Genomic Diversity and Structure of Copaifera langsdorffii Populations from a Transition Zone Between the Atlantic Forest and the Brazilian Savanna
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
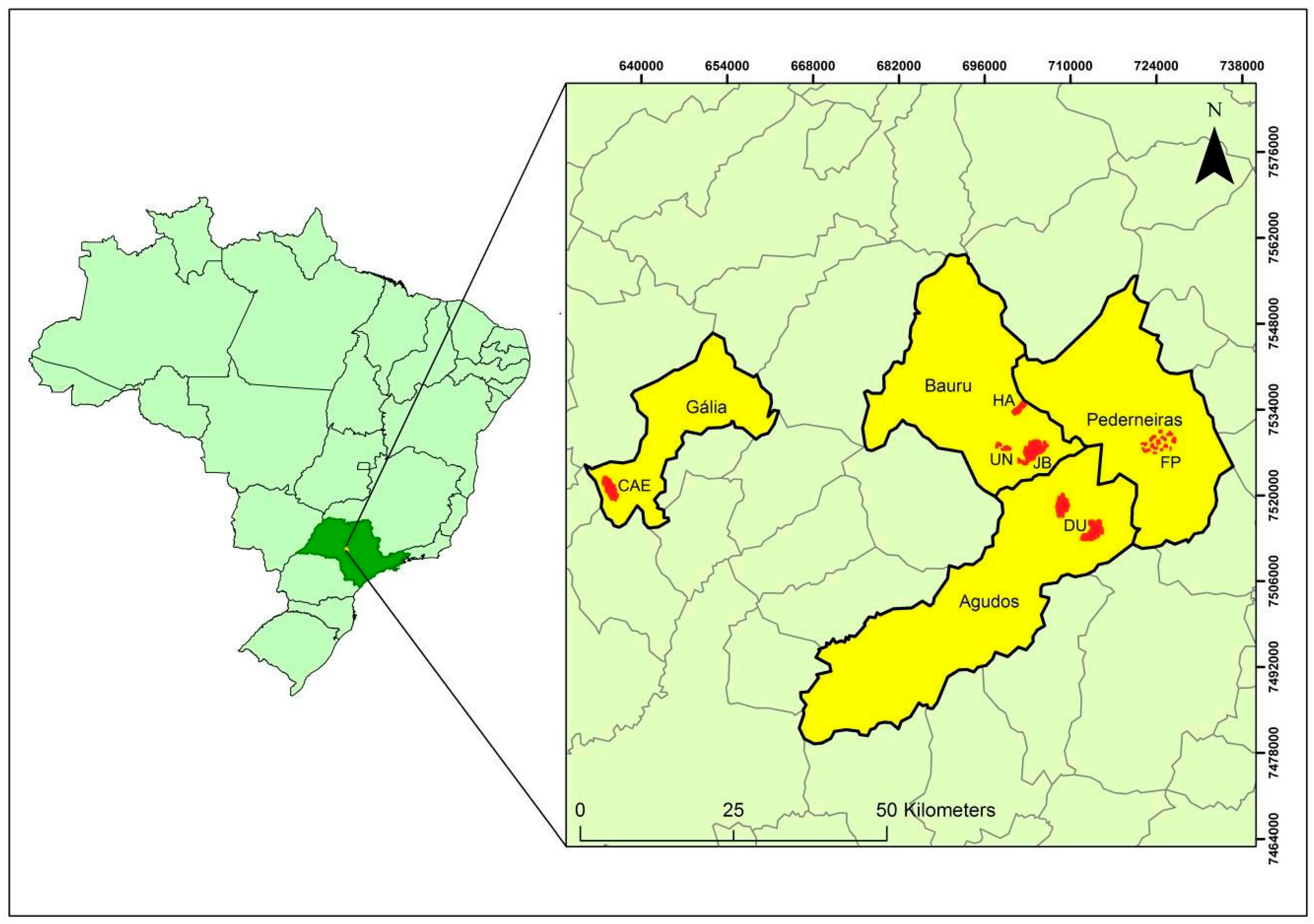
2.2. Sampling Sites
2.3. DNA Isolation and RADSeq Library Preparation
2.4. SNP Calling and Genotyping
2.5. Data Analysis
2.5.1. Genetic Diversity and Structure
2.5.2. FST Outlier Analysis
3. Results
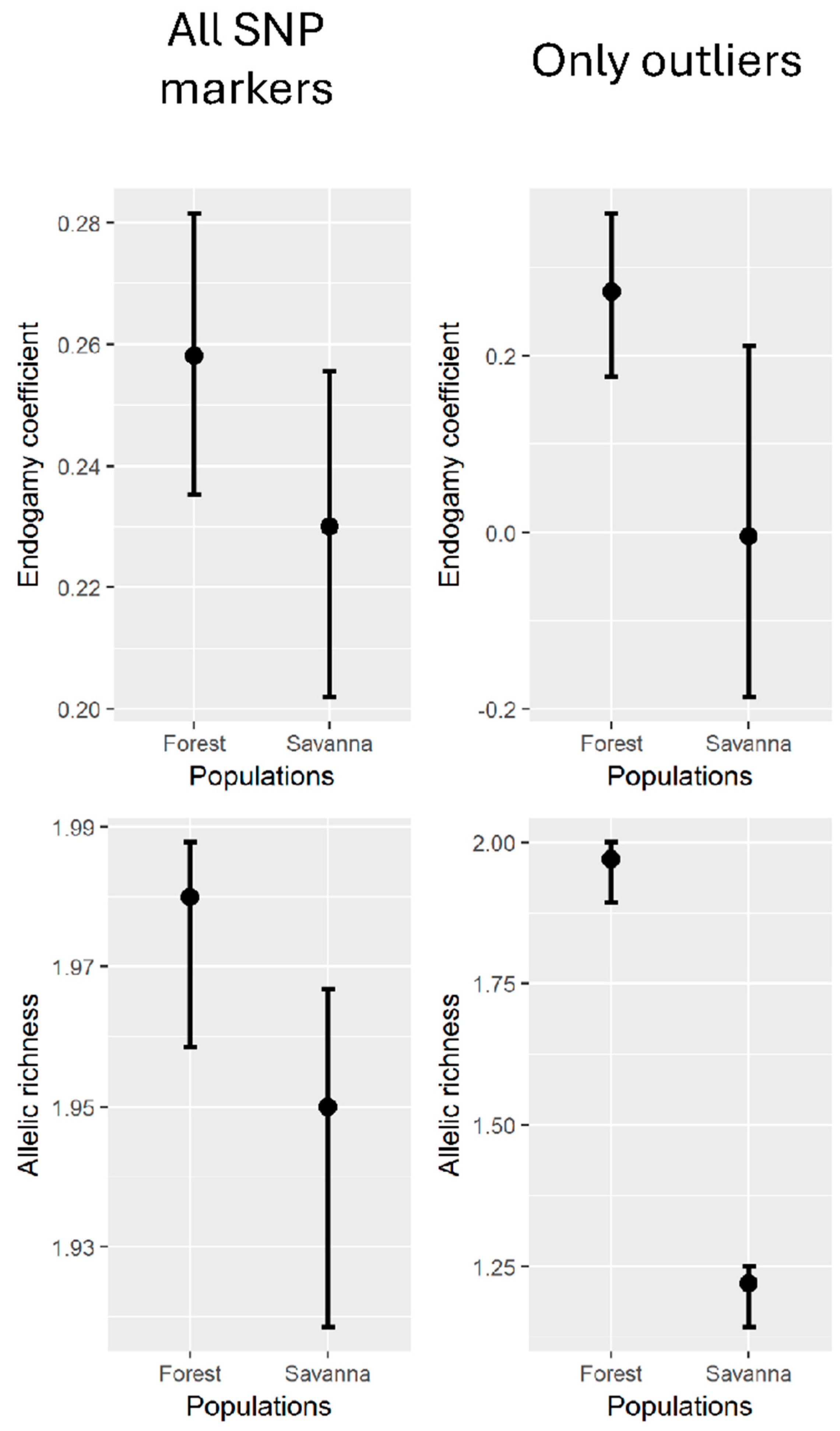
3.1. Genetic Diversity and Structure
3.2. Outlier FST Analysis
4. Discussion
4.1. Excess of Homozygotes in All Sites
4.2. Outcrossing Rates May Be High, but There Is Also Isolation by Distance
4.3. Population Structure Reflects the Transition Between Forests and Savannas
4.4. Outlier Loci Analysis Highlights Putative Local Adaptations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clare, E.; Garcia, C.; Bolan, S. Ecosystem functioning during biodiversity loss and recovery. Oikos 2024, 2024, e10154. [Google Scholar] [CrossRef]
- Flores, B.M.; Montoya, E.; Sakschewski, B.; Nascimento, N.; Staal, A. Critical transitions in the Amazon forest system. Nature 2023, 624, 65–72. [Google Scholar] [CrossRef]
- Jaureguiberry, P.; Titeux, N.; Wiemers, M.; Bowler, D.E.; Coscieme, L.; Golden, A.S.; Guerra, C.A.; Jacob, U.; Takahashi, Y.; Settele, J.; et al. The direct drivers of recent global anthropogenic biodiversity loss. Sci. Adv. 2022, 8, eabm9982. [Google Scholar] [CrossRef] [PubMed]
- de Lima, R.A.F.; Oliveira, A.A.; Pitta, G.R.; Gasper, A.L.; Vibrans, A.C.; Chave, J.; Steege, H.; Prado, P. The erosion of biodiversity and biomass in the Atlantic Forest biodiversity hotspot. Nat. Commun. 2020, 11, 6347. [Google Scholar] [CrossRef] [PubMed]
- Alencar, A.; Shimbo, J.Z.; Lenti, F.; Marques, C.B.; Zimbres, B.; Rosa, M.; Arruda, V.; Castro, I.; Ribeiro, J.P.F.M.; Varela, V.; et al. Mapping Three Decades of Changes in the Brazilian Savanna Native Vegetation Using Landsat Data Processed in the Google Earth Engine Platform. Remote Sens. 2020, 12, 924. [Google Scholar] [CrossRef]
- Aguiar, B.I.; Sebbenn, A.M.; Tarazi, R.; Vogado, N.A.; Morellato, L.P.C.; Tambarussi, E.V.; Moreno, M.A.; Pereira, L.C.S.M.; Montibeller, C.; Ferraz, E.M.; et al. Phenology, Seed Germination, and Genetics Explains the Reproductive Strategies of Diospyros lasiocalyx (Mart.) B. Wall. Trop. Plant Biol. 2020, 13, 23–35. [Google Scholar] [CrossRef]
- Rabeschini, G.; Persson, U.M.; West, C.; Kastner, T. Choosing fit-for-purpose biodiversity impact indicators for agriculture in the Brazilian Cerrado ecoregion. Nat. Commun. 2025, 16, 1799. [Google Scholar] [CrossRef]
- Resende, F.M.; Cimon-Morin, J.; Poulin, M.; Meyer, L.; Loyola, R. Consequences of delaying actions for safeguarding ecosystem services in the Brazilian Cerrado. Biol. Conserv. 2019, 234, 90–99. [Google Scholar] [CrossRef]
- Muniz, A.C.; Pimenta, R.J.G.; Cruz, M.V.; Rodrigues, J.G.; Buzatti, R.S.O.; Heuertz, M.; Lemos-Filho, J.P.; Lovato, M.B. Hybrid zone of a tree in a Cerrado/Atlantic Forest ecotone as a hotspot of genetic diversity and conservation. Ecol. Evol. 2022, 22, e8540. [Google Scholar] [CrossRef]
- Sano, E.E.; Rodrigues, A.A.; Martins, E.S.; Bettiol, G.M.; Bustamante, M.M.C.; Bezerra, A.S.; Couto, A.F.; Vasconcelos, V.; Schüler, J.; Bolfe, E.L. Cerrado ecoregions: A spatial framework to assess and prioritize Brazilian savanna environmental diversity for conservation. J. Environ. Manag. 2019, 232, 818–828. [Google Scholar] [CrossRef]
- Ramos, E.A.; Nuvoloni, F.M.; Lopes, E.R.N. Landscape Transformations and loss of Atlantic Forests: Challenges for conservation. J. Nat. Conserv. 2022, 66, 126152. [Google Scholar] [CrossRef]
- Nery, E.K.; Caddah, M.K.; Santos, M.F.; Nogueira, A. The evolution of ecological specialization underlies plant endemism in the Atlantic Forest. Ann. Bot. 2023, 131, 921–940. [Google Scholar] [CrossRef]
- Konzen, E.R. Towards conservation strategies for forest tree endangered species: The meaning of population genetic statistics. Adv. For. Sci. 2014, 1, 45–51. [Google Scholar] [CrossRef]
- Feng, J.; Dan, X.; Cui, Y.; Gong, Y.; Peng, M.; Sang, Y.; Ingvarsson, P.K.; Wang, J. Integrating evolutionary genomics of forest trees to inform future tree breeding amid rapid climate change. Plant Commun. 2024, 5, 101044. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.E.; Farquharson, K.A.; Bruford, M.W.; Coates, D.J.; Elliott, C.P.; Mergeay, J.; Ottewell, K.M.; Segelbacher, G.; Hoban, S.; Hvilsom, C.; et al. Global meta-analysis shows action is needed to halt genetic diversity loss. Nature 2025, 638, 704–710. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 13, 997. [Google Scholar] [CrossRef]
- Miller, M.R.; Dunham, J.P.; Amores, A.; Cresko, W.A.; Johnson, E.A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007, 17, 240–248. [Google Scholar] [CrossRef]
- Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct. Genom. 2010, 9, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Herrera, S.; Reyes-Herrera, P.H.; Shank, T.M. Predicting RAD-seq Marker Numbers across the Eukaryotic Tree of Life. Genome Biol. Evol. 2015, 3, 3207–3225. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, C.; Clavijo, C.; Rojas-Bonzi, V.; Miño, C.I.; González-Maya, J.F.; Bou, N.; Giraldo, A.; Martino, A.; Miyaki, C.Y.; Aguirre, L.F.; et al. Understanding the conservation-genetics gap in Latin America: Challenges and opportunities to integrate genetics into conservation practices. Front. Genet. 2024, 8, 1425531. [Google Scholar] [CrossRef]
- Lira, C.F. Neutral Genetic Diversity of Brazilian Native Flora: Current Approaches and Gaps. Environ. Earth Sci. Proc. 2024, 31, 7. [Google Scholar] [CrossRef]
- Martins, K.; Santos, J.D.; Gaiotto, F.A.; Moreno, M.A.; Kageyama, P.Y. Estrutura genética populacional de Copaifera langsdorffii Desf. (Leguminosae-Caesalpinioideae) em fragmentos florestais no Pontal do Paranapanema, SP, Brasil. Rev. Bras. Bot. 2008, 31, 61–69. [Google Scholar] [CrossRef]
- Carvalho, D.; Ingvarsson, P.K.; Joseph, J.; Suter, L.; Sedivy, C.; Macaya-Sanz, D.; Cottrell, J.; Heinze, B.; Schanzer, I.; Lexer, C. Admixture facilitates adaptation from standing variation in the European aspen (Populus tremula L.), a widespread forest tree. Mol. Ecol. 2010, 19, 1638–1650. [Google Scholar] [CrossRef] [PubMed]
- Tarazi, R.; Sebbenn, A.M.; Mollinari, M.; Vencovsky, R. Mendelian inheritance, linkage and linkage disequilibrium in microsatellite loci of Copaifera langsdorffii Desf. Conserv. Genet. Resour. 2010, 2, 201–204. [Google Scholar] [CrossRef]
- Manoel, R.O.; Cardin, L.T.; Moreira, J.P.; Silva, E.C.B.; Senna, S.N.; Kubota, T.Y.K.; Freitas, M.L.M.; Moraes, M.L.T.; Sebbenn, A.M. Sistema de reprodução, parentesco e tamanho efetivo em sementes de polinização livre de populações fragmentadas de Copaifera langsdorffii Desf. por análise de locos microssatélites. Sci. For. 2012, 40, 145–155. [Google Scholar]
- Gonela, A.; Sebbenn, A.M.; Soriani, H.H.; Mestriner, M.A.; Martinez, C.A.; Alzate-Marin, A.L. Genetic diversity and mating system of Copaifera langsdorffii (Leguminosae/Caesalpinioideae). Genet. Mol. Res. 2013, 12, 569–580. [Google Scholar] [CrossRef]
- Antiqueira, L.M.O.R.; Souza, R.G.V.D.C.; Bajay, M.M.; Kageyama, P.Y. Genetic structure and diversity of Copaifera langsdorffii Desf. in Cerrado fragments of the São Paulo State, Brazil. Rev. Árvore 2014, 38, 667–675. [Google Scholar] [CrossRef]
- Carvalho, D.; Oliveira, A.F. Genetic structure of Copaifera langsdorffii Desf. natural populations. Cerne 2015, 10, 137–153. [Google Scholar]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A.; et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Index Kewensis Supplement XX; Clarendon Press: Oxford, UK, 1996.
- Veiga Junior, V.; Pinto, A.C. O gênero Copaifera L. Química Nova 2002, 25, 273–286. [Google Scholar] [CrossRef]
- Carvalho, P.E.R. Espécies Arbóreas Brasileiras; Embrapa Informação Tecnológica: Brasília, Brazil, 2003; Volume 1. [Google Scholar]
- Oliveira, A.F.; Carvalho, D.; Rosado, S.C.S. Taxa de cruzamento e sistema reprodutivo de uma população natural de Copaifera langsdorffii Desf. na região de Lavras (MG) por meio de isoenzimas. Rev. Bras. Bot. 2002, 25, 331–338. [Google Scholar] [CrossRef]
- Pedroni, F.; Sanchez, M.; Santos, F.A.M. Fenologia da copaíba (Copaifera langsdorffii Desf.-Leguminosae, Caesalpinioideae) em uma floresta semidecídua no sudeste do Brasil. Rev. Bras. Bot. 2002, 25, 183–194. [Google Scholar] [CrossRef]
- Oliveira, E.C.P.; Lameira, O.A.; Zoghbi, M.G.B. Identificação da época de coleta do óleo-resina de copaíba (Copaifera spp.) no município de Moju, PA. Rev. Bras. Plantas Med. 2006, 8, 14–23. [Google Scholar]
- DeVore, J. The mind of the copaíba tree: Notes on extractivism, animism and ontology from Southern Bahia. J. R. Anthropol. Inst. 2017, 23, 234–252. [Google Scholar] [CrossRef]
- Almeida, S.D.; Proença, C.E.B.; Sano, S.M.; Ribeiro, J.F. Cerrado: Espécies Vegetais Úteis; Embrapa-CPAC: Planaltina, Brazil, 1998. [Google Scholar]
- Siqueira, M.V.B.M.; Silvério, G.H.; Carlos, J.S.; Toledo, J.A.M.; da Silva, C.J.; de Paula-Souza, J.; Galastri, N.A. Phenotypic plasticity in Copaifera langsdorffii Desf. An. Acad. Bras. Ciências 2023, 95, e20220048. [Google Scholar]
- Tarazi, R.; Sebbenn, A.; Kageyama, P.; Vencovsky, R. Long-distance dispersal in a fire- and livestock-protected savanna. Ecol. Evol. 2013, 3, 1003–1015. [Google Scholar] [CrossRef]
- Trindade, R.; da Silva, J.K.; Setzer, W.N. Copaifera of the Neotropics: A Review of the Phytochemistry and Pharmacology. Int. J. Mol. Sci. 2018, 19, 1511. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Russello, M.A.; Waterhouse, M.D.; Etter, P.D.; Johnson, E.A. From promise to practice: Pairing non-invasive sampling with genomics in conservation. PeerJ 2015, 3, e1106. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; McCarthy, A.S. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics 2017, 33, 2037–2039. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Resour. 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 22 August 2023).
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Kans. Univ. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2010. [Google Scholar]
- Caye, K.; Deist, T.M.; Martins, H.; Michel, O.; François, O. TESS3: Fast inference of spatial population structure and genome scans for selection. Mol. Ecol. Resour. 2016, 16, 540–548. [Google Scholar] [CrossRef]
- Hijmans, R.J.; van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A. Raster: Geographic Data Analysis and Modeling. R Package Version 2.5-2. 2015. Available online: https://CRAN.R-project.org/package=raster (accessed on 7 June 2019).
- South, A. rworldmap: A new R package for mapping global data. R J. 2011, 3, 35–43. [Google Scholar] [CrossRef]
- Beaumont, M.A.; Nichols, R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 1996, 263, 1619–1626. [Google Scholar] [CrossRef]
- Antão, T.; Lopes, A.; Lopes, R.J.; Beja-Pereira, A.; Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinform. 2008, 9, 323. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Audigeos, D.; Brousseau, L.; Traissac, S.; Scotti-Saintagne, C.; Scotti, I. Molecular divergence in tropical tree populations occupying environmental mosaics. J. Evol. Biol. 2013, 26, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Agashe, D.; Sane, M.; Singhal, S. Revisiting the Role of Genetic Variation in Adaptation. Am. Nat. 2023, 202, 486–502. [Google Scholar] [CrossRef]
- Meger, J.; Ulaszewski, B.; Chmura, D.J.; Burczyk, J. Signatures of local adaptation to current and future climate in phenology-related genes in natural populations of Quercus robur. BMC Genom. 2024, 25, 28. [Google Scholar] [CrossRef]
- Kremer, A.; Delcamp, A.; Lesur, I.; Wagner, S.; Rellstab, C.; Guichoux, E.; Leroy, T. Whole-genome screening for near-diagnostic genetic markers for four western European white oak species identification. Ann. For. Sci. 2024, 81, 21. [Google Scholar] [CrossRef]
- Sebbenn, A.M.; Carvalho, A.C.M.; Freitas, M.L.M.; Moraes, S.M.B.; Gaino, A.P.S.C.; Silva, J.M.; Jolivet, C.; Moraes, M.L.T. Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity 2011, 106, 134–145. [Google Scholar] [CrossRef]
- Pinto, S.I.C.; Souza, A.M.; Carvalho, D. Variabilidade genética por iso-enzimas em populações de Copaifera langsdorffii Desf. em dois fragmentos de mata ciliar. Sci. For. 2004, 65, 40–48. [Google Scholar]
- Wright, S. The interpretation of population structure by f-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Silva, E.S.; Mathias, C.; Lima, M.C.F.; Veiga Junior, V.F.; Rodrigues, D.P.; Clement, C.R. Análise físico-química do óleo-resina e variabilidade genética de copaíba na Floresta Nacional do Tapajós. Pesqui. Agropecu. Bras. 2012, 47, 1621–1628. [Google Scholar] [CrossRef]
- Vieira, F.A.; Carvalho, D. Genetic differentiation of the neotropical tree species Protium spruceanum (Benth.) Engler (Burseraceae) between fragments and vegetation corridors in Brazilian Atlantic forest. Acta Bot. Bras. 2009, 23, 1180–1185. [Google Scholar] [CrossRef]
- Farwig, N.; Braun, C.; Böhning-Gaese, K. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv. Genet. 2008, 9, 317–326. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. B Biol. Sci. 1996, 351, 1291–1298. [Google Scholar] [CrossRef]
- Jordano, P.; Godoy, J.Á. RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal-dispersed tree. Mol. Ecol. 2000, 9, 1293–1305. [Google Scholar] [CrossRef]
- Dick, C.W.; Hardy, O.J.; Jones, F.A.; Petit, R.J. Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop. Plant Biol. 2008, 1, 20–33. [Google Scholar] [CrossRef]
- Cavassan, O.; Weiser, V.L. Vascular flora of the cerrado of Bauru-SP. Biota Neotrop. 2015, 15, e20140093. [Google Scholar] [CrossRef]
- Faraco, A.G. Composição Florística e Fitossociologia de Uma Área de Cerrado na Reserva Legal do Campus de Bauru da UNESP–SP. Repositório Institucional UNESP, São Paulo, Brazil. 2007. Available online: https://repositorio.unesp.br/handle/11449/88136 (accessed on 12 August 2019).
- Cavassan, O. Bauru: Terra de cerrado ou floresta? Ciênc. Geogr. 2013, 17, 46–54. [Google Scholar]
- Serra Filho, R.; Cavalli, A.C.; Guillaumon, J.R.; Chiarini, J.V.; Nogueira, F.P.; Ivancko, C.M.A.M.; Barbieri, J.L.; Donzeli, P.L.; Coelho, A.G.S.; Bittencourt, I. Levantamento da cobertura natural e do reflorestamento no cerrado estado de São Paulo. Bol. Técnico Do Inst. Florest. 1974, 11, 1–5. [Google Scholar]
- Beheregaray, L.B.; Cooke, G.M.; Chao, N.L.; Landguth, E.L. Ecological speciation in the tropics: Insights from comparative genetic studies in Amazonia. Front. Genet. 2014, 5, 477. [Google Scholar] [CrossRef] [PubMed]




| Population | N | HO | HE | FIS |
|---|---|---|---|---|
| JB | 29 | 0.205 | 0.257 | 0.203 * |
| UN | 20 | 0.189 | 0.247 | 0.235 * |
| HA | 11 | 0.187 | 0.244 | 0.235 * |
| FP | 14 | 0.203 | 0.245 | 0.173 * |
| DU | 11 | 0.178 | 0.238 | 0.251 * |
| CAE | 10 | 0.196 | 0.233 | 0.158 * |
| Mean | - | 0.197 | 0.264 | 0.209 |
| Pop. | JB | UN | HA | FP | DU | CAE |
|---|---|---|---|---|---|---|
| JB | _ | 6 | 14 | 23 | 19 | 94 |
| UN | 0.0068 | _ | 17 | 24 | 14 | 90 |
| HA | 0.0172 | 0.0128 | _ | 31 | 30 | 92 |
| FP | 0.0234 * | 0.0166 | 0.0255 | _ | 38 | 112 |
| DU | 0.0260 | 0.0184 | 0.0226 | 0.0245 | _ | 101 |
| CAE | 0.0432 * | 0.0327 * | 0.0315 * | 0.0501 * | 0.0428 | _ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siqueira, M.V.B.M.; Carlos, J.S.; Orcini, W.; Bajay, M.M.; Martins, K.; Melo, A.T.d.O.; Veasey, E.A.; Tambarussi, E.V.; Konzen, E.R. Genomic Diversity and Structure of Copaifera langsdorffii Populations from a Transition Zone Between the Atlantic Forest and the Brazilian Savanna. Plants 2025, 14, 2858. https://doi.org/10.3390/plants14182858
Siqueira MVBM, Carlos JS, Orcini W, Bajay MM, Martins K, Melo ATdO, Veasey EA, Tambarussi EV, Konzen ER. Genomic Diversity and Structure of Copaifera langsdorffii Populations from a Transition Zone Between the Atlantic Forest and the Brazilian Savanna. Plants. 2025; 14(18):2858. https://doi.org/10.3390/plants14182858
Chicago/Turabian StyleSiqueira, Marcos Vínicius Bohrer Monteiro, Juliana Sanchez Carlos, Wilson Orcini, Miklos Maximiliano Bajay, Karina Martins, Arthur Tavares de Oliveira Melo, Elizabeth Ann Veasey, Evandro Vagner Tambarussi, and Enéas Ricardo Konzen. 2025. "Genomic Diversity and Structure of Copaifera langsdorffii Populations from a Transition Zone Between the Atlantic Forest and the Brazilian Savanna" Plants 14, no. 18: 2858. https://doi.org/10.3390/plants14182858
APA StyleSiqueira, M. V. B. M., Carlos, J. S., Orcini, W., Bajay, M. M., Martins, K., Melo, A. T. d. O., Veasey, E. A., Tambarussi, E. V., & Konzen, E. R. (2025). Genomic Diversity and Structure of Copaifera langsdorffii Populations from a Transition Zone Between the Atlantic Forest and the Brazilian Savanna. Plants, 14(18), 2858. https://doi.org/10.3390/plants14182858







