DNA Barcoding Provides Taxonomic Clues for Identifying Five Endangered Phoebe Species in Southern China
Abstract
1. Introduction
2. Results
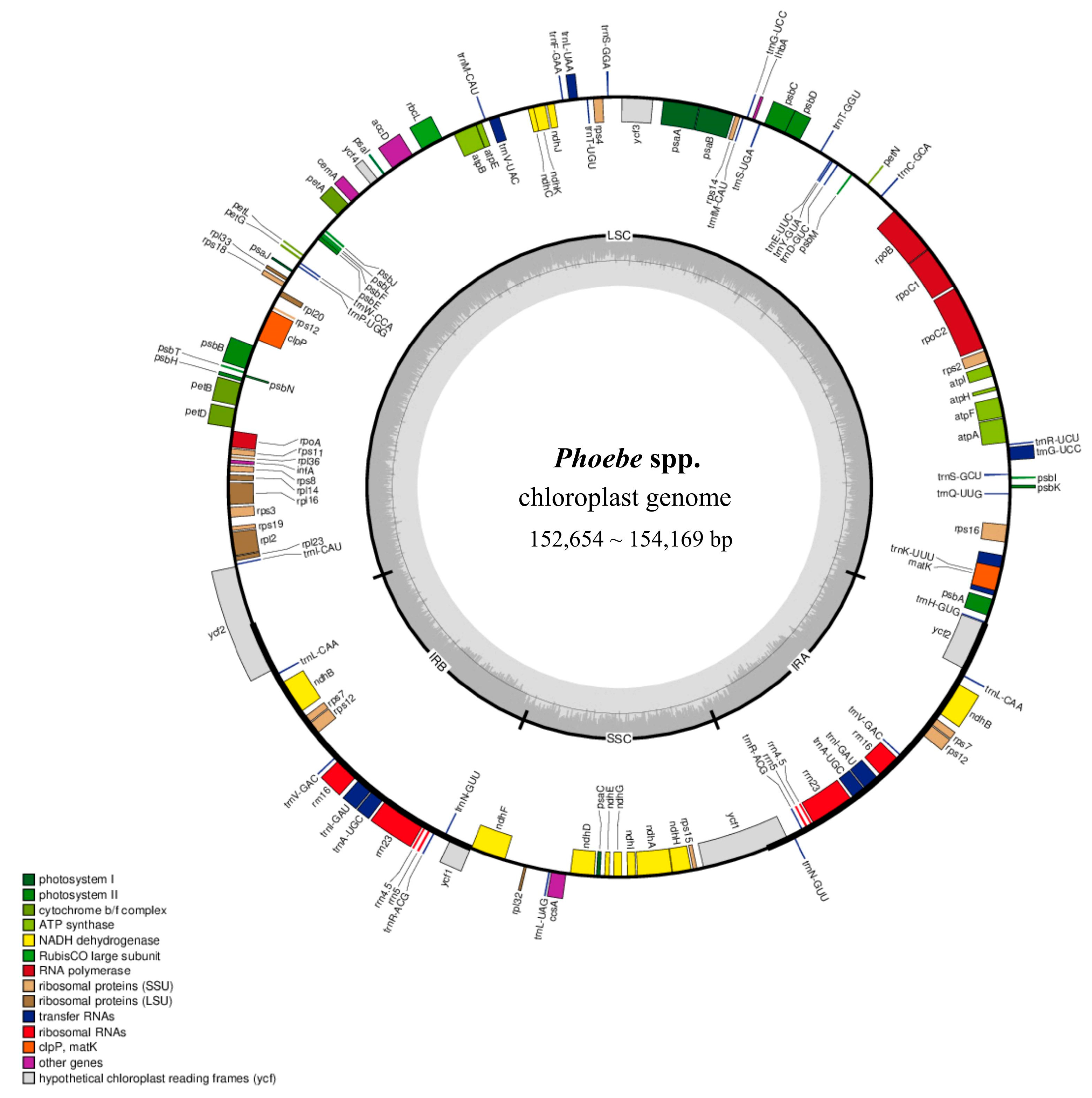
2.1. Comparison of Characteristic and Structure of Plastomes
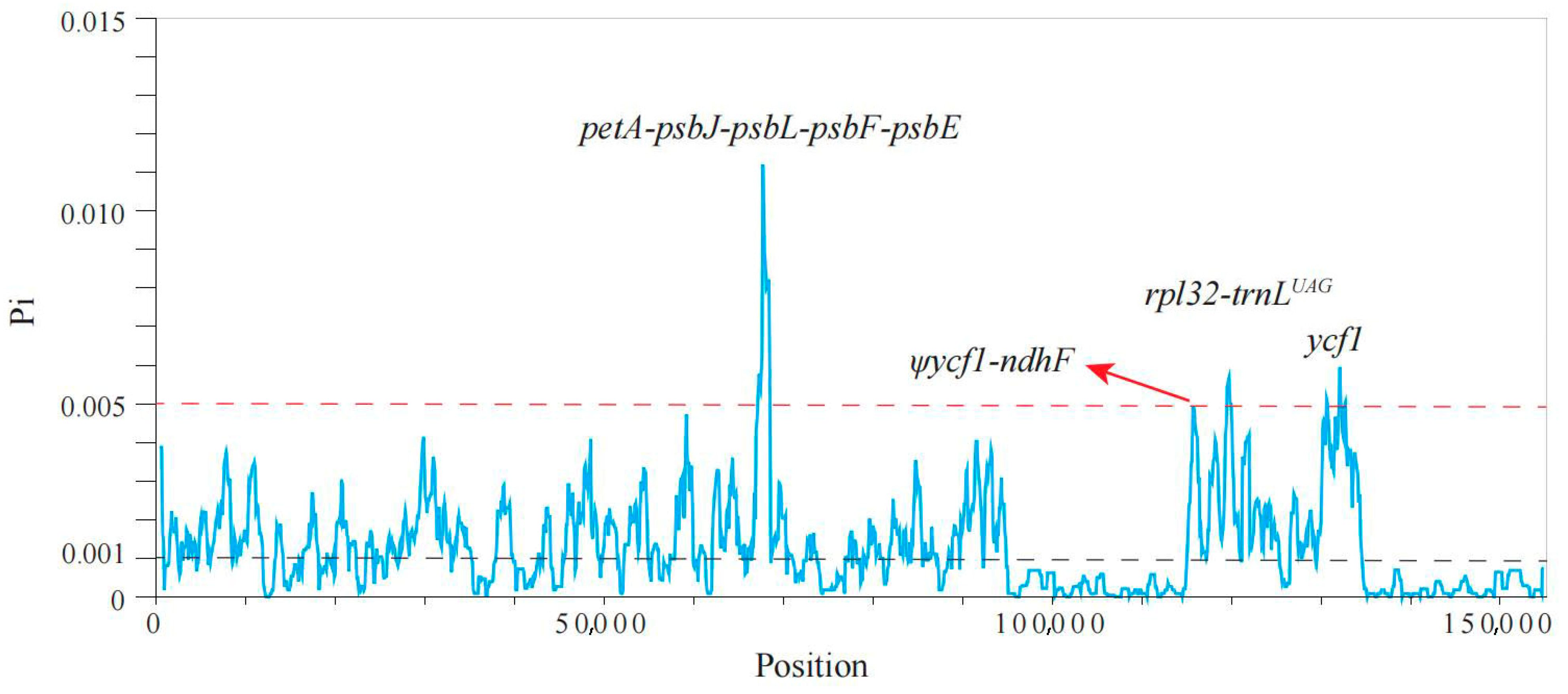
2.2. Candidate Hotspot Regions
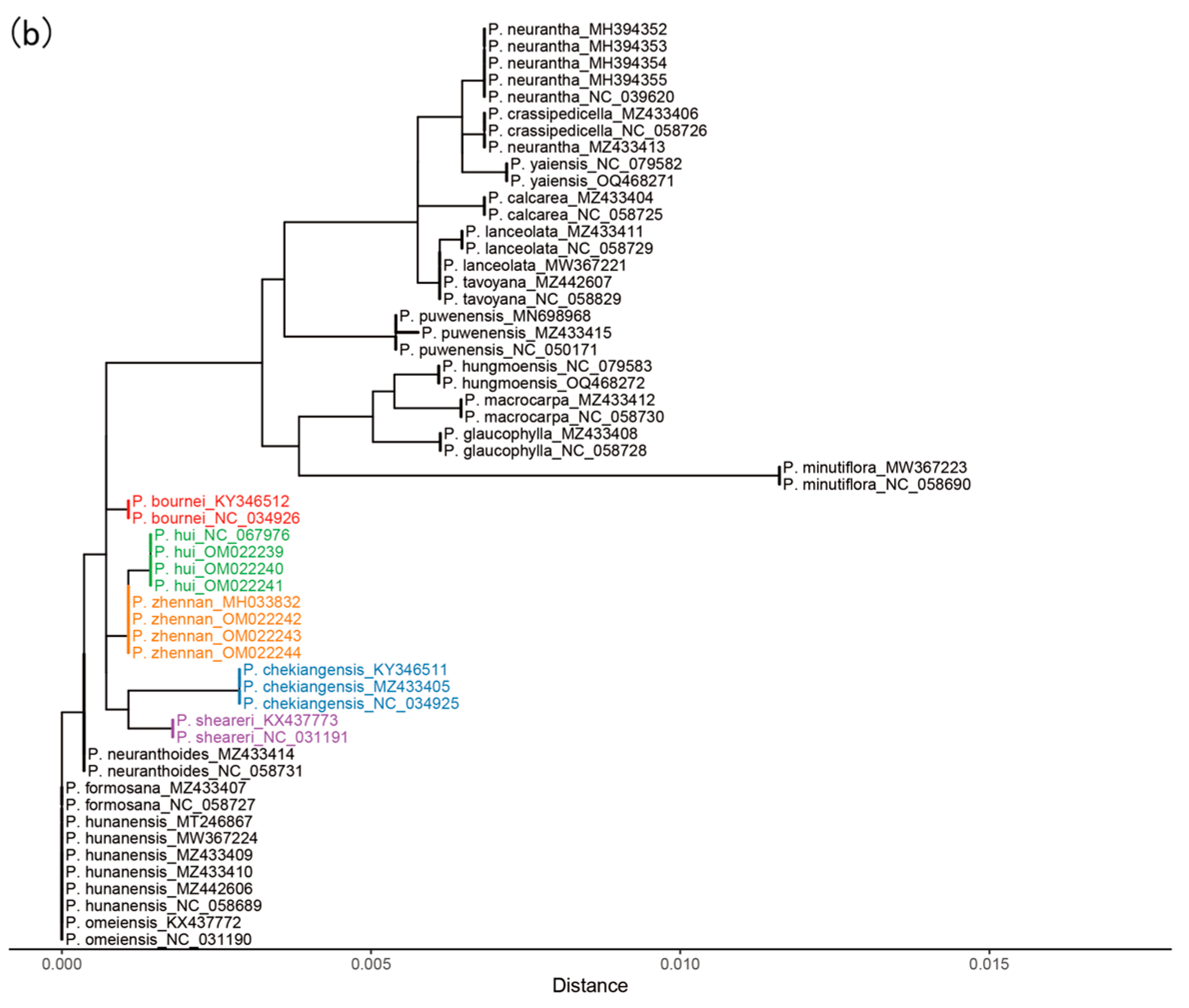
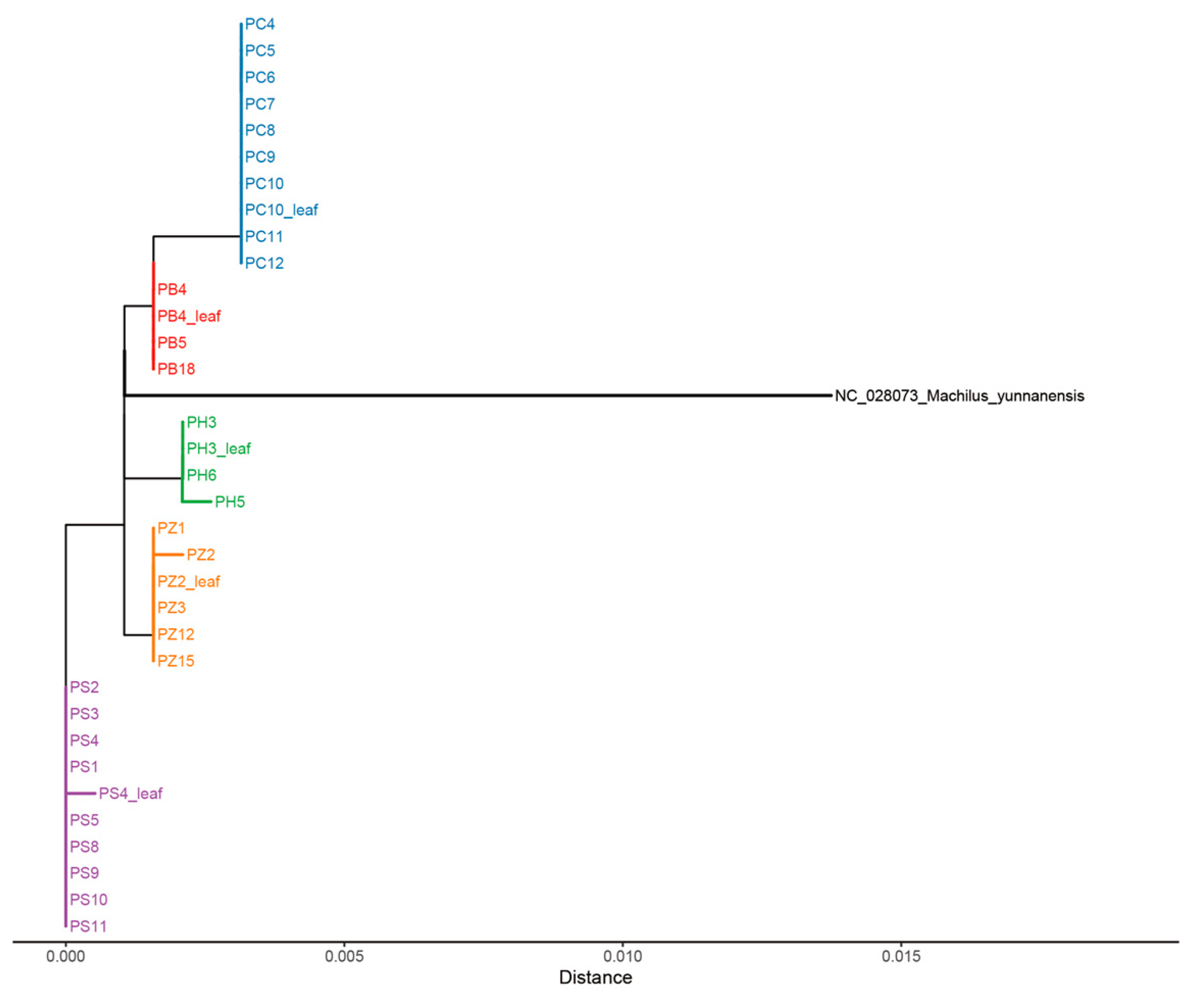
2.3. Phylogeny Analysis
3. Discussion
4. Materials and Methods
4.1. Sampling of Materials
4.2. Candidate Divergent Hotspot Regions
4.3. DNA Extraction
4.4. PCR Amplification and Sequencing
4.5. Analysis of Phylogeny
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiao, L.; Lu, Y.; Zhang, M.; Chen, Y.; Wang, Z.; Guo, Y.; Xu, C.; Guo, J.; He, T.; Ma, L.; et al. Ancient Plastid Genomes Solve the Tree Species Mystery of the Imperial Wood “Nanmu” in the Forbidden City, the Largest Existing Wooden Palace Complex in the World. Plants People Planet 2022, 4, 696–709. [Google Scholar] [CrossRef]
- Ding, X.; Xiao, J.H.; Li, L.; Conran, J.G.; Li, J. Congruent Species Delimitation of Two Controversial Gold-thread Nanmu Tree Species Based on Morphological and Restriction Site-associated DNA Sequencing Data. J. Syst. Evol. 2019, 57, 234–246. [Google Scholar] [CrossRef]
- Lan, Y. Imperial Wood Procurement in Ming and Qing Dynasties. Hist. Res. 1994, 6, 86–98. [Google Scholar]
- IUCN, The Iucn Red List of Threatened Species. Version 2023-1. 2023. Available online: https://www.iucnredlist.org (accessed on 27 March 2024).
- National Forestry and Grassland Administration, List of National Key Protected Wild Plants in China. 2021. Available online: https://www.forestry.gov.cn/c/www/gkml/11057.jhtml (accessed on 27 March 2024).
- Liu, J.; Lindenmayer, D.B.; Yang, W.; Ren, Y.; Campbell, M.J.; Wu, C.; Luo, Y.; Zhong, L.; Yu, M. Diversity and Density Patterns of Large Old Trees in China. Sci. Total Environ. 2019, 655, 255–262. [Google Scholar] [CrossRef]
- Liu, Z.F.; Ci, X.Q.; Li, L.; Li, H.-W.; Conran, J.G.; Li, J. DNA Barcoding Evaluation and Implications for Phylogenetic Relationships in Lauraceae from China. PLoS ONE 2017, 12, e0175788. [Google Scholar] [CrossRef]
- Liu, Z.F.; Ma, H.; Ci, X.Q.; Li, L.; Song, Y.; Liu, B.; Li, H.W.; Wang, S.L.; Qu, X.J.; Hu, J.L.; et al. Can Plastid Genome Sequencing Be Used for Species Identification in Lauraceae? Bot. J. Linn. Soc. 2021, 197, 1–14. [Google Scholar] [CrossRef]
- Yang, S.; Huang, J.; Qu, Y.; Zhang, D.; Tan, Y.; Wen, S.; Song, Y. Phylogenetic Incongruence in an Asiatic Species Complex of the Genus Caryodaphnopsis (Lauraceae). BMC Plant Biol. 2024, 24, 616. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Julia, S.; Soepadmo, E.; Yahud, W. Problem in the Generic Delimitation between Alseodaphne, Dehaasia and Nothaphoebe (Lauraceae) in Borneo. Blumea-Biodivers. Evol. Biogeogr. Plants 2009, 54, 192–197. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Rohwer, J.G.; Van Der Werff, H.; Wang, Z.; Li, H. Molecular Phylogenetic Analysis of the Persea Group (Lauraceae) and Its Biogeographic Implications on the Evolution of Tropical and Subtropical Amphi-pacific Disjunctions. Am. J. Bot. 2011, 98, 1520–1536. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yu, W.; Tan, Y.; Jin, J.; Wang, B.; Yang, J.; Liu, B.; Corlett, R.T. Plastid Phylogenomics Improve Phylogenetic Resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Van Der Werff, H.; Richter, H.G. Toward an Improved Classification of Lauraceae. Ann. Mo. Bot. Gard. 1996, 83, 409. [Google Scholar] [CrossRef]
- Xu, B.; Zhu, T.; Li, J.; Liu, S. Identification of Wood between Phoebe Zhennan and Machilus Pingii Using the Gas Chromatography-Mass Spectrometry Direct Injection Technique. Eur. J. Mass. Spectrom. 2013, 19, 187–193. [Google Scholar] [CrossRef]
- Li, Y.; Xu, W.; Zou, W.; Jiang, D.; Liu, X. Complete Chloroplast Genome Sequences of Two Endangered Phoebe (Lauraceae) Species. Bot. Stud. 2017, 58, 37. [Google Scholar] [CrossRef]
- Liu, C.; Chen, H.H.; Tang, L.Z.; Khine, P.K.; Han, L.H.; Song, Y.; Tan, Y.H. Plastid Genome Evolution of a Monophyletic Group in the Subtribe Lauriineae (Laureae, Lauraceae). Plant Divers. 2022, 44, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Song, W.; Chen, Z.; Cai, H.; Gong, Q.; Liu, J.; Shi, C.; Wang, S. Comparative Chloroplast Genome Analyses of Diverse Phoebe (Lauraceae) Species Endemic to China Provide Insight into Their Phylogeographical Origin. PeerJ 2023, 11, e14573. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yao, X.; Tan, Y.; Gan, Y.; Yang, J.; Corlett, R.T. Comparative Analysis of Complete Chloroplast Genome Sequences of Two Subtropical Trees, Phoebe Sheareri and Phoebe Omeiensis (Lauraceae). Tree Genet. Genomes 2017, 13, 120. [Google Scholar] [CrossRef]
- Liu, B.; Yang, Y.; Ferguson, D.K. Phylogeny and Taxonomy of Cinnamomum (Lauraceae). Ecol. Evol. 2022, 12, e9378. [Google Scholar] [CrossRef]
- Cai, C.; Ma, H.; Ci, X.; Conran, J.G.; Li, J. Comparative Phylogenetic Analyses of Chinese Horsfieldia (Myristicaceae) Using Complete Chloroplast Genome Sequences. J. Syst. Evol. 2021, 59, 504–514. [Google Scholar] [CrossRef]
- Tian, Y.; Zhou, J.; Zhang, Y.; Wang, S.; Wang, Y.; Liu, H.; Wang, Z. Research Progress in Plant Molecular Systematics of Lauraceae. Biology 2021, 10, 391. [Google Scholar] [CrossRef]
- Xiong, Y.; Xiong, Y.; Jia, S.; Ma, X. The Complete Chloroplast Genome Sequencing and Comparative Analysis of Reed Canary Grass (Phalaris arundinacea) and Hardinggrass (P. aquatica). Plants 2020, 9, 748. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA Barcoding: From Gene to Genome. Biol. Rev. 2015, 90, 157–166. [Google Scholar] [CrossRef]
- Lowe, A.J.; Cross, H.B. The Application of DNA Methods to Timber Tracking and Origin Verificat Ion. IAWA J. 2011, 32, 251–262. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Janzen, D.H.; Burns, J.M.; Hallwachs, W.; Hebert, P.D.N. DNA Barcodes Distinguish Species of Tropical Lepidoptera. Proc. Natl. Acad. Sci. USA 2006, 103, 968–971. [Google Scholar] [CrossRef]
- Hu, J.; Ci, X.; Liu, Z.; Dormontt, E.E.; Conran, J.G.; Lowe, A.J.; Li, J. Assessing Candidate DNA Barcodes for Chinese and Internationally Traded Timber Species. Mol. Ecol. Resour. 2022, 22, 1478–1492. [Google Scholar] [CrossRef]
- Lendvay, B.; Hartmann, M.; Brodbeck, S.; Nievergelt, D.; Reinig, F.; Zoller, S.; Parducci, L.; Gugerli, F.; Büntgen, U.; Sperisen, C. Improved Recovery of Ancient DNA from Subfossil Wood—Application to the World’s Oldest Late Glacial Pine Forest. New Phytol. 2018, 217, 1737–1748. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, L.; Wang, H.; Feng, S. Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy 2024, 14, 933. [Google Scholar] [CrossRef]
- China Plant BOL Group; Li, D.Z.; Gao, L.M.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative Analysis of a Large Dataset Indicates That Internal Transcribed Spacer (ITS) Should Be Incorporated into the Core Barcode for Seed Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Qimike, A.; Yang, C.; Chen, J.; Wang, Q. Testing Four Barcoding Markers for Species Identification of Potamogetonaceae. J. Syst. Evol. 2011, 49, 246–251. [Google Scholar] [CrossRef]
- Fu, Y.; Jiang, W.; Fu, C. Identification of Species within Tetrastigma (Miq.) Planch. (Vitaceae) Based on DNA Barcoding Techniques. J. Syst. Evol. 2011, 49, 237–245. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Li, X.W. Taxonomic Revision of Five Species of the Genus Phoebe (Lauraceae) from China. Plant Divers. 2011, 33, 157–160. [Google Scholar]
- Xu, J.; Zhang, H.; Yang, F.; Zhu, W.; Li, Q.; Cao, Z.; Song, Y.; Xin, P. Phylogeny of Camphora and Cinnamomum (Lauraceae) Based on Plastome and Nuclear Ribosomal DNA Data. Int. J. Mol. Sci. 2025, 26, 1370. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.S.; Ayres, K.L.; Toomey, N.; Haider, N.; Van Alphen Stahl, J.; Kelly, L.J.; Wikström, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of Candidate Coding DNA Barcoding Regions for Use on Land Plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef]
- Hernández-León, S.; Gernandt, D.S.; Pérez De La Rosa, J.A.; Jardón-Barbolla, L. Phylogenetic Relationships and Species Delimitation in Pinus Section Trifoliae Inferrred from Plastid DNA. PLoS ONE 2013, 8, e70501. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA Barcodes to Identify Flowering Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA Barcodes and a Community Phylogeny of a Tropical Forest Dynamics Plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [PubMed]
- Little, D.P.; Knopf, P.; Schulz, C. DNA Barcode Identification of Podocarpaceae—The Second Largest Conifer Family. PLoS ONE 2013, 8, e81008. [Google Scholar] [CrossRef]
- Pang, X.; Liu, C.; Shi, L.; Liu, R.; Liang, D.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnH–psbA Intergenic Spacer Region and Its Combinations as Plant DNA Barcodes: A Meta-Analysis. PLoS ONE 2012, 7, e48833. [Google Scholar] [CrossRef]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, F.; Yan, H.; Hao, G.; Hu, C.; Ge, X. Testing DNA Barcoding in Closely Related Groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 2012, 12, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.H.; Cai, D.; Li, J.Q. ITS1: A DNA Barcode Better than ITS2 in Eukaryotes? Mol. Ecol. Resour. 2015, 15, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Cheng, T.; Li, C.; Xu, C.; Long, P.; Chen, C.; Zhou, S. Discriminating Plants Using the DNA Barcode rbcLb: An Appraisal Based on a Large Data Set. Mol. Ecol. Resour. 2014, 14, 336–343. [Google Scholar] [CrossRef]
- Group, C.P.W. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Yu, M.; Jiao, L.; Guo, J.; Wiedenhoeft, A.C.; He, T.; Jiang, X.; Yin, Y. DNA Barcoding of Vouchered Xylarium Wood Specimens of Nine Endangered Dalbergia Species. Planta 2017, 246, 1165–1176. [Google Scholar] [CrossRef]
- Cannon, C.H.; Manos, P.S. Phylogeography of the Southeast Asian Stone Oaks (Lithocarpus). J. Biogeogr. 2003, 30, 211–226. [Google Scholar] [CrossRef]
- Jiao, L.; Yin, Y.; Cheng, Y.; Jiang, X. DNA Barcoding for Identification of the Endangered Species Aquilaria Sinensis: Comparison of Data from Heated or Aged Wood Samples. Holzforschung 2014, 68, 487–494. [Google Scholar] [CrossRef]
- Tanaka, S.; Ito, M. DNA Barcoding for Identification of Agarwood Source Species Using trnL-trnF and matK DNA Sequences. J. Nat. Med. 2020, 74, 42–50. [Google Scholar] [CrossRef]
- Jiao, L.; Yu, M.; Wiedenhoeft, A.C.; He, T.; Li, J.; Liu, B.; Jiang, X.; Yin, Y. DNA Barcode Authentication and Library Development for the Wood of Six Commercial Pterocarpus Species: The Critical Role of Xylarium Specimens. Sci. Rep. 2018, 8, 1945. [Google Scholar] [CrossRef]
- Ohyama, M.; Baba, K.; Itoh, T. Wood Identification of Japanese Cyclobalanopsis Species (Fagaceae) Based on DNA Polymorphism of the Intergenic Spacer between trnT and trnL 5′ Exon. J. Wood Sci. 2001, 47, 81–86. [Google Scholar] [CrossRef]
- Pizzolato, T.D. A Tannic Acid-Ferric Chloride-Toluidine Blue Stain for Wood Amyloplasts Embedded in Epoxy Resin. For. Sci. 1978, 24, 49–51. [Google Scholar]
- Deguilloux, M.; Pemonge, M.; Petit, R. Novel Perspectives in Wood Certification and Forensics: Dry Wood as a Source of DNA. Proc. R. Soc. Lond. B 2002, 269, 1039–1046. [Google Scholar] [CrossRef]
- Jiao, L.; Lu, Y.; He, T.; Guo, J.; Yin, Y. DNA Barcoding for Wood Identification: Global Review of the Last Decade and Future Perspective. IAWA J. 2020, 41, 620–643. [Google Scholar] [CrossRef]
- Winter, H.; Robinson, D.G.; Heldt, H.W. Subcellular Volumes and Metabolite Concentrations in Spinach Leaves. Planta 1994, 193, 530–535. [Google Scholar] [CrossRef]
- Abe, H.; Watanabe, U.; Yoshida, K.; Kuroda, K.; Zhang, C. Changes in Organelle and DNA Quality, Quantity, and Distribution in the Wood of Cryptomeria Japonica over Long-Term Storage. IAWA J. 2011, 32, 263–272. [Google Scholar] [CrossRef]
- Jiao, L.; Yin, Y.; Xiao, F.; Sun, Q.; Song, K.; Jiang, X. Comparative Analysis of Two DNA Extraction Protocols from Fresh and Dried Wood of Cunninghamia Lanceolata (Taxodiaceae). IAWA J. 2012, 33, 441–456. [Google Scholar] [CrossRef]
- Lu, Y.; Jiao, L.; Sun, G.; Wang, J.; Liu, S.; Li, R.; Zhang, Y.; Guo, Y.; Guo, J.; Jiang, X.; et al. Preservation Status and Microbial Community of Waterlogged Archaeological Woods over 7800 Years Old at the Jingtoushan Site, China. Wood Sci. Technol. 2023, 57, 537–556. [Google Scholar] [CrossRef]
- Rogers, S.O.; Kaya, Z. DNA from Ancient Cedar Wood from King Midas’ Tomb, Turkey, and al-Aksa Mosque, Israel. Silvae Genet. 2006, 55, 54–62. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Lee, S.Y.; Ng, W.L.; Mahat, M.N.; Nazre, M.; Mohamed, R. DNA Barcoding of the Endangered Aquilaria (Thymelaeaceae) and Its Application in Species Authentication of Agarwood Products Traded in the Market. PLoS ONE 2016, 11, e0154631. [Google Scholar] [CrossRef]
- Rachmayanti, Y.; Leinemann, L.; Gailing, O.; Finkeldey, R. Extraction, Amplification and Characterization of Wood DNA from Dipterocarpaceae. Plant Mol. Biol. Rep. 2006, 24, 45–55. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. ElasticBLAST: Accelerating Sequence Search via Cloud Computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]



| DNA Barcodes | Intraspecific Distance | Interspecific Distance | ISR | ||||
|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | ||
| P | 0.0000 | 0.0083 | 0.0011 | 0.0000 | 0.0077 | 0.0007 | 16.36% |
| R | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0109 | 0.0015 | 40.00% |
| Y | 0.0000 | 0.0010 | 0.0002 | 0.0000 | 0.0106 | 0.0012 | 56.36% |
| F | 0.0000 | 0.0030 | 0.0005 | 0.0000 | 0.0089 | 0.0013 | 67.27% |
| P + R | 0.0000 | 0.0045 | 0.0006 | 0.0000 | 0.0102 | 0.0014 | 50.91% |
| P + Y | 0.0000 | 0.0042 | 0.0007 | 0.0000 | 0.0096 | 0.0014 | 69.09% |
| P + F | 0.0000 | 0.0048 | 0.0007 | 0.0000 | 0.0075 | 0.0009 | 49.09% |
| R + Y | 0.0000 | 0.0005 | 0.0001 | 0.0000 | 0.0112 | 0.0015 | 70.91% |
| F + R | 0.0000 | 0.0006 | 0.0001 | 0.0000 | 0.0087 | 0.0011 | 49.09% |
| F + Y | 0.0000 | 0.0011 | 0.0002 | 0.0000 | 0.0091 | 0.0010 | 56.36% |
| P + R + Y | 0.0000 | 0.0029 | 0.0005 | 0.0000 | 0.0110 | 0.0016 | 78.18% |
| P + F + R | 0.0000 | 0.0032 | 0.0005 | 0.0000 | 0.0094 | 0.0013 | 65.45% |
| P + F + Y | 0.0000 | 0.0031 | 0.0005 | 0.0000 | 0.0090 | 0.0012 | 63.64% |
| F + R + Y | 0.0000 | 0.0007 | 0.0001 | 0.0000 | 0.0097 | 0.0013 | 70.91% |
| P + F + R + Y | 0.0000 | 0.0023 | 0.0004 | 0.0000 | 0.0102 | 0.0014 | 78.18% |
| Phoebe Primers | Type | Primer Sequences (5′–3′) |
|---|---|---|
| rpl32-trnLUAG | Forward | GCGAGATGGGGGTTGTAACT |
| Reverse | AGTATCATGGCAGGGGGTCA | |
| ycf1 | Forward | TGACCCCTTAACCAGTTTTTCCA |
| Reverse | CTGAAACCCTGGCGCAAATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, W.; Du, C.; Zhang, X.; Zhang, W.; Wu, W.; Fang, C.; Xiao, X.; Zhu, J.; Yang, F.; Zhang, M. DNA Barcoding Provides Taxonomic Clues for Identifying Five Endangered Phoebe Species in Southern China. Plants 2025, 14, 2895. https://doi.org/10.3390/plants14182895
Yin W, Du C, Zhang X, Zhang W, Wu W, Fang C, Xiao X, Zhu J, Yang F, Zhang M. DNA Barcoding Provides Taxonomic Clues for Identifying Five Endangered Phoebe Species in Southern China. Plants. 2025; 14(18):2895. https://doi.org/10.3390/plants14182895
Chicago/Turabian StyleYin, Wenxiu, Chungui Du, Xiaofeng Zhang, Wenbiao Zhang, Wenwu Wu, Chongrong Fang, Xingcui Xiao, Jiawei Zhu, Fei Yang, and Mingzhe Zhang. 2025. "DNA Barcoding Provides Taxonomic Clues for Identifying Five Endangered Phoebe Species in Southern China" Plants 14, no. 18: 2895. https://doi.org/10.3390/plants14182895
APA StyleYin, W., Du, C., Zhang, X., Zhang, W., Wu, W., Fang, C., Xiao, X., Zhu, J., Yang, F., & Zhang, M. (2025). DNA Barcoding Provides Taxonomic Clues for Identifying Five Endangered Phoebe Species in Southern China. Plants, 14(18), 2895. https://doi.org/10.3390/plants14182895






