Optimization of Somatic Embryogenesis and Transcriptomic Analysis of the Early Stage of Callus Redifferentiation in Quercus suber L.
Abstract
1. Introduction
2. Results
2.1. Transdifferentiation to Form Callus
2.1.1. Effects of Different Explants on Transdifferentiation to Form Callus of Q. suber
2.1.2. Effects of Sampling Time on Transdifferentiation to Form Callus of Q. suber
2.1.3. Effects of Basic Mediums, Light Conditions and PGRs on Transdifferentiation to Form Callus of Q. suber
2.2. Callus Proliferation
2.3. Induction, Maturation, and Germination in Somatic Embryogenesis
2.4. Cytological Observation
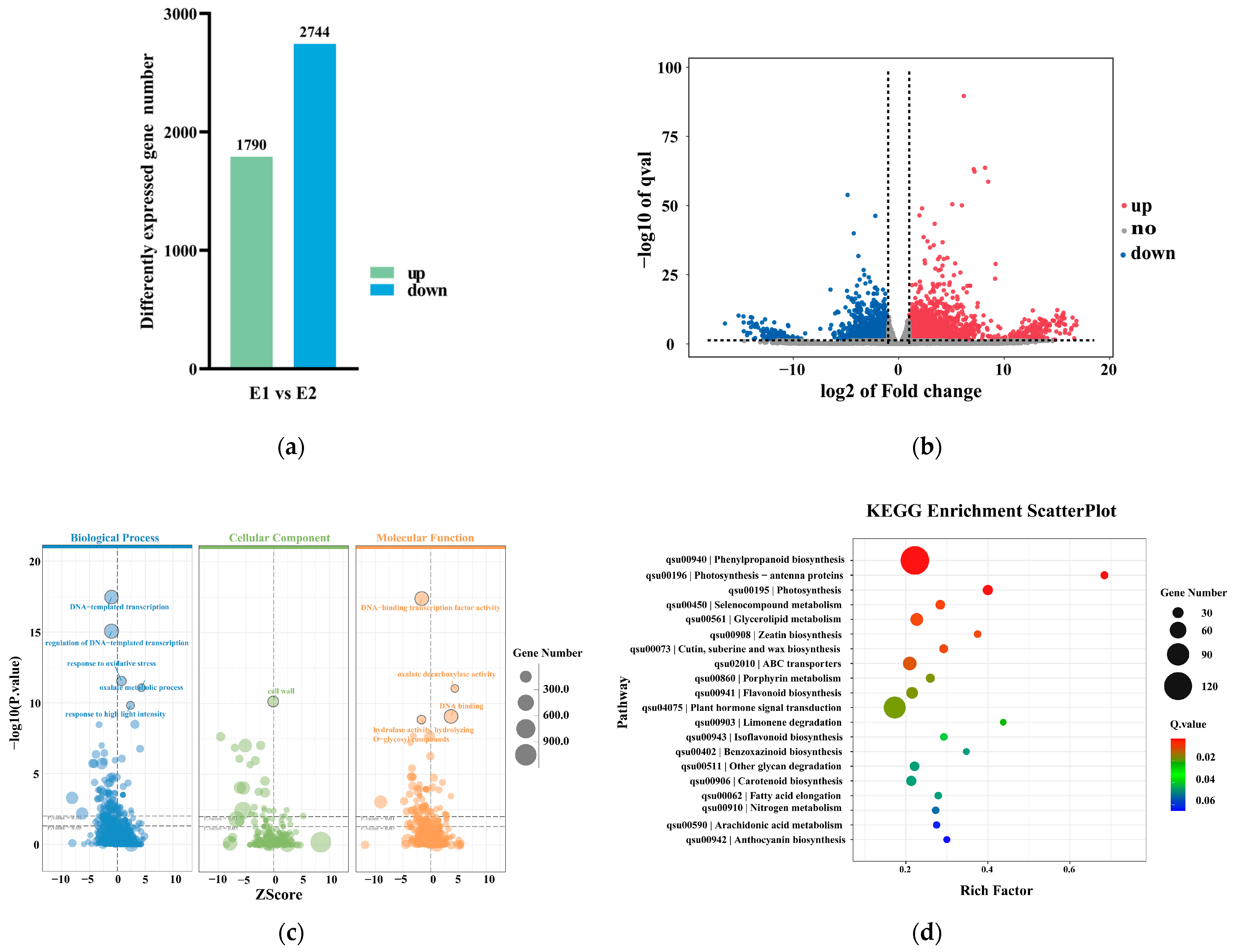
2.5. Transcriptomic Analysis at the Early Stage of Callus Redifferentiation
2.5.1. Analysis and Functional Enrichment of DEGs During the Early Stage of Callus Redifferentiation
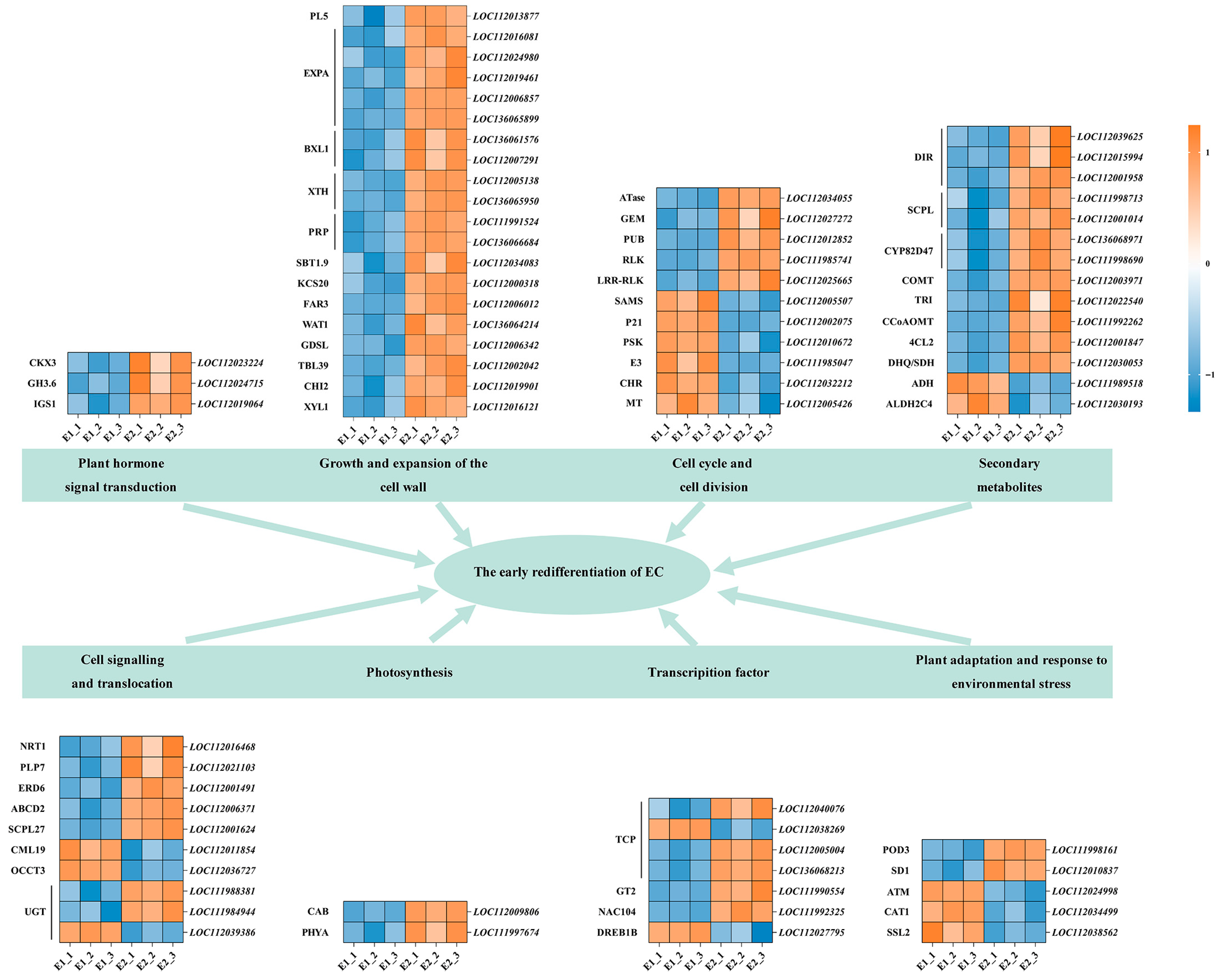
2.5.2. Key Genes and TFs Associated with the Early Stage of Callus Redifferentiation and Their Functional Analysis
2.5.3. Selection of Reference Genes and Quantitative Real-Time PCR (qRT-PCR) Validation of DEGs
3. Discussion
3.1. Transdifferentiation to Form Callus of Q. suber
3.2. Callus Proliferation
3.3. Somatic Embryo Induction Embryo, Maturation and Germination
3.4. Transcriptomics
4. Materials and Methods
4.1. Plant Material
4.2. Medium and Culture Conditions
4.2.1. Callus Induction
4.2.2. Callus Proliferation
4.2.3. Somatic Embryo Induction
4.2.4. Embryo Maturation and Germination
4.3. Various Factors Effect on Transdifferentiation to Form Callus
explants) × 100%
4.3.1. Explant Types Effect on Transdifferentiation to Form Callus
4.3.2. Sampling Time Effect on Transdifferentiation to Form Callus
4.3.3. Type of Basic Medium Effect on Transdifferentiation to Form Callus
4.3.4. Light Conditions Effect on Transdifferentiation to Form Callus
4.3.5. PGRs on Transdifferentiation to Form Callus
4.4. Various Factors Effect on Proliferation
weight)/Initial inoculated embryogenic callus weight
4.4.1. PGRs
4.4.2. Culture Method
4.5. PGRs Effect on Somatic Embryo Induction
embryogenic callus) × 100%
4.6. Cytological Observation
4.7. Transcriptome Sequencing and DEGs Analysis
4.8. Selection of Reference Genes and Gene Expression Validation via qRT-PCR
4.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SE | somatic embryogenesis |
| 6-BA | 6-benzyladenine |
| NAA | 1-Naphthaleneacetic acid |
| 2,4-D | 2,4-Dichlorophenoxyacetic acid |
| ZT | zeatin |
| IAA | indole-3-acetic acid |
| IBA | indole-3-butyric Acid |
| MS | Murashige and Skoog |
| SH | Schenk and Hildebrandt |
| MSSH | MS major elements + SH minor elements and vitamins |
| DEGs | differentially expressed genes |
| E1 | embryogenic callus |
| E2 | globular embryos |
| GO enrichment | Gene Ontology enrichment |
| KEGG enrichment | Kyoto Encyclopedia of Genes and Genomes enrichment |
| qRT-PCR | quantitative real-timepolymerase chain reaction |
| PGRs | plant growth regulators |
| TFs | transcription factors |
| LEC | LEAFY COTYLEDON |
| BBM | BABY BOOM |
| SERK | Somatic embryogenesis receptor-like kinase |
| CYP82D47 | cytochrome P450 |
| POD3 | peroxidase 3 |
| EXPA | expansin A |
| XTH | xyloglucan endotransglucosylase/hydrolase protein |
| PRP | repetitive proline-rich cell wall protein |
| SAMS | S-adenosylmethionine synthase |
| PSK | phytosulfokines |
| P21 | protein P21 |
| E3 | E3 ubiquitin-protein ligase |
| COMT | caffeic acid 3-O-methyltransferase |
| 4CL2 | 4-coumarate—CoA ligase 2 |
| ADH | alcohol dehydrogenase |
| CAT1 | cationic amino acid transporter 1 |
| SSL2 | protein strictosidine synthase-like 2 |
| SD1 | receptor-like serine/threonine-protein kinase |
| NRT1 | Nitrate transporter |
| ERD6 | sugar transporter |
| OCCT3 | organic cation/carnitine transporter |
| CML19 | the calcium sensor putative calcium-binding protein |
| CAB | chlorophyll a-b binding protein |
| PHY-A | phytochrome A |
| NCA104 | NAC domain-containing protein 104 |
| GT2 | Trihelix |
| DREB1B | dehydrationresponsiveelemen-tbindingprotein |
| QsRPII | RNA polymerase II |
| QsTUB | β-tubulin |
| QsEIF-5A | Eukaryotic translation initiation factor 5A |
| (QsCACs)C | Clathrin adaptor complexes medium subunit family protein |
Appendix A
| Gene | Primers | Amplicon Size (bp) | Temperature (°C) |
|---|---|---|---|
| Actin | Forward: 5′-TGGATTCTGGTGATGGTGTGAGTC-3′ | 162 | 62 |
| Reverse: 5′-CAATTTCCCGTTCAGCAGTAGTGG-3′ | |||
| QsRPII | Forward: 5′-GACATAGATCCCGTTACCCA-3′ | 168 | 54 |
| Reverse: 5′-TTTGATTGCACCAGTAGATTC-3′ | |||
| QsTUB | Forward: 5′-GCTCACTACCCCAAGCTTT-3′ | 187 | 58 |
| Reverse: 5′-GGAACCTCTGGAGGTTAAA-3′ | |||
| QsEIF-5A | Forward: 5′-GCCATGTCCGACGAGGAG-3′ | 86 | 62 |
| Reverse: 5′-CGGATGGTTCCGGCTTGC-3′ | |||
| Qs(CACs)C | Forward: 5′-TCTGGGAGAAGAGTGGCTACA-3′ | 175 | 57 |
| Reverse: 5′-GAGCCACCATTCAAATCCT-3′ | |||
| CKX3 | Forward: 5′-GCAATGGCTCCTAATGGGGT-3′ | 87 | 61 |
| Reverse: 5′-TGGGGTTCCAGAGACAGTGA-3′ | |||
| GH3.6 | Forward: 5′-AGACCATCCCAGGCCACTAT-3′ | 165 | 62 |
| Reverse: 5′-TCTCAAGGGGCCCAATTGAC-3′ | |||
| PL5 | Forward: 5′-CAAGGGCATGCAGGTCACTA-3′ | 155 | 61 |
| Reverse: 5′-TGATGGTTGGGGAAGCACTC-3′ | |||
| POD3 | Forward: 5′-CTTGCTCTCAGGTGCTCACA-3′ | 104 | 61 |
| Reverse: 5′-ggT CTAGAGCTGGGTCCTGA-3′ | |||
| CYP82D47 | Forward: 5′-GGTTGGACTTGGGAGGCTAC-3′ | 107 | 60 |
| Reverse: 5′-ACCTGAAATTTTTGCGCGCT-3′ | |||
| TCP4 | Forward: 5′-TGGGGGCTGAGATGACCATA-3′ | 84 | 60 |
| Reverse: 5′-ACCGAGTGTTGTCGATGCTT-3′ |
References
- Ramos, M.; Rocheta, M.; Carvalho, L.; Inácio, V.; Graça, J.; Morais-Cecilio, L. Expression of DNA methyltransferases is involved in Quercus suber cork quality. Tree Genet. Genomes 2013, 9, 1481–1492. [Google Scholar] [CrossRef][Green Version]
- Soler, M.; Serra, O.; Molinas, M.; García-Berthou, E.; Caritat, A.; Figueras, M. Seasonal variation in transcript abundance in cork tissue analyzed by real time RT-PCR. Tree Physiol. 2008, 28, 743–751. [Google Scholar] [CrossRef]
- Chaves, I.; Lin, Y.C.; Pinto-Ricardo, C.; Peer, Y.V.; Minguel, C. miRNA profiling in leaf and cork tissues of Quercus suber reveals novel miRNAs and tissue-specific expression patterns. Tree Genet. Genomes 2014, 10, 721–737. [Google Scholar] [CrossRef]
- Prasetia, D.; Purusatama, B.D.; Kim, J.H.; Yang, G.U.; Jang, J.H.; Park, S.Y.; Lee, S.H.; Kim, N.H. Quantitative Anatomical Characteristics of Virgin Cork in Quercus variabilis Grown in Korea. Forests 2022, 13, 1711. [Google Scholar] [CrossRef]
- He, S.A.; Wu, H.J. Introduction of Eucalyptus and European Cork Oak in Nanjing. For. Sci. 1964, 2, 88–90. [Google Scholar]
- She, Y.G. Germination Test of European Cork Oak Seeds. Econ. For. Res. 2000, 2, 28–29. [Google Scholar]
- Peng, J.G. European Cork Oak. Sichuan For. Sci. Technol. 1986, 2, 74. [Google Scholar]
- Wang, M.X. Improvement of Cork Oak Species in China. For. Sci. 1963, 4, 347–353. [Google Scholar]
- Hae, N.K.; Hye, Y.J.; Myeong, J.K.; Inkyin, K.; Ha, N.Y.; Tae, Y.L.; Tai, H.A.; Su, Y.W. Why does Quercus suber species decline in Mediterranean areas? J. Asia-Pac. Biodivers. 2017, 10, 337–341. [Google Scholar]
- Guo, H.H.; Qi, X.S.; Li, T.T.; Fan, Y.J.; Huo, H.Q.; Yu, Q.Y.; Zeng, F.C. Editorial: Molecular basis of asexual reproduction and its application in crops. Plant Sci. 2022, 13, 1033212. [Google Scholar] [CrossRef] [PubMed]
- Vieitez, A.M.; Corredoira, E.; Martínez, M.T.; San-José, M.C.; Sánchez, C.; Valladares, S.; Vidal, N.; Ballester, A. Application of biotechnological tools to Quercus improvement. Eur. J. For. Res. 2012, 131, 519–539. [Google Scholar] [CrossRef]
- Finer, J.J. Direct Somatic Embryogenesis. In Plant Cell, Tissue and Organ Culture, Springer Lab Manual; Gamborg, O.L., Phillips, G.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Guo, H.; Guo, H.; Zhang, L.; Fan, Y.; Wu, J.; Tang, Z.; Zhang, Y.; Fan, Y.; Zeng, F. Dynamic Transcriptome Analysis Reveals Uncharacterized Complex Regulatory Pathway Underlying Genotype-Recalcitrant Somatic Embryogenesis Transdifferentiation in Cotton. Genes 2020, 11, 519. [Google Scholar] [CrossRef]
- Fehér, A. Callus, Dedifferentiation, Totipotency, Somatic Embryogenesis: What These Terms Mean in the Era of Molecular Plant Biology? Front. Plant Sci. 2019, 10, 536. [Google Scholar] [CrossRef]
- Sugimoto, K.; Gordon, S.P.; Meyerowitz, E.M. Regeneration in plants and animals: Dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011, 21, 212–218. [Google Scholar] [CrossRef]
- Chadipiralla, K.; Gayathri, P.; Rajani, V.; Reddy, P.V.B. Plant Tissue Culture and Crop Improvement. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer: Cham, Switzerland, 2020; pp. 391–412. [Google Scholar]
- Awada, R.; Campa, C.; Gibault, E.; Déchamp, E.; Georget, F.; Lepelley, M.; Abdallah, C.; Erban, A.; Martinez-Seidel, F.; Kopka, J. Unravelling the Metabolic and Hormonal Machinery During Key Steps of Somatic Embryogenesis: A Case Study in Coffee. Int. J. Mol. Sci. 2019, 20, 4665. [Google Scholar] [CrossRef]
- Guan, Y.; Li, S.G.; Fan, X.F.; Su, Z.H. Application of Somatic Embryogenesis in Woody Plants. Front. Plant Sci. 2016, 7, 938. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Mosqueda, M.A. Overview of Somatic Embryogenesis. In Somatic Embryogenesis; Ramírez-Mosqueda, M.A., Ed.; Methods in Molecular Biology; Humana: New York, NY, USA, 2022; Volume 2527, pp. 1–8. [Google Scholar]
- Zimmerman, J.L. Somatic Embryogenesis: A Model for Early Development in Higher Plants. Plant Cell 1993, 5, 1411–1423. [Google Scholar] [CrossRef]
- Sivanesan, I.; Nayeem, S.; Venkidasamy, B.; Kuppuraj, S.P.; RN, C.; Samynathan, R. Genetic and epigenetic modes of the regulation of somatic embryogenesis: A review. Biol. Futur. 2022, 73, 259–277. [Google Scholar] [CrossRef]
- Kumaravel, M.; Uma, S.; Backiyarani, S.; Saraswathi, M.S. Proteomic analysis of somatic embryo development in Musa spp. cv. Grand Naine (AAA). Sci. Rep. 2020, 10, 4501. [Google Scholar] [CrossRef]
- Wang, F.X.; Shang, G.D.; Wu, L.Y.; Xu, Z.G.; Zhao, X.Y.; Wang, W.J. Chromatin Accessibility Dynamics and a Hierarchical Transcriptional Regulatory Network Structure for Plant Somatic Embryogenesis. Dev. Cell 2020, 54, 742–757. [Google Scholar] [CrossRef]
- Pre, A.; Obert, B. Flax (Linum usitatisimum L.)—A plant system for study of embryogenesis. Acta Biol. Cracoviensia Ser. Bot. 2003, 451, 15–18. [Google Scholar]
- Zeng, F.C.; Zhang, X.L.; Jin, S.X.; Cheng, L.; Liang, S.G.; Hu, L.S.; Guo, X.P.; Nie, Y.C.; Cao, J.L. Chromatin reorganization and endogenous auxin/cytokinin dynamic activity during somatic embryogenesis of cultured cotton cell. Plant Cell Tissue Organ Cult. 2007, 90, 63–70. [Google Scholar] [CrossRef]
- Testillano, P.S.; Gómez-Garay, A.; Pintos, B.; Risueño, M.C. Somatic Embryogenesis of Quercus suber L. from Immature Zygotic Embryos. In Plant Cell Culture Protocols. Methods in Molecular Biology; Loyola-Vargas, V., Ochoa-Alejo, N., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1815, pp. 247–256. [Google Scholar]
- Mauri, P.; Manzanera, J. Induction, maturation and germination of holm oak (Quercus ilex L.) somatic embryos. Plant Cell Tissue Organ Cult. 2003, 74, 229–235. [Google Scholar]
- Lebtahi, F.; Errahmani, M.; Nadia, B. Propagation of Cork oak (Quercus suber L.) by axillary shoot and somatic embryogenesis. Propag. Ornam. Plants 2015, 15, 113–122. [Google Scholar]
- Hernández, I.; Celestino, C.; Toribio, M. Vegetative propagation of Quercus suber L. by somatic embryogenesis. Plant Cell Rep. 2003, 21, 759–764. [Google Scholar]
- Pinto, G.; Valentim, H.; Costa, A.; Castro, S.; Santos, C. Somatic embryogenesis in leaf callus from a mature Quercus suber L. Tree. Vitr. Cell. Dev. Biol.-Plant 2002, 38, 569–572. [Google Scholar]
- Sané, D.; Aberlenc-Bertossi, F.; Diatta, L.; Guèye, B.; Daher, A.; Sagna, M.; Duval, Y.; Borgel, A. Influence of Growth Regulators on Callogenesis and Somatic Embryo Development in Date Palm (Phoenix dactylifera L.) Sahelian Cultivars. Sci. World J. 2012, 2012, 837395. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.; Edwards, M.; Kiernan, R.; Davey, C.; Blakesley, D.; Henshaw, G. Development of friable embryogenic callus and embryogenic suspension culture systems in cassava (Manihot esculenta Crantz). Nat. Biotechnol. 1996, 14, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Jeon, H.; Kim, M. Optimization of a mature embryo-based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tissue Organ Cult. 2002, 71, 237–244. [Google Scholar] [CrossRef]
- Carman, J.G.; Jefferson, N.E.; Campbell, W.F. Induction of embryogenic Triticum aestivum L. calli. I. Quantification of genotype and culture medium effects. Plant Cell Tissue Organ Cult. 1987, 10, 101–113. [Google Scholar] [CrossRef]
- Gomes da Cunha, A.C.; Fernandes Ferreira, M. Somatic embryogenesis, organogenesis and callus growth kinetics of flax. Plant Cell Tissue Organ Cult. 1996, 47, 1–8. [Google Scholar] [CrossRef]
- Guo, G.; Jeong, B.R. Explant, Medium, and Plant Growth Regulator (PGR) Affect Induction and Proliferation of Callus in Abies koreana. Forests 2021, 12, 1388. [Google Scholar] [CrossRef]
- Cheng, W.H.; Wang, F.L.; Cheng, X.Q.; Zhu, Q.H.; Sun, Y.Q.; Zhu, H.G.; Sun, J. Polyamine and Its Metabolite H2O2 Play a Key Role in the Conversion of Embryogenic Callus into Somatic Embryos in Upland Cotton (Gossypium hirsutum L.). Front. Plant Sci. 2015, 6, 1063. [Google Scholar] [CrossRef]
- Haddadi, P.; Moieni, A.; Karimzadeh, G.; Abdollahi, M.R. Effects of Gibberellin, Abscisic Acid and Embryo Desiccation on Normal Plantlet Regeneration, Secondary Embryogenesis and Callogenesis in Microspore Culture of Brassica napus L. cv. PF704. Int. J. Plant Prod. 2008, 2, 153–162. [Google Scholar]
- Loureiro, J.; Pinto, G.; Lopes, T.; Doležel, J.; Santos, C. Assessment of ploidy stability of the somatic embryogenesis process in Quercus suber L. using flow cytometry. Planta 2005, 221, 815–822. [Google Scholar] [CrossRef]
- Maâtaoui, M.; Espagnac, H.; Michaux-Ferrière, N. Histology of Callogenesis and Somatic Embryogenesis Induced in Stem Fragments of Cork Oak (Quercus suber) Cultured In Vitro. Ann. Bot. 1990, 66, 183–190. [Google Scholar] [CrossRef]
- Conner, A.J.; Searle, H.; Jacobs, J.M.E. Rejuvenation of chicory and lettuce plants following phase change in tissue culture. BMC Biotechnol. 2019, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Karami, O.; Aghavaisi, B.; Mahmoudi-Pour, A. Molecular aspects of somatic-to-embryogenic transition in plants. J. Biol. Chem. 2009, 2, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Gulzar, B.; Mujib, A.; Malik, M.Q.; Sayeed, R.; Mamgain, J.; Ejaz, B. Genes, proteins and other networks regulating somatic embryogenesis in plants. J. Genet. Eng. Biotechnol. 2022, 18, 31. [Google Scholar] [CrossRef]
- Meireles, B.; Usié, A.; Barbosa, P.; Fortes, A.M.; Folgado, A.; Chaves, I.; Carrasquinho, I.; Costa, R.L.; Gonçalves, S.; Teixeira, R.T. Characterization of the cork formation and production transcriptome in Quercus cerris × suber hybrids. Physiol. Mol. Biol. Plants 2018, 24, 535–549. [Google Scholar] [CrossRef]
- Sobral, R.; Costa, M.M.R. Role of floral organ identity genes in the development of unisexual flowers of Quercus suber L. Sci. Rep. 2017, 7, 10368. [Google Scholar]
- Capote, T.; Usié, A.; Barbosa, P.; Romos, M.; Morais-Cecílio, L.; Gonçalves, S. Transcriptome dynamics of cork oak (Quercus suber) somatic embryogenesis reveals active gene players in transcription regulation and phytohormone homeostasis of embryo development. Tree Genet. Genomes 2019, 15, 52. [Google Scholar] [CrossRef]
- Zhang, B.X.; Zhang, H.L.; Xia, Y.J. Harnessing spatial transcriptomics for advancing plant regeneration research. Trends Plant Sci. 2024, 29, 718–720. [Google Scholar] [CrossRef] [PubMed]
- Czernicka, M.; Chłosta, I.; Kęska, K.; Kozieradzka-Kiszkurno, M.; Abdullah, M.; Popielarska-Konieczna, M. Protuberances are organized distinct regions of long-term callus: Histological and transcriptomic analyses in kiwifruit. Plant Cell Rep. 2021, 40, 637–665. [Google Scholar] [CrossRef] [PubMed]
- Chalupa, V. Somatic Embryogenesis in Oak (Quercus spp.). In Somatic Embryogenesis in Woody Plants; Jain, S.M., Gupta, P.K., Newton, R.J., Eds.; Forestry Sciences; Springer: Dordrecht, The Netherlands, 1995; Volume 2, pp. 67–87. [Google Scholar]
- Salaün, C.; Lepiniec, L.; Dubreucq, B. Genetic and Molecular Control of Somatic Embryogenesis. Plants 2021, 10, 1467. [Google Scholar] [CrossRef]
- Li, Y.; Yan, L.J.; Wang, H.H.; Tang, S.J. Plant regeneration through somatic embryogenesis in Tilia cordata. Plant Cell Tissue Organ Cult. 2025, 160, 48. [Google Scholar] [CrossRef]
- Bhat, A.Y.; Shahzad, A.; Kausar, A.; Rashid, A. Integrating leaf and root induced shoot regeneration and embryogenesis for the conservation of Atropa acuminata Royle ex Lindl—An endangered Himalayan herb. Plant Cell Tissue Organ Cult. 2024, 159, 73. [Google Scholar] [CrossRef]
- Liang, H.Z.; Xiong, Y.P.; Guo, B.Y.; Yan, H.F.; Jian, S.G.; Ren, H.; Zhang, X.H.; Li, Y.; Zeng, S.J.; Wu, K.L. Shoot organogenesis and somatic embryogenesis from leaf and root explants of Scaevola sericea. Sci. Rep. 2020, 10, 11343. [Google Scholar] [CrossRef]
- Akkenapally, S.; Mudalkar, S.; Bodiga, S.; Sundaram, R.; Bokam, A.K. In vitro callus generation and somatic embryogenesis from leaf explant of Madhuca longifolia var latifolia (Roxb.) A. Chev. Vegetos 2024, 9, 1–7. [Google Scholar] [CrossRef]
- Ku, S.S.; Woo, H.A.; Shin, M.J.; Jie, E.Y.; Kim, H.; Kim, H.S.; Cho, H.S.; Jeong, W.J.; Lee, M.S.; Min, S.R. Efficient Plant Regeneration System from Leaf Explant Cultures of Daphne genkwa via Somatic Embryogenesis. Plants 2023, 12, 2175. [Google Scholar] [CrossRef]
- Zhai, M.; Dai, Q.; Zhao, Y.; Zhang, S.Y.; Yang, Y.L. Induction of callus and establishment of suspension culture system in Cassia mimosoides herb. Vitr. Cell. Dev. Biol.-Plant 2025, 61, 229–238. [Google Scholar] [CrossRef]
- Duong, T.N.; Nguyen, T.M.; Pham, Q.T.; Le, T.M.; Nguyen, T.H.; Ngo, C.C.; Nguyen, H.N.; Vinh, D.N. Liquid culture as a positive condition to induce and enhance quality and quantity of somatic embryogenesis of Lilium longiflorum. Sci. Hortic. 2006, 110, 93–97. [Google Scholar] [CrossRef]
- Ibraheem, Y.; Pinker, I.; Böhme, M. A comparative study between solid and liquid cultures relative to callus growth and somatic embryo formation in Phoenix dactylifera L. cv. Zaghlool. Emir. J. Food Agric. 2013, 25, 883–898. [Google Scholar] [CrossRef]
- Naouar, B.A.; Rajae, B.; Safaa, R.; Ouafaa, H.; Ibtissam, B.; Mustapha, H.; Latifa, A.; Alain, B.; Patrick, M.; Ahmed, L. Influence of exogenous polyamines on the secondary somatic embryogenesis of cork oak (Quercus suber L.). Bioengineered 2023, 14, 2288354. [Google Scholar] [CrossRef]
- Fernández-Guijarro, B.; Celestino, C.; Toribio, M. Influence of external factors on secondary embryogenesis and germination in somatic embryos from leaves of Quercus suber. Plant Cell Tissue Organ Cult. 1995, 41, 99–106. [Google Scholar] [CrossRef]
- Fan, Y.P.; Tang, Z.M.; Wei, J.M.; Yu, X.M.; Guo, H.H.; Li, T.T.; Guo, H.X.; Zhang, L.; Fan, Y.J.; Zhang, C.Y.; et al. Dynamic Transcriptome Analysis Reveals Complex Regulatory Pathway Underlying Induction and Dose Effect by Different Exogenous Auxin IAA and 2,4-D During in vitro Embryogenic Redifferentiation in Cotton. Front. Plant Sci. 2022, 13, 931105. [Google Scholar] [CrossRef]
- Zhang, X.L.; Wang, Y.L.; Yan, Y.Y.; Peng, H.; Long, Y.; Zhang, Y.C.; Jiang, Z.; Liu, P.; Zou, C.Y.; Peng, H.W. Transcriptome sequencing analysis of maize embryonic callus during early redifferentiation. BMC Genomics 2019, 20, 159. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Chwiałkowska, K.; Nowak, K.; Citerne, S.; Morończyk, J.; Wójcik, A.M.; Kiwior-Wesołowska, A.; Francikowski, J.; Kwaśniewski, M.; Gaj, M.D. Transcriptomic profiling reveals histone acetylation-regulated genes involved in somatic embryogenesis in Arabidopsis thaliana. BMC Genomics 2024, 25, 788. [Google Scholar] [CrossRef]
- Avilez-Montalvo, J.R.; Quintana-Escobar, A.O.; Méndez-Hernández, H.A.; Aguilar-Hernández, V.; Brito-Argáez, L.; Galaz-Ávalos, R.M.; Uc-Chuc, M.A.; Loyola-Vargas, V.M. Auxin-Cytokinin Cross Talk in Somatic Embryogenesis of Coffea canephora. Plants 2022, 11, 2013. [Google Scholar] [CrossRef]
- Méndez-Hernández, H.A.; Quintana-Escobar, A.O.; Uc-Chuc, M.A.; Loyola-Vargas, V.M. Genome-Wide Analysis, Modeling, and Identification of Amino Acid Binding Motifs Suggest the Involvement of GH3 Genes during Somatic Embryogenesis of Coffea canephora. Plants 2021, 10, 2034. [Google Scholar] [CrossRef]
- Yu, X.N.; Lu, M.J.; Zhou, M.; Wang, H.Y.; Feng, J.Y.; Wen, Y.Q. Reduction of pectin may decrease the embryogenicity of grapevine (Vitis vinifera) pro-embryonic masses after 10 years of in vitro culture. Sci. Hortic. 2023, 309, 111690. [Google Scholar] [CrossRef]
- Pérez-Pérez, Y.; Carneros, E.; Berenguer, E.; Solís, M.-T.; Bárány, I.; Pintos, B.; Gómez-Garay, A.; Risueño, M.C.; Testillano, P.S. Pectin De-methylesterification and AGP Increase Promote Cell Wall Remodeling and Are Required During Somatic Embryogenesis of Quercus suber. Plant Sci. 2019, 9, 1915. [Google Scholar] [CrossRef]
- Subrahmanyeswari, T.; Gantait, S.; Sarkar, R.; Kamble, S.N.; Singh, S.; Bhattacharyya, S. Polyamines- and growth inducers-mediated enhanced mono-phasic in vitro regeneration of sugar leaf plant (Stevia rebaudiana Bert.) in liquid medium. S. Afr. J. Botan. 2024, 173, 34–45. [Google Scholar] [CrossRef]
- Sanaa, N.; Frédéric, L.; Françoise, G.; Dominique, L.; Marc, F.; Alain, D.; Annie, J. Polyamine Content and Somatic Embryogenesis in Papaver somniferum Cells Transformed with sam-1 Gene. J. Plant Physiol. 1999, 56, 729–734. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, X.; Xu, X.; Li, Y.; Zhao, P.; Chen, X.; Shen, X.; Zhang, Z.; Chen, Y.; Liu, S. Genome-wide identification, evolution analysis of cytochrome P450 monooxygenase multigene family and their expression patterns during the early somatic embryogenesis in Dimocarpus longan Lour. Gene 2022, 826, 146453. [Google Scholar] [CrossRef]
- Lan, J.Q.; Wang, N.; Wang, Y.T.; Jiang, Y.D.; Yu, H.; Cao, X.F.; Qin, G.J. Arabidopsis TCP4 transcription factor inhibits high temperature-induced homeotic conversion of ovules. Nat. Commun. 2023, 14, 5673. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Zhang, D.P.; Shi, P.; Htwe, Y.M.; Yu, Q.; Huang, L.Y.; Zhou, H.Q.; Liu, L.Y.; Wang, Y. Cell wall lignification may be necessary for somatic embryogenesis of areca palm (Areca catechu). Sci. Hortic. 2023, 307, 111538. [Google Scholar] [CrossRef]
- Xie, Q.; Ahmed, U.; Qi, C.; Du, K.; Luo, J.; Wang, P.; Zheng, B.; Shi, X. A protocol for identifying universal reference genes within a genus based on RNA-Seq data: A case study of poplar stem gene expression. For. Res. 2024, 4, e021. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.P.; Zhang, C.J.; Liu, Q.H.; Zhang, Z.; Zheng, B.; Bao, M.Z. De novo comparative transcriptome analysis provides new insights into sucrose induced somatic embryogenesis in camphor tree (Cinnamomum camphora L.). BMC Genomics 2016, 17, 26. [Google Scholar] [CrossRef]
- Teng, M.; Love, M.I.; Davis, C.A.; Djebali, S.; Dobin, A.; Graveley, B.R.; Li, S.; Mason, C.E.; Olson, S.; Pervouchine, D.; et al. A benchmark for RNA-seq quantification pipelines. Genome Biol. 2016, 17, 74. [Google Scholar] [CrossRef]
- Wang, Z.; Lyu, Z.; Pan, L.; Zeng, G.; Randhawa, P. Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue. BMC Med. Genomics 2019, 12, 86. [Google Scholar]
- Chen, Y.; Li, X.; Su, L.; Chen, X.; Zhang, S.T.; Xu, X.P.; Zhang, Z.H.; Chen, Y.K.; Xu, X.H.; Lin, Y.L.; et al. Genome-wide identification and characterization of long non-coding RNAs involved in the early somatic embryogenesis in Dimocarpus longan Lour. BMC Genomics 2018, 19, 805. [Google Scholar] [CrossRef]
- Gao, Y.; Cui, Y.; Zhao, R.; Chen, X.; Zhang, J.; Zhao, J.; Kong, L. Cryo-Treatment Enhances the Embryogenicity of Mature Somatic Embryos via the lncRNA–miRNA–mRNA Network in White Spruce. Int. J. Mol. Sci. 2022, 23, 1111. [Google Scholar] [PubMed]
- Ok, R.L.; Ngoc, Q.N.; Kwang, H.L.; Young, C.K.; Jiho, S. Cytohistological study of the leaf structures of Panax ginseng Meyer and Panax quinquefolius L. J. Ginseng Res. 2017, 41, 463–468. [Google Scholar]
- Li, X.; Huang, X.; Wen, M.; Wen, Y.; Chen, Y.M.; Liu, Y.L.; Liu, X.D. Cytological observation and RNA-seq analysis reveal novel miRNAs high expression associated with the pollen fertility of neo-tetraploid rice. BMC Plant Biol. 2023, 23, 434. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 September 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar]
- Ebadzad, G.; Cravador, A. Quantitative RT-PCR analysis of differentially expressed genes in Quercus suber in response to Phytophthora cinnamomi infection. SpringerPlus 2014, 3, 613. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Winston, W. Microsoft Excel 2010 Data Analysis and Business Modeling; Pearson Education: Upper Saddle River, NJ, USA, 2011; Volume 1. [Google Scholar]
- Sweet, S.A.; Karen, G.M. Data Analysis with SPSS.; Allyn & Bacon: Boston, MA, USA, 1999; Volume 1. [Google Scholar]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Edwards, P.M. Origin 7.0: Scientific graphing and data analysis software. J. Chem. Inf. Comput. Sci. 2002, 42, 1270–1271. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]

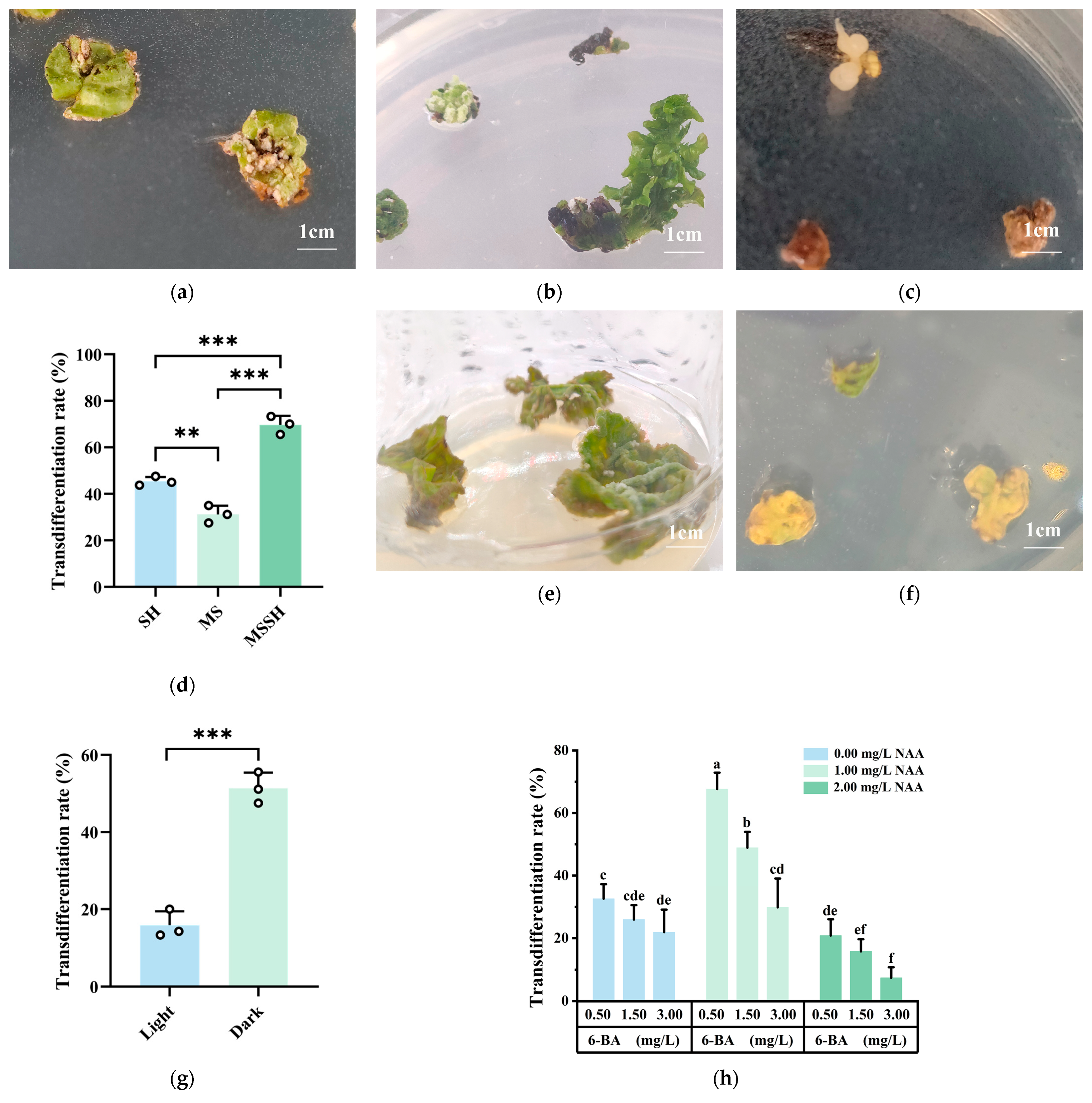
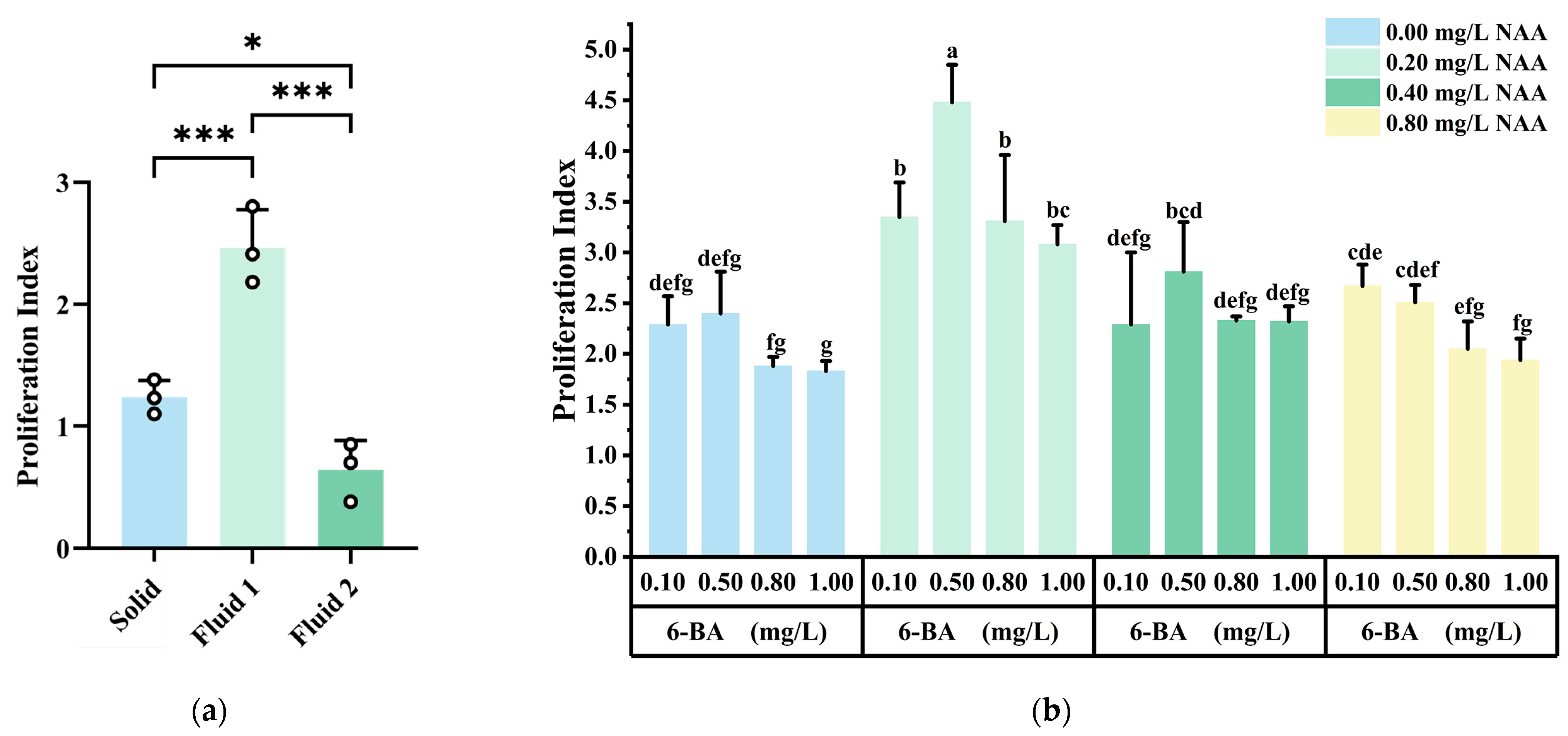
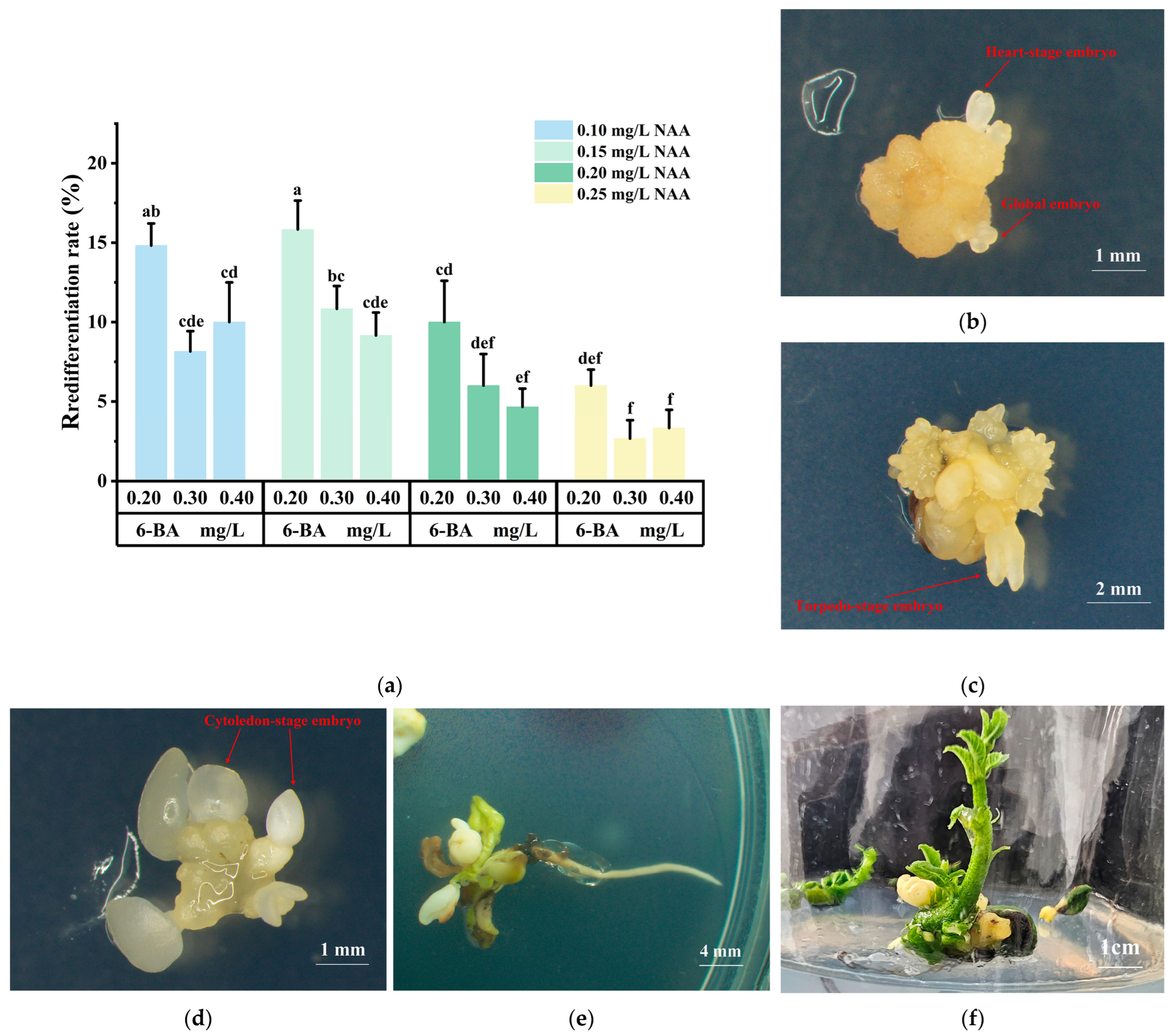
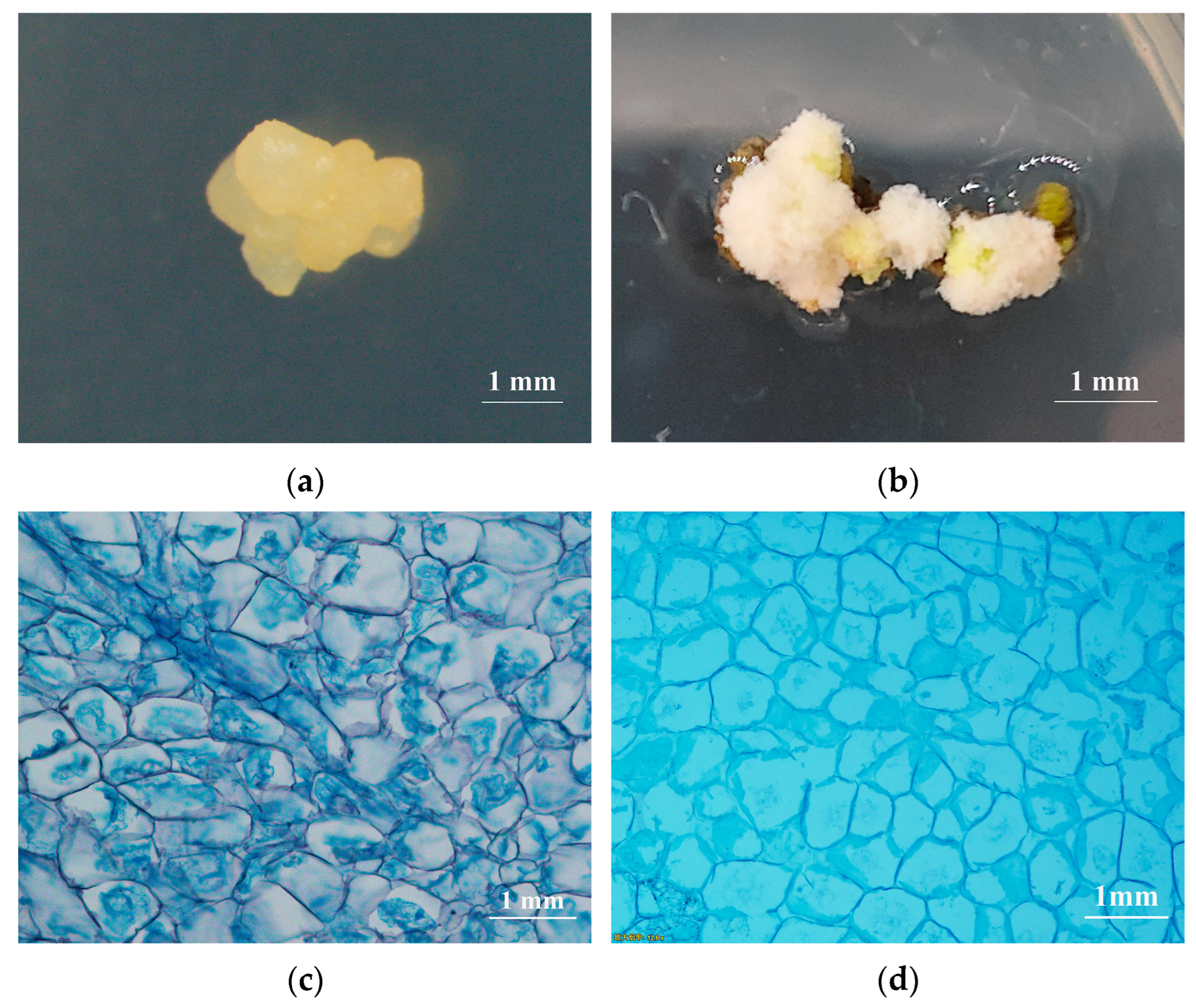

| Sampling Time | Transdifferentiation Rate (%) | Callus Growth Status |
|---|---|---|
| 10 May | 62.22 ± 4.45 a | The callus appeared yellowish-white, with a large quantity and vigorous growth. |
| 10 September | 38.52 ± 6.78 b | The explants were light purple in color, with the callus growing vigorously and showing slight browning. |
| 10 November | 27.41 ± 5.59 c | The explants were purple-black in color, with the callus mostly white and in small quantities. |
| 10 January of the following year | 7.41 ± 2.57 d | The explants exhibited severe browning, turning blackish-purple in color, and the callus formed in small quantities. |
| The Early Stage of Callus Redifferentiation | Genes | ||||
|---|---|---|---|---|---|
| Actin | QsRPII | QsTUB | QsEIF-5A | Qs(CACs)C | |
| E1 | 29.27 ± 1.40 | 31.89 ± 2.50 | 32.61 ± 0.34 | 19.74 ± 0.19 | 33.19 ± 1.28 |
| E2 | 28.43 ± 1.36 | 31.46 ± 2.84 | 32.46 ± 0.30 | 19.59 ± 0.17 | 33.52 ± 1.25 |
| Genes | Algorithm | ||
|---|---|---|---|
| GeNorm_M | NormFinder_Var | BestKeeper_SD | |
| Actin | 0.067090 | 0.030921 | 0.223572 |
| QsRPII | 0.068351 | 0.023950 | 0.270710 |
| QsTUB | 0.084357 | 0.798149 | 1.404111 |
| QsEIF-5A | 0.091297 | 0.499828 | 1.760916 |
| Qs(CACs)C | 0.101352 | 1.819655 | 2.198074 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Hou, Y.; Zhang, W.; Gong, H.; Liu, B.; Song, X.; Li, T.; Yang, Y.; Zhu, J. Optimization of Somatic Embryogenesis and Transcriptomic Analysis of the Early Stage of Callus Redifferentiation in Quercus suber L. Plants 2025, 14, 2855. https://doi.org/10.3390/plants14182855
Yu X, Hou Y, Zhang W, Gong H, Liu B, Song X, Li T, Yang Y, Zhu J. Optimization of Somatic Embryogenesis and Transcriptomic Analysis of the Early Stage of Callus Redifferentiation in Quercus suber L. Plants. 2025; 14(18):2855. https://doi.org/10.3390/plants14182855
Chicago/Turabian StyleYu, Xinran, Yaru Hou, Wan Zhang, Han Gong, Baoxuan Liu, Xiaozhou Song, Tiezhu Li, Yun Yang, and Jingle Zhu. 2025. "Optimization of Somatic Embryogenesis and Transcriptomic Analysis of the Early Stage of Callus Redifferentiation in Quercus suber L." Plants 14, no. 18: 2855. https://doi.org/10.3390/plants14182855
APA StyleYu, X., Hou, Y., Zhang, W., Gong, H., Liu, B., Song, X., Li, T., Yang, Y., & Zhu, J. (2025). Optimization of Somatic Embryogenesis and Transcriptomic Analysis of the Early Stage of Callus Redifferentiation in Quercus suber L. Plants, 14(18), 2855. https://doi.org/10.3390/plants14182855







