriptional regulation of transcription factor genes WRI1 and LAFL during Brassica napus seed development. Plant Physiol. 2024, 197, kiae378. [Google Scholar] [CrossRef]
- Tajima, D.; Kaneko, A.; Sakamoto, M.; Ito, Y.; Hue, N.T.; Miyazaki, M.; Ishibashi, Y.; Yuasa, T.; Iwaya-Inoue, M. Wrinkled 1 (WRI1) homologs, AP2-type transcription factors involving master regulation of seed storage oil synthesis in castor bean (Ricinus communis L.). Am. J. Plant Sci. 2013, 4, 7. [Google Scholar] [CrossRef]
- Sun, R.; Ye, R.; Gao, L.; Zhang, L.; Wang, R.; Mao, T.; Zheng, Y.; Li, D.; Lin, Y. Characterization and ectopic expression of CoWRI1, an AP2/EREBP domain-containing transcription factor from coconut (Cocos nucifera L.) endosperm, changes the seeds oil content in transgenic Arabidopsis thaliana and Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, Y.; Dong, Z.; Meng, F.; Sun, X.; Fan, X.; Zhang, Y.; Wang, M.; Wang, S. Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genom. 2018, 293, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, A.; Cao, X.; Dabbs, P.B.; Sung, H.-J.; Rahman, M.M.; Thrower, N.; Zynda, G.; Podicheti, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; et al. Oil biosynthesis in a basal angiosperm: Transcriptome analysis of Persea americana mesocarp. BMC Plant Biol. 2015, 15, 203. [Google Scholar] [CrossRef]
- Mano, F.; Aoyanagi, T.; Kozaki, A. Atypical splicing accompanied by skipping conserved micro-exons produces unique WRINKLED1, an AP2 domain transcription factor in rice plants. Plants 2019, 8, 207. [Google Scholar] [CrossRef]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 2013, 25, 2253–2264. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Grix, M.; Mantyla, J.J.; Yang, Y.; Benning, C.; Ohlrogge, J.B. Deletion of a C–terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015, 83, 864–874. [Google Scholar] [CrossRef]
- Kim, M.J.; Jang, I.-C.; Chua, N.-H. The mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol. 2016, 171, 1951–1964. [Google Scholar] [CrossRef]
- Qu, J.; Ye, J.; Geng, Y.-F.; Sun, Y.-W.; Gao, S.-Q.; Zhang, B.-P.; Chen, W.; Chua, N.-H. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 2012, 160, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Huang, L.; Liu, H.; Garg, V.; Gangurde, S.S.; Li, H.; Chitikineni, A.; Guo, D.; Pandey, M.K.; Li, S.; et al. A genomic variation map provides insights into peanut diversity in China and associations with 28 agronomic traits. Na. Genet. 2024, 56, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Yeap, W.-C.; Lee, F.-C.; Shabari Shan, D.K.; Musa, H.; Appleton, D.R.; Kulaveerasingam, H. WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 2017, 91, 97–113. [Google Scholar] [CrossRef]
- Kong, Q.; Ma, W.; Yang, H.; Ma, G.; Mantyla, J.J.; Benning, C. The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot. 2017, 68, 4627–4634. [Google Scholar] [CrossRef]
- Hao, S.; Ma, Y.; Zhao, S.; Ji, Q.; Zhang, K.; Yang, M.; Yao, Y. McWRI1, a transcription factor of the AP2/SHEN family, regulates the biosynthesis of the cuticular waxes on the apple fruit surface under low temperature. PLoS ONE 2017, 12, e0186996. [Google Scholar] [CrossRef]
- Kumar, V.; Jha, P.; Van Staden, J. LEAFY COTYLEDONs (LECs): Master regulators in plant embryo development. Plant Cell Tiss. Organ Cult. 2020, 140, 475–487. [Google Scholar] [CrossRef]
- An, D.; Suh, M.C. Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 2015, 9, 137–148. [Google Scholar] [CrossRef]
- Wu, X.-L.; Liu, Z.-H.; Hu, Z.-H.; Huang, R.-Z. BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J. Integr. Plant Biol. 2014, 56, 582–593. [Google Scholar] [CrossRef]
- Ito, Y.; Thirumurugan, T.; Serizawa, A.; Hiratsu, K.; Ohme-Takagi, M.; Kurata, N. Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Sci. 2011, 181, 105–110. [Google Scholar] [CrossRef]
- van Erp, H.; Kelly, A.A.; Menard, G.; Eastmond, P.J. Multigene Engineering of Triacylglycerol Metabolism Boosts Seed Oil Content in Arabidopsis. Plant Physiol. 2014, 165, 30–36. [Google Scholar] [CrossRef]
- Valliyodan, B.; Cannon, S.B.; Bayer, P.E.; Shu, S.; Brown, A.V.; Ren, L.; Jenkins, J.; Chung, C.Y.-L.; Chan, T.-F.; Daum, C.G.; et al. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019, 100, 1066–1082. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.; Ye, W.; Kirkbride, R.C.; Jenkins, J.; et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G.; et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010, 28, 951–956. [Google Scholar] [CrossRef]
- Motamayor, J.C.; Mockaitis, K.; Schmutz, J.; Haiminen, N.; Iii, D.L.; Cornejo, O.; Findley, S.D.; Zheng, P.; Utro, F.; Royaert, S.; et al. The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 2013, 14, r53. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Ventre, G.; Caldelari, D. Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol. 1999, 121, 1359–1366. [Google Scholar] [CrossRef] [PubMed]





| A. thaliana | A. hypogaea | A. monticola | A. duranensis | A. ipaensis | ||||
|---|---|---|---|---|---|---|---|---|
| Gene Name | PGR | Tifrunner | BaileyII | Line8 v1.3 | ||||
| AtLEC1 AtL1L | AhLEC1a | AH01G29730 | Ah01g388600 | chr01G4145 | AhLine8.01G229500 | EVM0044897 | — | — |
| AhLEC1b | AH11G27150 | Ah11g380300 | chr11G3925 | AhLine8.11G179600 | EVM0020102 | — | — | |
| AtFUS3 | AhFUS3a | AH06G23930 | Ah06g316900 | chr06G3370 | AhLine8.06G179300 | EVM0043936 | Aradu.EI7Y8 | — |
| AhFUS3b | AH16G29570 | Ah16g401200 | chr16G3980 | AhLine8.16G197300 | EVM0071796 | — | Araip.3K0PR | |
| AtWRI1 | AhWRI1a | AH08G25830 | Ah08g309800 | chr08G3455 | AhLine8.08G210300 | EVM0023005 | Aradu.EJI6Q | — |
| AhWRI1b | AH18G30790 | Ah18g415600 | chr18G4478 | AhLine8.18G216000 | — | — | — | |
| AhWRI1c | AH15G12290 | Ah15g212800 | chr15G2176 | AhLine8.15G128400 | — | — | Araip.SZ63C | |
| AhWRI1d | — | Ah10g334400 | IDmodified-mrna-2114 | AhLine8.10G185300 | EVM0064695 | Aradu.387PF | — | |
| AhWRI1e | AH20G31430 | Ah20g425400 | chr20G4248 | AhLine8.20G202100 | EVM0058654 | — | Araip.L05NW | |
| AhWRI1f | AH04G29530 | Ah04g393900 | chr04G4066 | AhLine8.04G202100 | EVM0014666 | Aradu.G1CDJ | — | |
| Ah04g396000 | ||||||||
| Ah04g394100 | ||||||||
| AhWRI1g | AH14G34420 | Ah14g462800 | IDmodified-mrna-77 | AhLine8.14G213200 | — | — | Araip.34E5E | |
| AhWRI1h | AH03G41970 | Ah03g520600 | chr03G5733 | AhLine8.03G308300 | EVM0009822 | — | Araip.N6C0B | |
| Ah03g520700 | ||||||||
| AhWRI1i | AH13G44600 | Ah13g549900 | chr13G6111 | AhLine8.13G320200 | EVM0034741 | Aradu.EN58B | — | |
| AtWRI2 | AhWRI2a | AH06G02150 | Ah06g106000 | chr06G1256 | AhLine8.06G083900 | — | Aradu.0KI78 | — |
| AhLine8.06G084200 | ||||||||
| AhWRI2b | AH16G04650 | Ah16g055500 | chr16G566 | AhLine8.16G038500 | EVM0053674 | — | Araip.6J28H | |
| EVM0007664 | ||||||||
| AtWRI3 AtWRI4 | AhWRI3a | AH01G23420 | Ah01g316300 | IDmodified-mrna-7040 | AhLine8.01G177800 | EVM0007256 | Aradu.A4KG4 | — |
| AhWRI3b | AH11G34510 | Ah11g474800 | chr11G4916 | AhLine8.11G236600 | — | — | — | |
| AhWRI3c | AH09G00410 | Ah09g004900 | IDmodified-mrna-2659 | AhLine8.09G003900 | — | Aradu.P6UBG | — | |
| AhWRI3d | AH19G01070 | Ah19g015300 | chr19G149 | AhLine8.19G012100 | EVM0074703 | — | Araip.S3MIZ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Zhao, J.; Pan, L.; Wang, E.; Chen, N.; Xu, J.; Jiang, X.; Zhao, X.; Ma, J.; Li, S.; et al. Genome-Wide Identification of Arachis hypogaea LEC1s, FUS3s, and WRIs and Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a and AhWRI1d Increased Oil Content in Arabidopsis Seeds. Plants 2025, 14, 2910. https://doi.org/10.3390/plants14182910
Yin X, Zhao J, Pan L, Wang E, Chen N, Xu J, Jiang X, Zhao X, Ma J, Li S, et al. Genome-Wide Identification of Arachis hypogaea LEC1s, FUS3s, and WRIs and Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a and AhWRI1d Increased Oil Content in Arabidopsis Seeds. Plants. 2025; 14(18):2910. https://doi.org/10.3390/plants14182910
Chicago/Turabian StyleYin, Xiangzhen, Jianxin Zhao, Lijuan Pan, Enqi Wang, Na Chen, Jing Xu, Xiao Jiang, Xuhong Zhao, Junqing Ma, Shouhui Li, and et al. 2025. "Genome-Wide Identification of Arachis hypogaea LEC1s, FUS3s, and WRIs and Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a and AhWRI1d Increased Oil Content in Arabidopsis Seeds" Plants 14, no. 18: 2910. https://doi.org/10.3390/plants14182910
APA StyleYin, X., Zhao, J., Pan, L., Wang, E., Chen, N., Xu, J., Jiang, X., Zhao, X., Ma, J., Li, S., Xie, H., Yang, Z., Yu, S., & Chi, X. (2025). Genome-Wide Identification of Arachis hypogaea LEC1s, FUS3s, and WRIs and Co-Overexpression of AhLEC1b, AhFUS3b, AhWRI1a and AhWRI1d Increased Oil Content in Arabidopsis Seeds. Plants, 14(18), 2910. https://doi.org/10.3390/plants14182910






