Advances in Functional Genomics for Exploring Abiotic Stress Tolerance Mechanisms in Cereals
Abstract
1. Introduction
| Abiotic Stress | Crop Species | Genotypes | Environmental Condition/Context | Outcome/Key Findings | Reference |
|---|---|---|---|---|---|
| Transcriptomics | |||||
| Drought and salinity | Wheat | Drought/salt tolerant JM22 and salt sensitive YM20 | Genome-wide transcriptome analysis of wheat cultivars’ response to drought (10% soil moisture) and salinity (100 mM NaCl) stresses. | 10 DEGs, annotated to cellular process, metabolic process, osmotic regulation, and MAPK signalling pathway, were co-identified as drought and salinity tolerance-associated DEGs contributing to better stress tolerance in JM22. | [45] |
| Drought | Maize | Drought-tolerant CML69 and susceptible LX9801 inbred lines | Comparative transcriptomic and physiological analyses of maize seedling leaf tissues response to 3–5 days of drought treatment. | Among other key results, the tolerant line showed significantly higher leaf RWC, and lower electrolyte leakage and MDA levels than the susceptible line, which possibly contributed to its better drought tolerance. | [73] |
| Waterlogging | Maize | Tropical line Suwan-2 and temperate line Cim-3 | Comparative physiological and transcriptomic analysis of maize inbred line tolerance to waterlogging stress. | Crucial waterlogging-responsive DEGs in Suwan-2 were related to TF modulation, cellular redox homeostasis maintenance, and plant hormone biosynthesis regulation. | [74] |
| Salinity | Rice | Salt-tolerant HH11 and salt-sensitive IR29 cultivars | Transcriptome analysis of rice response to 200 mM NaCl salt for 0 h, 6 h, 24 h and 48 h at the 3 leaf stage. | HH11 showed more favourable antioxidant and osmotic adjustments than IR29 upon salt stress exposure, thus, better salt tolerance. | [75] |
| Heat stress (HS) | Wheat | Heat-tolerant genotype WH-730 | Transcriptomic analysis of a heat-tolerant wheat genotype response to control and heat treatment conditions. | 5610 heat-responsive DEGs were identified, and participate in HS response pathways, e.g., HSPs, antioxidant defence, and metabolic adjustments. Among them, peroxidase was dominant, enhancing HS tolerance, possibly via regulation of lignin biosynthesis. | [76] |
| Cold | Rice | Cold-tolerant cultivar Huaidao5 and Cold-sensitive cultivar Huaidao9 | Differential expression and co-expression network analyses of rice panicle and flag leaf transcriptomes under reproductive-stage cold stress. | Huaidao5 showed better panicle tolerance to cold stress due to higher expression levels of cold-responsive genes in related pathways, e.g., MAPK signalling pathway, glutathione metabolism, plant hormone signal transduction, etc. | [77] |
| Cadmium | Rice | ZZ143 (low grain Cd) and YX409 (high grain Cd) | Genotypes subjected to 100 μmol/L Cd stress for 10 days. | ZZ143 showed higher root Cd tolerance than the susceptible genotype, possibly due to its greater root sulphur assimilation, and higher number of Cd-responsive DEGs and pathways, e.g., secondary metabolites biosynthesis, MAPK signalling, etc. | [78] |
| Low nitrogen | Sorghum | N-efficient (398B) and the N-inefficient (CS3541) inbred lines | Comparative phenotypic and transcriptome analysis of sorghum genotypes under low N hydroponic and field conditions | 398B exhibited superior low N tolerance than CS3541 under both field and hydroponic conditions, due to its higher photosynthetic performance and sustenance of N metabolism-related enzyme activities. | [79] |
| Proteomics | |||||
| Drought | Wheat | Tolerant BW35695 and drought-sensitive BW4074 | Physiological, biochemical, and iTRAQ leaf proteome analyses of wheat responses to drought. | Tolerant variety showed greater osmotic adjustment, antioxidant capacity, and high upregulation of protein synthesis-related proteins, contributing to better stress tolerance. | [80] |
| Drought | Maize | Drought-tolerant YE8112 and drought-sensitive MO17 | Physiological and iTRAQ leaf proteome analyses of maize responses to drought. | A total of 721 DAPs were identified. Most DAPs in YE8112 were associated with photosynthesis antenna proteins pathway, and YE8112 had better tolerance due to its activation of photosynthesis proteins related to balancing light capture and utilisation. | [81] |
| Heat | Rice | Heat-tolerant variety 9311 and sensitive variety Guangluai4 (GLA4) | Phosphoproteomic analysis of high temp (30–38 °C for 1 to 9 days)-induced changes in indica rice developing grains. | A total of 9994 phosphosites from 3216 phosphoproteins were identified in all endosperm samples. Several HS-induced consensus phosphorylation motifs were identified, and revealed a core set of HS-responsive protein kinases, splicing factors, and regulatory factors, especially those involved in starch metabolism. | [82] |
| DS and elevated temp (ET) | Barley | 7 spring barley RILs (hybrids of European and Syrian accessions) | LC-MS based proteomic analysis of barley flag leaf response to drought and ET (20/30 °C night/day). | Several protein accumulation changes under DS, ET and combined stresses were identified, including for photosynthetic apparatus-related proteins. Dehydrins were found among universally stress-responsive proteins. | [83] |
| Waterlogging | Wheat | Tolerant XM 55 and sensitive genotypes YM 158 | iTRAQ proteomic analysis of wheat responses to waterlogging stress. | Of the 7710 DAPs identified, 16 were distinct between the 2 cultivars under stress; 11 DAPs were upregulated and 5 were downregulated. 9 DAPs, including DEAD-box ATP-dependent RNA helicase 3, responded to waterlogging with non-cultivar specificity. | [47] |
| Salinity | Pearl millet (Pennisetum glaucum) | Tolerant (Tol) and sensitive (Sen) accessions | 2DE-based whole proteome analysis of pearl millet response to 150 mm NaCl treatment | 295 and 315 protein spots were identified in tolerant and sensitive accessions, respectively. Salinity tolerance of the tolerant accession was attributed to its higher upregulation of stress-responsive proteins. | [84] |
| Salinity | Wheat | Kharchia-65 salt-tolerant) and PBW-373 (salt-sensitive) | LC–MS/MS based proteomic analysis of wheat responses to 0 and 300 mM NaCl treatment for 48 h. | 21,863 proteins and 5133 protein groups were identified. There was higher upregulation of stress-responsive proteins, e.g., auxin-responsive, peroxidase, etc., in tolerant genotype and comparative downregulation in susceptible genotype. | [85] |
| Low temperature (LT) | Maize | LT-tolerant Gurez local and LT-sensitive GM6 | 2D-PAGE based proteomic analysis of maize leaf responses to low temp (6 °C) exposure for 12 h at 3-leaf stage. | 19 and 10 proteins were identified in Gurez local and GM6, respectively, including 3 novel abiotic stress- and LT-responsive proteins (e.g., nodulin-like protein) identified from Gurez local. | [86] |
| Aluminium (Al) | Barley | Al-sensitive barley cultivar ZU9 | TMT-based quantitative proteomic analysis of barley response to aluminium stress under phosphorus-Piriformospora indica interaction | DEPs were mostly enriched in the phenylpropanoid biosynthesis pathway, among which peroxidases were prominent. P. indica in combination with P helped barley plants to endure Al-induced stress by modulating antioxidative defence system. | [87] |
| Low inorganic phosphorus (Pi) | Wheat | Higher PUE genotype TM98 and a lower PUE genotype H4399 | Label-free quantitative proteomic analysis of wheat leaf response to low Pi. | 2110 high-confidence proteins were identified, among them 244 and 133 DAPs under Pi deficiency in H4399 and TM98, respectively. Abundance of energy metabolism-related proteins was decreased by Pi deficiency in H4399 shoots, but not in TM98. | [88] |
| Drought | Sorghum | Drought-sensitive S4 and S4-1, and drought-resistant T33 and T14 | nano-LC-MS/MS-based leaf proteome analysis of sorghum response to drought. | A total of 3927 proteins were quantified, with 46, 36, 35, and 102 DAPs identified in S4, S4-1, T14, and T33 varieties, respectively. Tolerant genotypes showed enhanced TCA cycle and influenced aminoacyl-tRNA biosynthesis. | [89] |
| Metabolomics | |||||
| Low temp (LT) | Rice | Varieties 02428 (japonica) and YZX (indica) | LC–MS/MS-based metabolomics analysis of rice response to LT (15 °C for 4 days) at germination. | A total of 730 metabolites were detected by LC-MS/MS method. 7 key LT-responsive metabolites were identified, and these metabolites were observed to participate in biosynthesis of amino acids and phenylpropanoids, as well as metabolism of glutathione and inositol phosphate. | [48] |
| Low nitrogen | Wheat | Zheng Mai 366 and Ai Kang58, dominant species in Henan. | UPLC-QTOF-based analysis of wheat flag leaf response to low N stress. | Chemical analyses identified 11 secondary metabolites, considered biomarkers of low N stress. Most of these secondary metabolites were flavonoids and their related derivatives, such as iso-vitexin, iso-orientin, etc. | [90] |
| Low nitrogen | Sorghum | 10 diverse entries (including inbreds and hybrids) | UPLC-MS/MS-based analysis of sorghum roots’ response to low N. | Roots from plants with N stress contained reduced phenylalanine, a precursor for salicylic acid, providing evidence for compromised metabolic capacity for defence response under low N conditions. | [91] |
| Drought | Barley | German variety Maresi and Syrian breeding line Cam/B1//CI08887/CI05761 | Untargeted GC-MS based metabolomics profiling of barley leaf and root responses to drought. | Compatible solutes and osmolytes were the major group of compounds accumulated under drought, and revealed changes in accumulation of some metabolites, e.g., proline and other amino acids, CHOs or carboxylic acids were considered a basic plant strategy for acquiring drought stress tolerance. | [92] |
| Drought | Wheat | Drought-tolerant T13 and drought-susceptible T2 | Integrated transcriptome and metabolomics analyses of wheat responses to drought. | Flavonoids and phenolic acids metabolism were associated with wheat seedlings’ drought tolerance, with their biosynthesis-related DEMs and genes possibly being key factors underlining the difference in drought tolerance. | [93] |
| Low phosphorus (LP) | Wheat | G28 (LP-tolerant) and L143 (LP-sensitive) varieties | Metabolomics and transcriptomics analysis of wheat response to 72 h of LP stress. | A total of 181 and 163 DAMs were detected in G28LP and L143LP under LP stress, respectively. Additionally, joint metabolomics and transcriptomic analysis revealed that wheat LP tolerance was closely related to 15 metabolites and 18 key genes in the sugar and amino acid metabolism pathways. | [94] |
| Salt and heat | Wheat | Warm-adapted Fahng60 and heat-sensitive Samerng2 cultivars | Physiological and metabolomics analysis of seedlings’ response to salt (150 mM NaCl) and HS (42 °C for 4 h) treatments. | Amino acids, sugars, and sugar derivatives were the major responsive metabolites in leaves under the stress. Additionally, in both genotypes, the ABC transporters, glucosinolate metabolism, aminoacyl-tRNA biosynthesis, etc., were the key overrepresented pathways under the stress combination. | [95] |
| Saline-alkaline | Rice | Saline–alkali-tolerant cultivar Tongxi926 | Integrated transcriptome and metabolomics analysis of rice response to high saline–alkali stress (pH > 9.5). | 9347 DEGs and 693 DAMs were identified. Among the DAMs, lipid and amino acid accumulation were greatly enhanced, and pathways related to ABC transporter, amino acid biosynthesis, glutathione metabolism, TCA cycle, etc., were significantly enriched. | [96] |
| Drought | Maize | Drought-tolerant line si287 and a drought-sensitive line X178 | Transcriptomic and metabolomics analysis of maize response to a 7-day drought at the 3-leaf stage | DEGs and DEMs were significantly enriched in flavonoid biosynthesis, starch and sucrose metabolism, and amino acids biosynthesis-related pathways. Joint analysis identified proline, tryptophan and phenylalanine as key stress-responsive amino acids. | [97] |
| Salinity | Barley | GN2 (salt-tolerant) and GN18 (salt-sensitive) | Proteomic and metabolomics analysis of barley response to salt stress at germination stage. | Besides the stress-responsive DAPs, a total of 187 salt-regulated metabolites were identified, which were mainly related to ABC transporters, amino acid metabolism, CHO metabolism and lipid metabolism. | [98] |
2. Overview of Abiotic Stress Tolerance Mechanisms in Cereals
3. Recent Advances in Crop Functional Genomics
3.1. Third Generation Sequencing, Long Reads and Pangenomes
3.2. Transcriptomics
3.3. Proteomics
3.4. Metabolomics
4. Genome Editing Technologies
5. Epigenomics
6. Integrating Novel Breeding Methods for Quick Trait Fixation and Optimization
7. Metagenomics
8. Challenges and Perspectives
9. Conclusions and Future Outlook
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic Strategies for Improving Crop Yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mittler, R.; Blumwald, E.; Zandalinas, S.I. Developing Climate-Resilient Crops: Improving Plant Tolerance to Stress Combination. Plant J. Cell Mol. Biol. 2022, 109, 373–389. [Google Scholar] [CrossRef]
- Dhankher, O.P.; Foyer, C.H. Climate Resilient Crops for Improving Global Food Security and Safety. Plant Cell Environ. 2018, 41, 877–884. [Google Scholar] [CrossRef]
- Mariem, S.B.; Soba, D.; Zhou, B.; Loladze, I.; Morales, F.; Aranjuelo, I. Climate Change, Crop Yields, and Grain Quality of C3 Cereals: A Meta-analysis of [CO2], Temperature, and Drought Effects. Plants 2021, 10, 1052. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global Warming, Climate Change, and Environmental Pollution: Recipe for a Multifactorial Stress Combination Disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef]
- Eckardt, N.A.; Ainsworth, E.A.; Bahuguna, R.N.; Broadley, M.R.; Busch, W.; Carpita, N.C.; Castrillo, G.; Chory, J.; DeHaan, L.R.; Duarte, C.M.; et al. Climate Change Challenges, Plant Science Solutions. Plant Cell 2022, 35, 24–66. [Google Scholar] [CrossRef]
- Xiong, W.; Reynolds, M.; Xu, Y. Climate Change Challenges Plant Breeding. Curr. Opin. Plant Biol. 2022, 70, 102308. [Google Scholar] [CrossRef] [PubMed]
- United Nations Department of Economic and Social Affairs, Population Division. World Population Prospects 2022: Summary of Results; United Nations: New York, NY, USA, 2022. [Google Scholar]
- Scheben, A.; Yuan, Y.; Edwards, D. Advances in Genomics for Adapting Crops to Climate Change. Curr. Plant Biol. 2016, 6, 2–10. [Google Scholar] [CrossRef]
- Hemathilake, D.M.K.S.; Gunathilake, D.M.C.C. Chapter 31—Agricultural Productivity and Food Supply to Meet Increased Demands. In Future Foods; Bhat, R., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 539–553. ISBN 978-0-323-91001-9. [Google Scholar]
- Umesha, S.; Manukumar, H.M.G.; Chandrasekhar, B. Chapter 3—Sustainable Agriculture and Food Security. In Biotechnology for Sustainable Agriculture; Singh, R.L., Mondal, S., Eds.; Woodhead Publishing: Cambridge, UK, 2018; pp. 67–92. ISBN 978-0-12-812160-3. [Google Scholar]
- Shahzad, A.; Ullah, S.; Dar, A.A.; Sardar, M.F.; Mehmood, T.; Tufail, M.A.; Shakoor, A.; Haris, M. Nexus on Climate Change: Agriculture and Possible Solution to Cope Future Climate Change Stresses. Environ. Sci. Pollut. Res. Int. 2021, 28, 14211–14232. [Google Scholar] [CrossRef]
- Anderson, J.; Song, B.-H. Plant Adaptation to Climate Change—Where Are We? J. Syst. Evol. 2020, 58, 533–545. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Farooq, A.; Farooq, N.; Akbar, H.; Hassan, Z.U.; Gheewala, S.H. A Critical Review of Climate Change Impact at a Global Scale on Cereal Crop Production. Agronomy 2023, 13, 162. [Google Scholar] [CrossRef]
- Fatima, Z.; Ahmed, M.; Hussain, M.; Abbas, G.; Ul-Allah, S.; Ahmad, S.; Ahmed, N.; Ali, M.A.; Sarwar, G.; Haque, E.U.; et al. The Fingerprints of Climate Warming on Cereal Crops Phenology and Adaptation Options. Sci. Rep. 2020, 10, 18013. [Google Scholar] [CrossRef] [PubMed]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic Resources in Plant Breeding for Sustainable Agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef]
- Santini, M.; Noce, S.; Antonelli, M.; Caporaso, L. Complex Drought Patterns Robustly Explain Global Yield Loss for Major Crops. Sci. Rep. 2022, 12, 5792. [Google Scholar] [CrossRef]
- Hendrawan, V.S.A.; Komori, D.; Kim, W. Possible Factors Determining Global-Scale Patterns of Crop Yield Sensitivity to Drought. PLoS ONE 2023, 18, e0281287. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Zandalinas, S.I.; Huck, C.; Fritschi, F.B.; Mittler, R. Meta-Analysis of Drought and Heat Stress Combination Impact on Crop Yield and Yield Components. Physiol. Plant. 2021, 171, 66–76. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-Morphological Traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [PubMed]
- Anzano, A.; Bonanomi, G.; Mazzoleni, S.; Lanzotti, V. Plant Metabolomics in Biotic and Abiotic Stress: A Critical Overview. Phytochem. Rev. 2022, 21, 503–524. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Pourkheirandish, M.; Golicz, A.A.; Bhalla, P.L.; Singh, M.B. Global Role of Crop Genomics in the Face of Climate Change. Front. Plant Sci. 2020, 11, 922. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing Naturally Stress-Resistant Crops for a Sustainable Agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef]
- Ndlovu, E.; van Staden, J.; Maphosa, M. Morpho-Physiological Effects of Moisture, Heat and Combined Stresses on Sorghum bicolor [Moench (L.)] and Its Acclimation Mechanisms. Plant Stress 2021, 2, 100018. [Google Scholar] [CrossRef]
- Saharan, B.S.; Brar, B.; Duhan, J.S.; Kumar, R.; Marwaha, S.; Rajput, V.D.; Minkina, T. Molecular and Physiological Mechanisms to Mitigate Abiotic Stress Conditions in Plants. Life 2022, 12, 1634. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.-Y. Engineering Drought and Salinity Tolerance Traits in Crops through CRISPR-Mediated Genome Editing: Targets, Tools, Challenges, and Perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Chang, Y.-N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.-K.; Duan, C.-G. Epigenetic Regulation in Plant Abiotic Stress Responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Jacques, C.; Salon, C.; Barnard, R.L.; Vernoud, V.; Prudent, M. Drought Stress Memory at the Plant Cycle Level: A Review. Plants 2021, 10, 1873. [Google Scholar] [CrossRef] [PubMed]
- Kole, C.; Muthamilarasan, M.; Henry, R.; Edwards, D.; Sharma, R.; Abberton, M.; Batley, J.; Bentley, A.; Blakeney, M.; Bryant, J.; et al. Application of Genomics-Assisted Breeding for Generation of Climate Resilient Crops: Progress and Prospects. Front. Plant Sci. 2015, 6, 563. [Google Scholar] [CrossRef]
- Adamski, N.M.; Borrill, P.; Brinton, J.; Harrington, S.A.; Marchal, C.; Bentley, A.R.; Bovill, W.D.; Cattivelli, L.; Cockram, J.; Contreras-Moreira, B.; et al. A Roadmap for Gene Functional Characterisation in Crops with Large Genomes: Lessons from Polyploid Wheat. eLife 2020, 9, e55646. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Dong, A.; Li, J.; Wang, Y.; Liu, X.; Wang, N.; Duan, H. Omics-Facilitated Crop Improvement for Climate Resilience and Superior Nutritive Value. Front. Plant Sci. 2021, 12, 774994. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Bansal, V.; Boucher, C. Sequencing Technologies and Analyses: Where Have We Been and Where Are We Going? iScience 2019, 18, 37–41. [Google Scholar] [CrossRef]
- Chaney, L.; Sharp, A.R.; Evans, C.R.; Udall, J.A. Genome Mapping in Plant Comparative Genomics. Trends Plant Sci. 2016, 21, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Meng, F.; Moore, B.M.; Shiu, S.-H. Impact of Short-Read Sequencing on the Misassembly of a Plant Genome. BMC Genom. 2021, 22, 99. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Michael, T.P.; VanBuren, R. Building Near-Complete Plant Genomes. Curr. Opin. Plant Biol. 2020, 54, 26–33. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J. Innovations in Plant Genetics Adapting Agriculture to Climate Change. Curr. Opin. Plant Biol. 2020, 56, 168–173. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Dong, A.; Duan, H. Advances in Cereal Crop Genomics for Resilience under Climate Change. Life 2021, 11, 502. [Google Scholar] [CrossRef]
- Dugasa, M.T.; Feng, X.; Wang, N.-H.; Wang, J.; Wu, F. Comparative Transcriptome and Tolerance Mechanism Analysis in the Two Contrasting Wheat (Triticum aestivum L.) Cultivars in Response to Drought and Salinity Stresses. Plant Growth Regul. 2021, 94, 101–114. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Wang, X.; Liu, G.; Jin, H.; Dong, A.; Yang, Y.; Duan, H. Key Maize Drought-Responsive Genes and Pathways Revealed by Comparative Transcriptome and Physiological Analyses of Contrasting Inbred Lines. Int. J. Mol. Sci. 2019, 20, 1268. [Google Scholar] [CrossRef]
- Yang, R.; Li, M.; Harrison, M.T.; Fahad, S.; Wei, M.; Li, X.; Yin, L.; Sha, A.; Zhou, M.; Liu, K. iTRAQ Proteomic Analysis of Wheat (Triticum aestivum L.) Genotypes Differing in Waterlogging Tolerance. Front. Plant Sci. 2022, 13, 890083. [Google Scholar] [CrossRef]
- Yang, M.; Yang, J.; Su, L.; Sun, K.; Li, D.; Liu, Y.; Wang, H.; Chen, Z.; Guo, T. Metabolic Profile Analysis and Identification of Key Metabolites during Rice Seed Germination under Low-Temperature Stress. Plant Sci. 2019, 289, 110282. [Google Scholar] [CrossRef]
- Pucker, B.; Irisarri, I.; de Vries, J.; Xu, B. Plant Genome Sequence Assembly in the Era of Long Reads: Progress, Challenges and Future Directions. Quant. Plant Biol. 2022, 3, e5. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shang, L.; Zhu, Q.-H.; Fan, L.; Guo, L. Twenty Years of Plant Genome Sequencing: Achievements and Challenges. Trends Plant Sci. 2022, 27, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Liu, Y.; Ma, X.; Liu, T.; Yang, X.; Wang, Z.; Liang, Q.; Liu, S.; Zhang, M.; Wang, Z.; et al. Pan-3D Genome Analysis Reveals Structural and Functional Differentiation of Soybean Genomes. Genome Biol. 2023, 24, 12. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Bohra, A.; Varshney, R.K. Pan-Genome for Pearl Millet That Beats the Heat. Trends Plant Sci. 2023, 28, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tian, Z.; Lai, J.; Huang, X. Plant Pan-Genomics and Its Applications. Mol. Plant 2023, 16, 168–186. [Google Scholar] [CrossRef]
- Yan, H.; Sun, M.; Zhang, Z.; Jin, Y.; Zhang, A.; Lin, C.; Wu, B.; He, M.; Xu, B.; Wang, J.; et al. Pangenomic Analysis Identifies Structural Variation Associated with Heat Tolerance in Pearl Millet. Nat. Genet. 2023, 55, 507–518. [Google Scholar] [CrossRef]
- Shivhare, R.; Lata, C. Exploration of Genetic and Genomic Resources for Abiotic and Biotic Stress Tolerance in Pearl Millet. Front. Plant Sci. 2017, 7, 2069. [Google Scholar] [CrossRef]
- Masters, A.; Kang, M.; McCaw, M.; Zobrist, J.D.; Gordon-Kamm, W.; Jones, T.; Wang, K. Agrobacterium-Mediated Immature Embryo Transformation of Recalcitrant Maize Inbred Lines Using Morphogenic Genes. J. Vis. Exp. JoVE 2020, e60782. [Google Scholar] [CrossRef]
- Chen, Z.; Debernardi, J.M.; Dubcovsky, J.; Gallavotti, A. Recent Advances in Crop Transformation Technologies. Nat. Plants 2022, 8, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.; Finley, T.; Chappell, H.; Veena, V.; Wang, K. An Improved Agrobacterium-Mediated Transformation and Genome-Editing Method for Maize Inbred B104 Using a Ternary Vector System and Immature Embryos. Front. Plant Sci. 2022, 13, 860971. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Wang, K. Strategies for Genotype-Flexible Plant Transformation. Curr. Opin. Biotechnol. 2023, 79, 102848. [Google Scholar] [CrossRef]
- Smith, D.T.; Potgieter, A.B.; Chapman, S.C. Scaling up High-Throughput Phenotyping for Abiotic Stress Selection in the Field. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2021, 134, 1845–1866. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D.; D’Auria, J.C.; Ferreira, A.C.S.; Gibon, Y.; Kruszka, D.; Mishra, P.; Zedde, R. van de High-Throughput Plant Phenotyping: A Role for Metabolomics? Trends Plant Sci. 2022, 27, 549–563. [Google Scholar] [CrossRef]
- Scossa, F.; Alseekh, S.; Fernie, A.R. Integrating Multi-Omics Data for Crop Improvement. J. Plant Physiol. 2021, 257, 153352. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Goldman, I.; Ceccarelli, S.; Ortiz, R. Chapter Three—Advanced Analytics, Phenomics and Biotechnology Approaches to Enhance Genetic Gains in Plant Breeding. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 162, pp. 89–142. [Google Scholar]
- Mohd Saad, N.S.; Neik, T.X.; Thomas, W.J.W.; Amas, J.C.; Cantila, A.Y.; Craig, R.J.; Edwards, D.; Batley, J. Advancing Designer Crops for Climate Resilience through an Integrated Genomics Approach. Curr. Opin. Plant Biol. 2022, 67, 102220. [Google Scholar] [CrossRef]
- Sidak, D.; Schwarzerová, J.; Weckwerth, W.; Waldherr, S. Interpretable Machine Learning Methods for Predictions in Systems Biology from Omics Data. Front. Mol. Biosci. 2022, 9, 926623. [Google Scholar] [CrossRef] [PubMed]
- Munaweera, T.I.K.; Jayawardana, N.U.; Rajaratnam, R.; Dissanayake, N. Modern Plant Biotechnology as a Strategy in Addressing Climate Change and Attaining Food Security. Agric. Food Secur. 2022, 11, 26. [Google Scholar] [CrossRef]
- Steinwand, M.A.; Ronald, P.C. Crop Biotechnology and the Future of Food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Demirer, G.S.; Silva, T.N.; Jackson, C.T.; Thomas, J.B.; Ehrhardt, D.W.; Rhee, S.Y.; Mortimer, J.C.; Landry, M.P. Nanotechnology to Advance CRISPR-Cas Genetic Engineering of Plants. Nat. Nanotechnol. 2021, 16, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Genome Engineering for Crop Improvement and Future Agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.B.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-Edited Crops for Improved Food Security of Smallholder Farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Singh, V.K.; Bohra, A.; Kumar, A.; Reif, J.C.; Varshney, R.K. Genomics and Breeding Innovations for Enhancing Genetic Gain for Climate Resilience and Nutrition Traits. TAG Theor. Appl. Genet. 2021, 134, 1829–1843. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Waititu, J.K.; Zhang, X.; Chen, T.; Zhang, C.; Zhao, Y.; Wang, H. Transcriptome Analysis of Tolerant and Susceptible Maize Genotypes Reveals Novel Insights about the Molecular Mechanisms Underlying Drought Responses in Leaves. Int. J. Mol. Sci. 2021, 22, 6980. [Google Scholar] [CrossRef]
- Yao, Q. Crucial Waterlogging-Responsive Genes and Pathways Revealed by Comparative Physiology and Transcriptome in Tropical and Temperate Maize (Zea mays L.) Inbred Lines. J. Plant Biol. 2021, 64, 313–325. [Google Scholar] [CrossRef]
- Fang, X.; Mo, J.; Zhou, H.; Shen, X.; Xie, Y.; Xu, J.; Yang, S. Comparative Transcriptome Analysis of Gene Responses of Salt-Tolerant and Salt-Sensitive Rice Cultivars to Salt Stress. Sci. Rep. 2023, 13, 19065. [Google Scholar] [CrossRef] [PubMed]
- Lamba, K.; Kumar, M.; Singh, V.; Chaudhary, L.; Gupta, V. Transcriptome Analysis for Heat Stress Related Genes in Wheat Genotype WH-730. Cereal Res. Commun. 2025, 53, 793–805. [Google Scholar] [CrossRef]
- Niu, Y.; Fan, S.; Cheng, B.; Li, H.; Wu, J.; Zhao, H.; Huang, Z.; Yan, F.; Qi, B.; Zhang, L. Comparative Transcriptomics and Co-Expression Networks Reveal Cultivar-Specific Molecular Signatures Associated with Reproductive-Stage Cold Stress in Rice. Plant Cell Rep. 2023, 42, 707–722. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Xiao, W.; Chen, D.; Hu, J.; Gao, N.; Huang, M.; Ye, X. Comparative Transcriptomic Analysis Reveals the Important Process in Two Rice Cultivars with Differences in Cadmium Accumulation. Ecotoxicol. Environ. Saf. 2023, 252, 114629. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gu, W.; Liu, C.; Shi, X.; Li, B.; Zhou, Y. Comparative Phenotypic and Transcriptomic Analysis Reveals Genotypic Differences in Nitrogen Use Efficiency in Sorghum. Plant Physiol. Biochem. 2024, 215, 109028. [Google Scholar] [CrossRef]
- Moloi, S.J.; Alqarni, A.O.; Brown, A.P.; Goche, T.; Shargie, N.G.; Moloi, M.J.; Gokul, A.; Chivasa, S.; Ngara, R. Comparative Physiological, Biochemical, and Leaf Proteome Responses of Contrasting Wheat Varieties to Drought Stress. Plants 2024, 13, 2797. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Wang, X.; Jin, H.; Liu, G.; Duan, H. Comparative Proteomic and Physiological Analyses of Two Divergent Maize Inbred Lines Provide More Insights into Drought-Stress Tolerance Mechanisms. Int. J. Mol. Sci. 2018, 19, 3225. [Google Scholar] [CrossRef]
- Pang, Y.; Hu, Y.; Bao, J. Comparative Phosphoproteomic Analysis Reveals the Response of Starch Metabolism to High-Temperature Stress in Rice Endosperm. Int. J. Mol. Sci. 2021, 22, 10546. [Google Scholar] [CrossRef]
- Mikołajczak, K.; Kuczyńska, A.; Krajewski, P.; Kempa, M.; Witaszak, N. Global Proteome Profiling Revealed the Adaptive Reprogramming of Barley Flag Leaf to Drought and Elevated Temperature. Cells 2023, 12, 1685. [Google Scholar] [CrossRef] [PubMed]
- Jha, S. Proteome Responses of Pearl Millet Genotypes under Salinity. Plant Gene 2022, 29, 100347. [Google Scholar] [CrossRef]
- Yadav, R.; Santal, A.R.; Singh, N.P. Comparative Root Proteome Analysis of Two Contrasting Wheat Genotypes Kharchia-65 (Highly Salt-Tolerant) and PBW-373 (Salt-Sensitive) for Salinity Tolerance Using LC–MS/MS Approach. Vegetos 2022, 35, 133–139. [Google Scholar] [CrossRef]
- Ramazan, S.; Jan, N.; John, R. Comparative Protein Analysis of Two Maize Genotypes with Contrasting Tolerance to Low Temperature. BMC Plant Biol. 2023, 23, 183. [Google Scholar] [CrossRef]
- Feng, Q.; Sehar, S.; Zhou, F.; Wei, D.; Askri, S.M.H.; Ma, Z.; Adil, M.F.; Shamsi, I.H. Physiological and TMT-Based Quantitative Proteomic Responses of Barley to Aluminium Stress under Phosphorus-Piriformospora Indica Interaction. Plant Physiol. Biochem. 2023, 196, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wang, R.; Zhou, P.; Pan, Y.; Shen, R.; Lan, P. Comparative Physiological and Proteomic Response to Phosphate Deficiency between Two Wheat Genotypes Differing in Phosphorus Utilization Efficiency. J. Proteom. 2023, 280, 104894. [Google Scholar] [CrossRef]
- Li, Y.; Tan, B.; Wang, D.; Mu, Y.; Li, G.; Zhang, Z.; Pan, Y.; Zhu, L. Proteomic Analysis Revealed Different Molecular Mechanisms of Response to PEG Stress in Drought-Sensitive and Drought-Resistant Sorghums. Int. J. Mol. Sci. 2022, 23, 13297. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, X.; Wang, X.; Liu, J.; Huang, B.; Guo, X.; Xiong, S.; La, G.-X. UPLC-QTOF Analysis Reveals Metabolomic Changes in the Flag Leaf of Wheat (Triticum aestivum L.) under Low-Nitrogen Stress. Plant Physiol. Biochem. 2017, 111, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Sheflin, A.M.; Chiniquy, D.; Yuan, C.; Goren, E.; Kumar, I.; Braud, M.; Brutnell, T.; Eveland, A.L.; Tringe, S.; Liu, P.; et al. Metabolomics of Sorghum Roots during Nitrogen Stress Reveals Compromised Metabolic Capacity for Salicylic Acid Biosynthesis. Plant Direct 2019, 3, e00122. [Google Scholar] [CrossRef] [PubMed]
- Swarcewicz, B.; Sawikowska, A.; Marczak, Ł.; Łuczak, M.; Ciesiołka, D.; Krystkowiak, K.; Kuczyńska, A.; Piślewska-Bednarek, M.; Krajewski, P.; Stobiecki, M. Effect of Drought Stress on Metabolite Contents in Barley Recombinant Inbred Line Population Revealed by Untargeted GC–MS Profiling. Acta Physiol. Plant. 2017, 39, 158. [Google Scholar] [CrossRef]
- Guo, X.; Lv, L.; Zhao, A.; Zhao, W.; Liu, Y.; Li, Z.; Li, H.; Chen, X. Integrated Transcriptome and Metabolome Analysis Revealed Differential Drought Stress Response Mechanisms of Wheat Seedlings with Varying Drought Tolerance. BMC Plant Biol. 2025, 25, 571. [Google Scholar] [CrossRef]
- Li, P.; Ma, X.; Wang, J.; Yao, L.; Li, B.; Meng, Y.; Si, E.; Yang, K.; Shang, X.; Zhang, X. Integrated Analysis of Metabolome and Transcriptome Reveals Insights for Low Phosphorus Tolerance in Wheat Seedling. Int. J. Mol. Sci. 2023, 24, 14840. [Google Scholar] [CrossRef]
- Shunkao, S.; Theerakulpisut, P.; Wanichthanarak, K.; Pongdontri, P.; Thitisaksakul, M. Integrative Physiological and Metabolomics Study Reveals Adaptive Strategies of Wheat Seedlings to Salt and Heat Stress Combination. Plant Growth Regul. 2023, 100, 181–196. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Wang, X.; Liu, K.; Li, Y.; Bu, Y.; Li, L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress. Int. J. Mol. Sci. 2023, 24, 4062. [Google Scholar] [CrossRef]
- Li, Y.; Su, Z.; Lin, Y.; Xu, Z.; Bao, H.; Wang, F.; Liu, J.; Hu, S.; Wang, Z.; Yu, X. Utilizing Transcriptomics and Metabolomics to Unravel Key Genes and Metabolites of Maize Seedlings in Response to Drought Stress. BMC Plant Biol. 2024, 24, 34. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Yao, L.; Li, B.; Ma, X.; Si, E.; Yang, K.; Li, C.; Shang, X.; Meng, Y. Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley. Int. J. Mol. Sci. 2022, 23, 10515. [Google Scholar] [CrossRef]
- Snowdon, R.J.; Wittkop, B.; Chen, T.-W.; Stahl, A. Crop Adaptation to Climate Change as a Consequence of Long-Term Breeding. TAG Theor. Appl. Genet. 2021, 134, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Chen, W.; Bao, G.; Sun, J.; Ding, X.; Fan, C. Physiological Response of Secale cereale L. Seedlings under Freezing-Thawing and Alkaline Salt Stress. Environ. Sci. Pollut. Res. Int. 2020, 27, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Aycan, M.; Baslam, M.; Asiloglu, R.; Mitsui, T.; Yildiz, M. Development of New High-Salt Tolerant Bread Wheat (Triticum aestivum L.) Genotypes and Insight into the Tolerance Mechanisms. Plant Physiol. Biochem. PPB 2021, 166, 314–327. [Google Scholar] [CrossRef]
- Frimpong, F.; Windt, C.W.; van Dusschoten, D.; Naz, A.A.; Frei, M.; Fiorani, F. A Wild Allele of Pyrroline-5-Carboxylate Synthase1 Leads to Proline Accumulation in Spikes and Leaves of Barley Contributing to Improved Performance Under Reduced Water Availability. Front. Plant Sci. 2021, 12, 633448. [Google Scholar] [CrossRef]
- Nefissi Ouertani, R.; Abid, G.; Karmous, C.; Ben Chikha, M.; Boudaya, O.; Mahmoudi, H.; Mejri, S.; Jansen, R.K.; Ghorbel, A. Evaluating the Contribution of Osmotic and Oxidative Stress Components on Barley Growth under Salt Stress. AoB Plants 2021, 13, plab034. [Google Scholar] [CrossRef]
- Zhang, M.-X.; Bai, R.; Nan, M.; Ren, W.; Wang, C.-M.; Shabala, S.; Zhang, J.-L. Evaluation of Salt Tolerance of Oat Cultivars and the Mechanism of Adaptation to Salinity. J. Plant Physiol. 2022, 273, 153708. [Google Scholar] [CrossRef]
- Reddy, P.S.; Dhaware, M.G.; Sivasakthi, K.; Divya, K.; Nagaraju, M.; Sri Cindhuri, K.; Kavi Kishor, P.B.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Pearl Millet Aquaporin Gene PgPIP2;6 Improves Abiotic Stress Tolerance in Transgenic Tobacco. Front. Plant Sci. 2022, 13, 820996. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.; Lv, Y.; Li, T.; Tang, J.; Yang, X.; Bai, J.; Jin, X.; Zhou, H. Effects of Drought Stress during Critical Periods on the Photosynthetic Characteristics and Production Performance of Naked Oat (Avena nuda L.). Sci. Rep. 2022, 12, 11199. [Google Scholar] [CrossRef]
- Goche, T.; Shargie, N.G.; Cummins, I.; Brown, A.P.; Chivasa, S.; Ngara, R. Comparative Physiological and Root Proteome Analyses of Two Sorghum Varieties Responding to Water Limitation. Sci. Rep. 2020, 10, 11835. [Google Scholar] [CrossRef]
- Liao, Q.; Chebotarov, D.; Islam, M.S.; Quintana, M.R.; Natividad, M.A.; De Ocampo, M.; Beredo, J.C.; Torres, R.O.; Zhang, Z.; Song, H.; et al. Aus Rice Root Architecture Variation Contributing to Grain Yield under Drought Suggests a Key Role of Nodal Root Diameter Class. Plant Cell Environ. 2022, 45, 854–870. [Google Scholar] [CrossRef]
- Ngcala, M.G.; Goche, T.; Brown, A.P.; Chivasa, S.; Ngara, R. Heat Stress Triggers Differential Protein Accumulation in the Extracellular Matrix of Sorghum Cell Suspension Cultures. Proteomes 2020, 8, 29. [Google Scholar] [CrossRef]
- Griffith, M.; Ala, P.; Yang, D.S.; Hon, W.C.; Moffatt, B.A. Antifreeze Protein Produced Endogenously in Winter Rye Leaves. Plant Physiol. 1992, 100, 593–596. [Google Scholar] [CrossRef]
- Yu, X.-M.; Griffith, M. Winter Rye Antifreeze Activity Increases in Response to Cold and Drought, but Not Abscisic Acid. Physiol. Plant. 2001, 112, 78–86. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 Signaling Module Confers Salt Tolerance through Maintaining ROS Homeostasis in Cereal Crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Punia, H.; Tokas, J.; Malik, A.; Bajguz, A.; El-Sheikh, M.A.; Ahmad, P. Ascorbate-Glutathione Oxidant Scavengers, Metabolome Analysis and Adaptation Mechanisms of Ion Exclusion in Sorghum under Salt Stress. Int. J. Mol. Sci. 2021, 22, 13249. [Google Scholar] [CrossRef]
- Tian, Q.; Shen, L.; Luan, J.; Zhou, Z.; Guo, D.; Shen, Y.; Jing, W.; Zhang, B.; Zhang, Q.; Zhang, W. Rice Shaker Potassium Channel OsAKT2 Positively Regulates Salt Tolerance and Grain Yield by Mediating K(+) Redistribution. Plant Cell Environ. 2021, 44, 2951–2965. [Google Scholar] [CrossRef]
- Alam, M.S.; Yang, Z.-K.; Li, C.; Yan, Y.; Liu, Z.; Nazir, M.M.; Xu, J.-H. Loss-of-Function Mutations of OsbHLH044 Transcription Factor Lead to Salinity Sensitivity and a Greater Chalkiness in Rice (Oryza sativa L.). Plant Physiol. Biochem. PPB 2022, 193, 110–123. [Google Scholar] [CrossRef]
- Sun, S.; Li, X.; Gao, S.; Nie, N.; Zhang, H.; Yang, Y.; He, S.; Liu, Q.; Zhai, H. A Novel WRKY Transcription Factor from Ipomoea Trifida, ItfWRKY70, Confers Drought Tolerance in Sweet Potato. Int. J. Mol. Sci. 2022, 23, 686. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Bao, L.; Huang, X.; Qian, X.; Chen, E.; Shen, B. OsABT Is Involved in Abscisic Acid Signaling Pathway and Salt Tolerance of Roots at the Rice Seedling Stage. Int. J. Mol. Sci. 2022, 23, 10656. [Google Scholar] [CrossRef]
- Siddiqui, M.N.; Léon, J.; Naz, A.A.; Ballvora, A. Genetics and Genomics of Root System Variation in Adaptation to Drought Stress in Cereal Crops. J. Exp. Bot. 2021, 72, 1007–1019. [Google Scholar] [CrossRef]
- Halder, T.; Choudhary, M.; Liu, H.; Chen, Y.; Yan, G.; Siddique, K.H.M. Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects. Proteomes 2022, 10, 17. [Google Scholar] [CrossRef]
- Ramegowda, V.; Senthil, A.; Senthil-Kumar, M. Stress Combinations and Their Interactions in Crop Plants. Plant Physiol. Rep. 2024, 29, 1–5. [Google Scholar] [CrossRef]
- Ru, C.; Hu, X.; Chen, D.; Wang, W.; Zhen, J.; Song, T. Individual and Combined Effects of Heat and Drought and Subsequent Recovery on Winter Wheat (Triticum aestivum L.) Photosynthesis, Nitrogen Metabolism, Cell Osmoregulation, and Yield Formation. Plant Physiol. Biochem. 2023, 196, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Ngara, R.; Ndimba, B.K. Understanding the Complex Nature of Salinity and Drought-Stress Response in Cereals Using Proteomics Technologies. Proteomics 2014, 14, 611–621. [Google Scholar] [CrossRef]
- Schmidt, J.; Tricker, P.J.; Eckermann, P.; Kalambettu, P.; Garcia, M.; Fleury, D. Novel Alleles for Combined Drought and Heat Stress Tolerance in Wheat. Front. Plant Sci. 2019, 10, 1800. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, X.; Wei, J.; Li, W.; Wang, H.; Xu, Y.; Yang, Z.; Xu, C.; Li, P. Primary Root Response to Combined Drought and Heat Stress Is Regulated via Salicylic Acid Metabolism in Maize. BMC Plant Biol. 2022, 22, 417. [Google Scholar] [CrossRef]
- Barua, D.; Mishra, A.; Kirti, P.B.; Barah, P. Identifying Signal-Crosstalk Mechanism in Maize Plants during Combined Salinity and Boron Stress Using Integrative Systems Biology Approaches. BioMed Res. Int. 2022, 2022, 1027288. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, Z.; Jiang, D.; Högy, P.; Fangmeier, A. Independent and Combined Effects of Elevated CO2 and Post-Anthesis Heat Stress on Protein Quantity and Quality in Spring Wheat Grains. Food Chem. 2019, 277, 524–530. [Google Scholar] [CrossRef]
- Adrees, M.; Khan, Z.S.; Rehman, M.Z.U.; Rizwan, M.; Ali, S. Foliar Spray of Silicon Nanoparticles Improved the Growth and Minimized Cadmium (Cd) in Wheat under Combined Cd and Water-Limited Stress. Environ. Sci. Pollut. Res. Int. 2022, 29, 77321–77332. [Google Scholar] [CrossRef]
- Itam, M.O.; Mega, R.; Gorafi, Y.S.A.; Yamasaki, Y.; Tahir, I.S.A.; Akashi, K.; Tsujimoto, H. Genomic Analysis for Heat and Combined Heat-Drought Resilience in Bread Wheat under Field Conditions. TAG Theor. Appl. Genet. 2022, 135, 337–350. [Google Scholar] [CrossRef]
- Da Costa, M.V.J.; Ramegowda, V.; Sreeman, S.; Nataraja, K.N. Targeted Phytohormone Profiling Identifies Potential Regulators of Spikelet Sterility in Rice under Combined Drought and Heat Stress. Int. J. Mol. Sci. 2021, 22, 11690. [Google Scholar] [CrossRef]
- Da Costa, M.V.J.; Ramegowda, V.; Ramakrishnan, P.; Nataraja, K.N.; Sheshshayee, M.S. Comparative Metabolite Profiling of Rice Contrasts Reveal Combined Drought and Heat Stress Signatures in Flag Leaf and Spikelets. Plant Sci. Int. J. Exp. Plant Biol. 2022, 320, 111262. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lim, F.-L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic Analysis of Sorghum Bicolor Responding to Combined Heat and Drought Stress. BMC Genom. 2014, 15, 456. [Google Scholar] [CrossRef]
- Mikołajczak, K.; Kuczyńska, A.; Krajewski, P.; Kempa, M.; Nuc, M. Transcriptome Profiling Disclosed the Effect of Single and Combined Drought and Heat Stress on Reprogramming of Genes Expression in Barley Flag Leaf. Front. Plant Sci. 2022, 13, 1096685. [Google Scholar] [CrossRef]
- Li, C.; Lin, F.; An, D.; Wang, W.; Huang, R. Genome Sequencing and Assembly by Long Reads in Plants. Genes 2017, 9, 6. [Google Scholar] [CrossRef]
- Sharma, T.R.; Devanna, B.N.; Kiran, K.; Singh, P.K.; Arora, K.; Jain, P.; Tiwari, I.M.; Dubey, H.; Saklani, B.; Kumari, M.; et al. Status and Prospects of Next Generation Sequencing Technologies in Crop Plants. Curr. Issues Mol. Biol. 2018, 27, 1–36. [Google Scholar] [CrossRef]
- Jung, H.; Winefield, C.; Bombarely, A.; Prentis, P.; Waterhouse, P. Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes. Trends Plant Sci. 2019, 24, 700–724. [Google Scholar] [CrossRef]
- Michael, T.P.; VanBuren, R. Progress, Challenges and the Future of Crop Genomes. Curr. Opin. Plant Biol. 2015, 24, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and Exploiting Pan-Genomics for Crop Improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef]
- Zanini, S.F.; Bayer, P.E.; Wells, R.; Snowdon, R.J.; Batley, J.; Varshney, R.K.; Nguyen, H.T.; Edwards, D.; Golicz, A.A. Pangenomics in Crop Improvement-from Coding Structural Variations to Finding Regulatory Variants with Pangenome Graphs. Plant Genome 2022, 15, e20177. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef]
- Mammadov, J.; Buyyarapu, R.; Guttikonda, S.K.; Parliament, K.; Abdurakhmonov, I.Y.; Kumpatla, S.P. Wild Relatives of Maize, Rice, Cotton, and Soybean: Treasure Troves for Tolerance to Biotic and Abiotic Stresses. Front. Plant Sci. 2018, 9, 886. [Google Scholar] [CrossRef]
- Tian, Z.; Wang, J.-W.; Li, J.; Han, B. Designing Future Crops: Challenges and Strategies for Sustainable Agriculture. Plant J. Cell Mol. Biol. 2021, 105, 1165–1178. [Google Scholar] [CrossRef]
- Bohra, A.; Tiwari, A.; Kaur, P.; Ganie, S.A.; Raza, A.; Roorkiwal, M.; Mir, R.R.; Fernie, A.R.; Smýkal, P.; Varshney, R.K. The Key to the Future Lies in the Past: Insights from Grain Legume Domestication and Improvement Should Inform Future Breeding Strategies. Plant Cell Physiol. 2022, 63, 1554–1572. [Google Scholar] [CrossRef]
- Yasir, M.; Turner, A.K.; Lott, M.; Rudder, S.; Baker, D.; Bastkowski, S.; Page, A.J.; Webber, M.A.; Charles, I.G. Long-Read Sequencing for Identification of Insertion Sites in Large Transposon Mutant Libraries. Sci. Rep. 2022, 12, 3546. [Google Scholar] [CrossRef]
- Method of the Year 2022: Long-Read Sequencing. Nat. Methods 2023, 20, 1. [CrossRef]
- Shi, J.; Ma, X.; Zhang, J.; Zhou, Y.; Liu, M.; Huang, L.; Sun, S.; Zhang, X.; Gao, X.; Zhan, W.; et al. Chromosome Conformation Capture Resolved near Complete Genome Assembly of Broomcorn Millet. Nat. Commun. 2019, 10, 464. [Google Scholar] [CrossRef]
- Depledge, D.P.; Srinivas, K.P.; Sadaoka, T.; Bready, D.; Mori, Y.; Placantonakis, D.G.; Mohr, I.; Wilson, A.C. Direct RNA Sequencing on Nanopore Arrays Redefines the Transcriptional Complexity of a Viral Pathogen. Nat. Commun. 2019, 10, 754. [Google Scholar] [CrossRef]
- Bohra, A.; Chand Jha, U.; Godwin, I.D.; Kumar Varshney, R. Genomic Interventions for Sustainable Agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent Advancements in Molecular Marker-Assisted Selection and Applications in Plant Breeding Programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd Allah, E.F. Systems Biology Approach in Plant Abiotic Stresses. Plant Physiol. Biochem. PPB 2017, 121, 58–73. [Google Scholar] [CrossRef]
- Perkel, J. SNP Genotyping: Six Technologies That Keyed a Revolution. Nat. Methods 2008, 5, 447–453. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Zhang, L.; Luo, J.; Zhao, H.; Zhang, J.; Wen, C. A New SNP Genotyping Technology Target SNP-Seq and Its Application in Genetic Analysis of Cucumber Varieties. Sci. Rep. 2020, 10, 5623. [Google Scholar] [CrossRef]
- Nepolean, T.; Kaul, J.; Mukri, G.; Mittal, S. Genomics-Enabled Next-Generation Breeding Approaches for Developing System-Specific Drought Tolerant Hybrids in Maize. Front. Plant Sci. 2018, 9, 361. [Google Scholar] [CrossRef]
- Wang, N.; Yuan, Y.; Wang, H.; Yu, D.; Liu, Y.; Zhang, A.; Gowda, M.; Nair, S.K.; Hao, Z.; Lu, Y.; et al. Applications of Genotyping-by-Sequencing (GBS) in Maize Genetics and Breeding. Sci. Rep. 2020, 10, 16308. [Google Scholar] [CrossRef]
- Du, H.; Qin, R.; Li, H.; Du, Q.; Li, X.; Yang, H.; Kong, F.; Liu, B.; Yu, D.; Wang, H. Genome-Wide Association Studies Reveal Novel Loci for Herbivore Resistance in Wild Soybean (Glycine soja). Int. J. Mol. Sci. 2022, 23, 8016. [Google Scholar] [CrossRef] [PubMed]
- Abdelraheem, A.; Thyssen, G.N.; Fang, D.D.; Jenkins, J.N.; McCarty, J.C.; Wedegaertner, T.; Zhang, J. GWAS Reveals Consistent QTL for Drought and Salt Tolerance in a MAGIC Population of 550 Lines Derived from Intermating of 11 Upland Cotton (Gossypium hirsutum) Parents. Mol. Genet. Genom. MGG 2021, 296, 119–129. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Beynon, S.; Srivastava, R.K.; Sehgal, D.; Ojulong, H.; Yadav, R. Genome-Wide Association Mapping and Comparative Genomics Identifies Genomic Regions Governing Grain Nutritional Traits in Finger Millet (Eleusine coracana L. Gaertn.). Plants People Planet 2020, 2, 649–662. [Google Scholar] [CrossRef]
- Li, L.; Mao, X.; Wang, J.; Chang, X.; Reynolds, M.; Jing, R. Genetic Dissection of Drought and Heat-Responsive Agronomic Traits in Wheat. Plant Cell Environ. 2019, 42, 2540–2553. [Google Scholar] [CrossRef]
- Yang, L.; Lei, L.; Liu, H.; Wang, J.; Zheng, H.; Zou, D. Whole-Genome Mining of Abiotic Stress Gene Loci in Rice. Planta 2020, 252, 85. [Google Scholar] [CrossRef]
- Raza, A.; Mubarik, M.S.; Sharif, R.; Habib, M.; Jabeen, W.; Zhang, C.; Chen, H.; Chen, Z.-H.; Siddique, K.H.M.; Zhuang, W.; et al. Developing Drought-Smart, Ready-to-Grow Future Crops. Plant Genome 2022, 16, e20279. [Google Scholar] [CrossRef]
- Lu, X.; Liu, J.; Ren, W.; Yang, Q.; Chai, Z.; Chen, R.; Wang, L.; Zhao, J.; Lang, Z.; Wang, H.; et al. Gene-Indexed Mutations in Maize. Mol. Plant 2018, 11, 496–504. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of Differentially Expressed Genes between Sorghum Genotypes with Contrasting Nitrogen Stress Tolerance by Genome-Wide Transcriptional Profiling. BMC Genom. 2014, 15, 179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Kong, X.; Xu, X.; Li, C.; Tian, H.; Ding, Z. Comparative Transcriptome Profiling of the Maize Primary, Crown and Seminal Root in Response to Salinity Stress. PLoS ONE 2015, 10, e0121222. [Google Scholar] [CrossRef]
- He, X.; Ma, H.; Zhao, X.; Nie, S.; Li, Y.; Zhang, Z.; Shen, Y.; Chen, Q.; Lu, Y.; Lan, H.; et al. Comparative RNA-Seq Analysis Reveals That Regulatory Network of Maize Root Development Controls the Expression of Genes in Response to N Stress. PLoS ONE 2016, 11, e0151697. [Google Scholar] [CrossRef]
- Tian, T.; Chen, L.; Ai, Y.; He, H. Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq. Genes 2022, 13, 224. [Google Scholar] [CrossRef] [PubMed]
- Poonia, A.K.; Mishra, S.K.; Sirohi, P.; Chaudhary, R.; Kanwar, M.; Germain, H.; Chauhan, H. Overexpression of Wheat Transcription Factor (TaHsfA6b) Provides Thermotolerance in Barley. Planta 2020, 252, 53. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zenda, T.; Dong, A.; Yang, Y.; Wang, N.; Duan, H. Global Transcriptome and Weighted Gene Co-Expression Network Analyses of Growth-Stage-Specific Drought Stress Responses in Maize. Front. Genet. 2021, 12, 645443. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Zhang, W.; Sun, L.; Zhao, A.; Zhang, Y.; Wang, L.; Liu, Y.; Li, Z.; Li, H.; Chen, X. Gene Co-Expression Network Analysis to Identify Critical Modules and Candidate Genes of Drought-Resistance in Wheat. PLoS ONE 2020, 15, e0236186. [Google Scholar] [CrossRef]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-Wide Association Analysis of Seedling Traits in Diverse Sorghum Germplasm under Thermal Stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Siddique, K.H.M.; Farooq, M.; Thornton, P.K.; Ortiz, R. Using Biotechnology-Led Approaches to Uplift Cereal and Food Legume Yields in Dryland Environments. Front. Plant Sci. 2018, 9, 1249. [Google Scholar] [CrossRef]
- Lin, F.; Zhang, Y.; Jiang, M.-Y. Alternative Splicing and Differential Expression of Two Transcripts of Nicotine Adenine Dinucleotide Phosphate Oxidase B Gene from Zea mays. J. Integr. Plant Biol. 2009, 51, 287–298. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Mastrangelo, A.M.; Marone, D.; Laidò, G.; De Leonardis, A.M.; De Vita, P. Alternative Splicing: Enhancing Ability to Cope with Stress via Transcriptome Plasticity. Plant Sci. Int. J. Exp. Plant Biol. 2012, 185–186, 40–49. [Google Scholar] [CrossRef]
- Mirdar Mansuri, R.; Azizi, A.-H.; Sadri, A.-H.; Shobbar, Z.-S. Long Non-Coding RNAs as the Regulatory Hubs in Rice Response to Salt Stress. Sci. Rep. 2022, 12, 21696. [Google Scholar] [CrossRef]
- De Quattro, C.; Pè, M.E.; Bertolini, E. Long Noncoding RNAs in the Model Species Brachypodium distachyon. Sci. Rep. 2017, 7, 11252. [Google Scholar] [CrossRef]
- Zhang, W.; Han, Z.; Guo, Q.; Liu, Y.; Zheng, Y.; Wu, F.; Jin, W. Identification of Maize Long Non-Coding RNAs Responsive to Drought Stress. PLoS ONE 2014, 9, e98958. [Google Scholar] [CrossRef] [PubMed]
- Huanca-Mamani, W.; Arias-Carrasco, R.; Cárdenas-Ninasivincha, S.; Rojas-Herrera, M.; Sepúlveda-Hermosilla, G.; Caris-Maldonado, J.C.; Bastías, E.; Maracaja-Coutinho, V. Long Non-Coding RNAs Responsive to Salt and Boron Stress in the Hyper-Arid Lluteño Maize from Atacama Desert. Genes 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Zhang, X.; Ma, X.; Zhao, J. Spatio-Temporal Transcriptional Dynamics of Maize Long Non-Coding RNAs Responsive to Drought Stress. Genes 2019, 10, 138. [Google Scholar] [CrossRef]
- Hu, X.; Wei, Q.; Wu, H.; Huang, Y.; Peng, X.; Han, G.; Ma, Q.; Zhao, Y. Identification and Characterization of Heat-Responsive lncRNAs in Maize Inbred Line CM1. BMC Genom. 2022, 23, 208. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, Y.; Zou, C.; Yang, C.; Pan, G.; Ma, L.; Shen, Y. Integrated Analysis of Long Non-Coding RNAs and mRNAs Reveals the Regulatory Network of Maize Seedling Root Responding to Salt Stress. BMC Genom. 2022, 23, 50. [Google Scholar] [CrossRef]
- Xuhui, L.; Weiwei, C.; Siqi, L.; Junteng, F.; Hang, Z.; Xiangbo, Z.; Yongwen, Q. Full-Length Transcriptome Analysis of Maize Root Tips Reveals the Molecular Mechanism of Cold Stress during the Seedling Stage. BMC Plant Biol. 2022, 22, 398. [Google Scholar] [CrossRef] [PubMed]
- De Quattro, C.; Mica, E.; Pè, M.E.; Bertolini, E. Brachypodium distachyon Long Noncoding RNAs: Genome-Wide Identification and Expression Analysis. Methods Mol. Biol. 2018, 1667, 31–42. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High Resolution Two-Dimensional Electrophoresis of Proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Paša-Tolić, L. High-Throughput Proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef]
- Andjelković, U.; Josić, D. Mass Spectrometry Based Proteomics as Foodomics Tool in Research and Assurance of Food Quality and Safety. Trends Food Sci. Technol. 2018, 77, 100–119. [Google Scholar] [CrossRef]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Ali, A.E.; Husselmann, L.H.; Tabb, D.L.; Ludidi, N. Comparative Proteomics Analysis between Maize and Sorghum Uncovers Important Proteins and Metabolic Pathways Mediating Drought Tolerance. Life 2023, 13, 170. [Google Scholar] [CrossRef]
- López-Cristoffanini, C.; Bundó, M.; Serrat, X.; San Segundo, B.; López-Carbonell, M.; Nogués, S. A Comprehensive Study of the Proteins Involved in Salinity Stress Response in Roots and Shoots of the FL478 Genotype of Rice (Oryza sativa L. ssp. Indica). Crop J. 2021, 9, 1154–1168. [Google Scholar] [CrossRef]
- Yadav, R.; Chakraborty, S.; Ramakrishna, W. Wheat Grain Proteomic and Protein–Metabolite Interactions Analyses Provide Insights into Plant Growth Promoting Bacteria–Arbuscular Mycorrhizal Fungi–Wheat Interactions. Plant Cell Rep. 2022, 41, 1417–1437. [Google Scholar] [CrossRef] [PubMed]
- Widodo; Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic Responses to Salt Stress of Barley (Hordeum vulgare L.) Cultivars, Sahara and Clipper, Which Differ in Salinity Tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Paul, S.; Gayen, D.; Datta, S.K.; Datta, K. Dissecting Root Proteome of Transgenic Rice Cultivars Unravels Metabolic Alterations and Accumulation of Novel Stress Responsive Proteins under Drought Stress. Plant Sci. 2015, 234, 133–143. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, Y.; He, Q.; Li, H.; Zhang, X.; Zhang, F. Comparative Proteomics Illustrates the Complexity of Drought Resistance Mechanisms in Two Wheat (Triticum aestivum L.) Cultivars under Dehydration and Rehydration. BMC Plant Biol. 2016, 16, 188. [Google Scholar] [CrossRef]
- Yousuf, P.Y.; Abd Allah, E.F.; Nauman, M.; Asif, A.; Hashem, A.; Alqarawi, A.A.; Ahmad, A. Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen. Genes 2017, 8, 356. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Prášil, I.T.; Klíma, M.; Nesvadba, Z.; Vítámvás, P.; Ovesná, J. Proteomics of Wheat and Barley Cereals in Response to Environmental Stresses: Current State and Future Challenges. J. Proteomics 2023, 282, 104923. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The Link between Genotypes and Phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Carrera, F.P.; Noceda, C.; Maridueña-Zavala, M.G.; Cevallos-Cevallos, J.M. Metabolomics, a Powerful Tool for Understanding Plant Abiotic Stress. Agronomy 2021, 11, 824. [Google Scholar] [CrossRef]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted Large-Scale Plant Metabolomics Using Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Ramalingam, A.P.; Mohanavel, W.; Premnath, A.; Muthurajan, R.; Prasad, P.V.V.; Perumal, R. Large-Scale Non-Targeted Metabolomics Reveals Antioxidant, Nutraceutical and Therapeutic Potentials of Sorghum. Antioxidants 2021, 10, 1511. [Google Scholar] [CrossRef]
- Alseekh, S.; Bermudez, L.; de Haro, L.A.; Fernie, A.R.; Carrari, F. Crop Metabolomics: From Diagnostics to Assisted Breeding. Metabolomics 2018, 14, 148. [Google Scholar] [CrossRef]
- Jendoubi, T. Approaches to Integrating Metabolomics and Multi-Omics Data: A Primer. Metabolites 2021, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, J.; Sade, N.; Wu, S.; Egbaria, A.; Fernie, A.R.; Yan, J.; Qin, F.; Chen, W.; Brotman, Y.; et al. Genomic Basis Underlying the Metabolome-Mediated Drought Adaptation of Maize. Genome Biol. 2021, 22, 260. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Radani, Y.; Ke, Y.; Chen, J.; Yang, L. CRISPR/Cas Genome Editing in Plants: Mechanisms, Applications, and Overcoming Bottlenecks. Funct. Integr. Genom. 2024, 24, 50. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Pandey, S.; Divakar, S.; Singh, A. Genome Editing Prospects for Heat Stress Tolerance in Cereal Crops. Plant Physiol. Biochem. 2024, 215, 108989. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, Z.; Li, J.; Wang, S.; Chen, Y.; Liu, Y.; Mao, D.; Luan, S.; Chen, L. bHLH57 Confers Chilling Tolerance and Grain Yield Improvement in Rice. Plant Cell Environ. 2023, 46, 1402–1418. [Google Scholar] [CrossRef]
- Nazir, R.; Mandal, S.; Mitra, S.; Ghorai, M.; Das, N.; Jha, N.K.; Majumder, M.; Pandey, D.K.; Dey, A. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-Associated Genome-Editing Toolkit to Enhance Salt Stress Tolerance in Rice and Wheat. Physiol. Plant. 2022, 174, e13642. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome Editing with Engineered Zinc Finger Nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome Engineering with Zinc-Finger Nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Wyman, C.; Kanaar, R. DNA Double-Strand Break Repair: All’s Well That Ends Well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Bin Moon, S.; Lee, J.M.; Kang, J.G.; Lee, N.-E.; Ha, D.-I.; Kim, D.Y.; Kim, S.H.; Yoo, K.; Kim, D.; Ko, J.-H.; et al. Highly Efficient Genome Editing by CRISPR-Cpf1 Using CRISPR RNA with a Uridinylate-Rich 3′-Overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef]
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient Targeted Mutagenesis of Rice and Tobacco Genomes Using Cpf1 from Francisella Novicida. Sci. Rep. 2016, 6, 38169. [Google Scholar] [CrossRef]
- Wang, M.; Mao, Y.; Lu, Y.; Tao, X.; Zhu, J.-K. Multiplex Gene Editing in Rice Using the CRISPR-Cpf1 System. Mol. Plant 2017, 10, 1011–1013. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Qin, R.; Li, H.; Li, D.; Li, L.; Wei, P.; Yang, J. Generation of Targeted Mutant Rice Using a CRISPR-Cpf1 System. Plant Biotechnol. J. 2017, 15, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 Mediated Targeting of a Stomatal Developmental Gene EPFL9 in Rice. Plant Cell Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K.; et al. Activities and Specificities of CRISPR/Cas9 and Cas12a Nucleases for Targeted Mutagenesis in Maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef]
- Singh, D.; Chaudhary, P.; Taunk, J.; Kumar Singh, C.; Sharma, S.; Singh, V.J.; Singh, D.; Chinnusamy, V.; Yadav, R.; Pal, M. Plant Epigenomics for Extenuation of Abiotic Stresses: Challenges and Future Perspectives. J. Exp. Bot. 2021, 72, 6836–6855. [Google Scholar] [CrossRef]
- Lloyd, J.P.B.; Lister, R. Epigenome Plasticity in Plants. Nat. Rev. Genet. 2022, 23, 55–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Andrews, H.; Eglitis-Sexton, J.; Godwin, I.; Tanurdžić, M.; Crisp, P.A. Epigenome Guided Crop Improvement: Current Progress and Future Opportunities. Emerg. Top. Life Sci. 2022, 6, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Perrone, A.; Martinelli, F. Plant Stress Biology in Epigenomic Era. Plant Sci. Int. J. Exp. Plant Biol. 2020, 294, 110376. [Google Scholar] [CrossRef]
- Kakoulidou, I.; Avramidou, E.V.; Baránek, M.; Brunel-Muguet, S.; Farrona, S.; Johannes, F.; Kaiserli, E.; Lieberman-Lazarovich, M.; Martinelli, F.; Mladenov, V.; et al. Epigenetics for Crop Improvement in Times of Global Change. Biology 2021, 10, 766. [Google Scholar] [CrossRef]
- Samantara, K.; Shiv, A.; de Sousa, L.L.; Sandhu, K.S.; Priyadarshini, P.; Mohapatra, S.R. A Comprehensive Review on Epigenetic Mechanisms and Application of Epigenetic Modifications for Crop Improvement. Environ. Exp. Bot. 2021, 188, 104479. [Google Scholar] [CrossRef]
- Crisp, P.A.; Bhatnagar-Mathur, P.; Hundleby, P.; Godwin, I.D.; Waterhouse, P.M.; Hickey, L.T. Beyond the Gene: Epigenetic and Cis-Regulatory Targets Offer New Breeding Potential for the Future. Curr. Opin. Biotechnol. 2022, 73, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Kim, K.D.; Cho, J. Harnessing Epigenetic Variability for Crop Improvement: Current Status and Future Prospects. Genes Genom. 2022, 44, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Villagómez-Aranda, A.L.; Feregrino-Pérez, A.A.; García-Ortega, L.F.; González-Chavira, M.M.; Torres-Pacheco, I.; Guevara-González, R.G. Activating Stress Memory: Eustressors as Potential Tools for Plant Breeding. Plant Cell Rep. 2022, 41, 1481–1498. [Google Scholar] [CrossRef]
- Gahlaut, V.; Zinta, G.; Jaiswal, V.; Kumar, S. Quantitative Epigenetics: A New Avenue for Crop Improvement. Epigenomes 2020, 4, 25. [Google Scholar] [CrossRef]
- Agius, D.R.; Kapazoglou, A.; Avramidou, E.; Baranek, M.; Carneros, E.; Caro, E.; Castiglione, S.; Cicatelli, A.; Radanovic, A.; Ebejer, J.-P.; et al. Exploring the Crop Epigenome: A Comparison of DNA Methylation Profiling Techniques. Front. Plant Sci. 2023, 14, 1181039. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-Coding RNAs and Their Roles in Stress Response in Plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and Their Roles in Plant Development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Wu, L.; Liu, S.; Qi, H.; Cai, H.; Xu, M. Research Progress on Plant Long Non-Coding RNA. Plants 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Zenda, T.; Wang, N.; Dong, A.; Zhou, Y.; Duan, H. Reproductive-Stage Heat Stress in Cereals: Impact, Plant Responses and Strategies for Tolerance Improvement. Int. J. Mol. Sci. 2022, 23, 6929. [Google Scholar] [CrossRef]
- Balyan, S.; Kumar, M.; Mutum, R.D.; Raghuvanshi, U.; Agarwal, P.; Mathur, S.; Raghuvanshi, S. Identification of miRNA-Mediated Drought Responsive Multi-Tiered Regulatory Network in Drought Tolerant Rice, Nagina 22. Sci. Rep. 2017, 7, 15446. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Xia, Y.; Zhang, H.; Li, R.; Bai, G.; Siddique, K.H.M.; Guo, P. Identification of Conserved and Novel miRNAs Responsive to Heat Stress in Flowering Chinese Cabbage Using High-Throughput Sequencing. Sci. Rep. 2019, 9, 14922. [Google Scholar] [CrossRef]
- Lu, Q.; Xu, Q.; Guo, F.; Lv, Y.; Song, C.; Feng, M.; Yu, J.; Zhang, D.; Cang, J. Identification and Characterization of Long Non-Coding RNAs as Competing Endogenous RNAs in the Cold Stress Response of Triticum aestivum. Plant Biol. 2020, 22, 635–645. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, Q.; Chen, T.; Xu, L.; Liu, J.; Zhang, X.; Han, G.; Ma, Q. Identification and Characterization of Heat-Responsive miRNAs and Their Regulatory Network in Maize. Plant Growth Regul. 2022, 96, 195–208. [Google Scholar] [CrossRef]
- Xu, J.; Hou, Q.-M.; Khare, T.; Verma, S.K.; Kumar, V. Exploring miRNAs for Developing Climate-Resilient Crops: A Perspective Review. Sci. Total Environ. 2019, 653, 91–104. [Google Scholar] [CrossRef]
- Tirnaz, S.; Batley, J. Epigenetics: Potentials and Challenges in Crop Breeding. Mol. Plant 2019, 12, 1309–1311. [Google Scholar] [CrossRef]
- Sun, C.; Ali, K.; Yan, K.; Fiaz, S.; Dormatey, R.; Bi, Z.; Bai, J. Exploration of Epigenetics for Improvement of Drought and Other Stress Resistance in Crops: A Review. Plants 2021, 10, 1226. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Saze, H. Epigenetic Inheritance and Plant Evolution. Popul. Ecol. 2020, 62, 17–27. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Akram, N.A.; Ashraf, M.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Plant Responses to Environmental Stresses-from Gene to Biotechnology. AoB Plants 2017, 9, plx025. [Google Scholar] [CrossRef]
- Choudhary, M.; Wani, S.H.; Kumar, P.; Bagaria, P.K.; Rakshit, S.; Roorkiwal, M.; Varshney, R.K. QTLian Breeding for Climate Resilience in Cereals: Progress and Prospects. Funct. Integr. Genom. 2019, 19, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Husaini, A.M. High-Value Pleiotropic Genes for Developing Multiple Stress-Tolerant Biofortified Crops for 21st-Century Challenges. Heredity 2022, 128, 460–472. [Google Scholar] [CrossRef]
- Li, Z.; Zhong, F.; Guo, J.; Chen, Z.; Song, J.; Zhang, Y. Improving Wheat Salt Tolerance for Saline Agriculture. J. Agric. Food Chem. 2022, 70, 14989–15006. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.; Edwards, D.; et al. Rapid Delivery Systems for Future Food Security. Nat. Biotechnol. 2021, 39, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing Genetic Gain through Genomic Selection: From Livestock to Plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef]
- He, T.; Li, C. Harness the Power of Genomic Selection and the Potential of Germplasm in Crop Breeding for Global Food Security in the Era with Rapid Climate Change. Crop J. 2020, 8, 688–700. [Google Scholar] [CrossRef]
- Crain, J.; DeHaan, L.; Poland, J. Genomic Prediction Enables Rapid Selection of High-Performing Genets in an Intermediate Wheatgrass Breeding Program. Plant Genome 2021, 14, e20080. [Google Scholar] [CrossRef]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.A.; Yu, D.; Bohra, A.; Ganie, S.A.; Varshney, R.K. Features and Applications of Haplotypes in Crop Breeding. Commun. Biol. 2021, 4, 1266. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-Forward Breeding for a Food-Secure World. Trends Genet. TIG 2021, 37, 1124–1136. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart Breeding Driven by Big Data, Artificial Intelligence, and Integrated Genomic-Enviromic Prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating Crop Genetic Gains with Genomic Selection. TAG Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D.C.; et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, R. Molecular Markers and Selection for Complex Traits in Plants: Learning from the Last 20 Years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic Selection Methods for Crop Improvement: Current Status and Prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Qian, L.; Hickey, L.T.; Stahl, A.; Werner, C.R.; Hayes, B.; Snowdon, R.J.; Voss-Fels, K.P. Exploring and Harnessing Haplotype Diversity to Improve Yield Stability in Crops. Front. Plant Sci. 2017, 8, 1534. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.J.W.; Zhang, Y.; Amas, J.C.; Cantila, A.Y.; Zandberg, J.D.; Harvie, S.L.; Batley, J. Innovative Advances in Plant Genotyping. In Plant Genotyping: Methods and Protocols; Shavrukov, Y., Ed.; Methods in Molecular Biology; Springer US: New York, NY, USA, 2023; pp. 451–465. ISBN 978-1-07-163024-2. [Google Scholar]
- Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S.; et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology 2022, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Samineni, S.; Sen, M.; Sajja, S.B.; Gaur, P.M. Rapid Generation Advance (RGA) in Chickpea to Produce up to Seven Generations per Year and Enable Speed Breeding. Crop J. 2020, 8, 164–169. [Google Scholar] [CrossRef]
- Swami, P.; Deswal, K.; Rana, V.; Yadav, D.; Munjal, R. Chapter 3—Speed Breeding—A Powerful Tool to Breed More Crops in Less Time Accelerating Crop Research. In Abiotic Stresses in Wheat; Khan, M.K., Pandey, A., Hamurcu, M., Gupta, O.P., Gezgin, S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 33–49. ISBN 978-0-323-95368-9. [Google Scholar]
- Sinha, P.; Singh, V.K.; Saxena, R.K.; Khan, A.W.; Abbai, R.; Chitikineni, A.; Desai, A.; Molla, J.; Upadhyaya, H.D.; Kumar, A.; et al. Superior Haplotypes for Haplotype-Based Breeding for Drought Tolerance in Pigeonpea (Cajanus cajan L.). Plant Biotechnol. J. 2020, 18, 2482–2490. [Google Scholar] [CrossRef]
- Brinton, J.; Ramirez-Gonzalez, R.H.; Simmonds, J.; Wingen, L.; Orford, S.; Griffiths, S.; Haberer, G.; Spannagl, M.; Walkowiak, S.; Pozniak, C.; et al. A Haplotype-Led Approach to Increase the Precision of Wheat Breeding. Commun. Biol. 2020, 3, 712. [Google Scholar] [CrossRef]
- Crossa, J.; Fritsche-Neto, R.; Montesinos-Lopez, O.A.; Costa-Neto, G.; Dreisigacker, S.; Montesinos-Lopez, A.; Bentley, A.R. The Modern Plant Breeding Triangle: Optimizing the Use of Genomics, Phenomics, and Enviromics Data. Front. Plant Sci. 2021, 12, 651480. [Google Scholar] [CrossRef]
- Friesen, M.L.; Porter, S.S.; Stark, S.C.; von Wettberg, E.J.; Sachs, J.L.; Martinez-Romero, E. Microbially Mediated Plant Functional Traits. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 23–46. [Google Scholar] [CrossRef]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Konwar, N.; Singh, K.N.; Narzary, D. Plant–Microbe Interactions in Combating Abiotic Stresses. In Plant Stress: Challenges and Management in the New Decade; Roy, S., Mathur, P., Chakraborty, A.P., Saha, S.P., Eds.; Advances in Science, Technology & Innovation; Springer International Publishing: Cham, Switzerland, 2022; pp. 217–234. ISBN 978-3-030-95365-2. [Google Scholar]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.C.; Bottini, R.; Pontin, M.; Berli, F.J.; Moreno, D.; Boccanlandro, H.; Travaglia, C.N.; Piccoli, P.N. Azospirillum Brasilense Ameliorates the Response of Arabidopsis Thaliana to Drought Mainly via Enhancement of ABA Levels. Physiol. Plant. 2015, 153, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Prasad, V.; Chauhan, P.S.; Lata, C. Bacillus Amyloliquefaciens Confers Tolerance to Various Abiotic Stresses and Modulates Plant Response to Phytohormones through Osmoprotection and Gene Expression Regulation in Rice. Front. Plant Sci. 2017, 8, 1510. [Google Scholar] [CrossRef]
- Ahmed, V.; Verma, M.K.; Gupta, S.; Mandhan, V.; Chauhan, N.S. Metagenomic Profiling of Soil Microbes to Mine Salt Stress Tolerance Genes. Front. Microbiol. 2018, 9, 159. [Google Scholar] [CrossRef]
- Kamran, M.; Imran, Q.M.; Ahmed, M.B.; Falak, N.; Khatoon, A.; Yun, B.-W. Endophyte-Mediated Stress Tolerance in Plants: A Sustainable Strategy to Enhance Resilience and Assist Crop Improvement. Cells 2022, 11, 3292. [Google Scholar] [CrossRef]
- Santos, L.F.; Olivares, F.L. Plant Microbiome Structure and Benefits for Sustainable Agriculture. Curr. Plant Biol. 2021, 26, 100198. [Google Scholar] [CrossRef]
- Omae, N.; Tsuda, K. Plant-Microbiota Interactions in Abiotic Stress Environments. Mol. Plant-Microbe Interact. MPMI 2022, 35, 511–526. [Google Scholar] [CrossRef]
- Chouhan, G.K.; Verma, J.P.; Jaiswal, D.K.; Mukherjee, A.; Singh, S.; de Araujo Pereira, A.P.; Liu, H.; Abd Allah, E.F.; Singh, B.K. Phytomicrobiome for Promoting Sustainable Agriculture and Food Security: Opportunities, Challenges, and Solutions. Microbiol. Res. 2021, 248, 126763. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P.; Egidi, E.; Macdonald, C.A.; Delgado-Baquerizo, M. Crop Microbiome and Sustainable Agriculture. Nat. Rev. Microbiol. 2020, 18, 601–602. [Google Scholar] [CrossRef]
- Mahmud, A.A.; Upadhyay, S.K.; Srivastava, A.K.; Bhojiya, A.A. Biofertilizers: A Nexus between Soil Fertility and Crop Productivity under Abiotic Stress. Curr. Res. Environ. Sustain. 2021, 3, 100063. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Babalola, O.O. Metagenomics Methods for the Study of Plant-Associated Microbial Communities: A Review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A Practical Guide to Amplicon and Metagenomic Analysis of Microbiome Data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-J.; Sang, M.K.; Pathiraja, D.; Park, B.; Choi, I.-G.; Kim, K.D. Draft Genome Sequence of Phosphate-Solubilizing Chryseobacterium Sp. Strain ISE14, a Biocontrol and Plant Growth-Promoting Rhizobacterium Isolated from Cucumber. Genome Announc. 2018, 6, e00612-18. [Google Scholar] [CrossRef] [PubMed]
- Ormeño-Orrillo, E.; Aguilar-Cuba, Y.; Zúñiga-Dávila, D. Draft Genome Sequence of Rhizobium Sophoriradicis H4, a Nitrogen-Fixing Bacterium Associated with the Leguminous Plant Phaseolus Vulgaris on the Coast of Peru. Genome Announc. 2018, 6, e00241-18. [Google Scholar] [CrossRef] [PubMed]
- Raza, A. Metabolomics: A Systems Biology Approach for Enhancing Heat Stress Tolerance in Plants. Plant Cell Rep. 2022, 41, 741–763. [Google Scholar] [CrossRef] [PubMed]
- Alternate Routes to Gene Functions. Nat. Plants 2024, 10, 1605–1606. [CrossRef] [PubMed]
- Nguyen, Q.-H.; Nguyen, H.; Oh, E.C.; Nguyen, T. Current Approaches and Outstanding Challenges of Functional Annotation of Metabolites: A Comprehensive Review. Brief. Bioinform. 2024, 25, bbae498. [Google Scholar] [CrossRef]
- Fan, B.-L.; Chen, L.-H.; Chen, L.-L.; Guo, H. Integrative Multi-Omics Approaches for Identifying and Characterizing Biological Elements in Crop Traits: Current Progress and Future Prospects. Int. J. Mol. Sci. 2025, 26, 1466. [Google Scholar] [CrossRef]
- Rane, J.; Singh, A.K.; Kumar, M.; Boraiah, K.M.; Meena, K.K.; Pradhan, A.; Prasad, P.V.V. The Adaptation and Tolerance of Major Cereals and Legumes to Important Abiotic Stresses. Int. J. Mol. Sci. 2021, 22, 12970. [Google Scholar] [CrossRef]
- Zenda, T.; Wang, N.; Yan, X.; Dong, A.; Yang, Q.; Zhong, Y.; Duan, H. Opportunities and Avenues for Achieving Crop Climate Resilience. Environ. Exp. Bot. 2023, 213, 105414. [Google Scholar] [CrossRef]
- Chen, V.; Yang, M.; Cui, W.; Kim, J.S.; Talwalkar, A.; Ma, J. Applying Interpretable Machine Learning in Computational Biology—Pitfalls, Recommendations and Opportunities for New Developments. Nat. Methods 2024, 21, 1454–1461. [Google Scholar] [CrossRef]
- Silva, T.N.; Thomas, J.B.; Dahlberg, J.; Rhee, S.Y.; Mortimer, J.C. Progress and Challenges in Sorghum Biotechnology, a Multipurpose Feedstock for the Bioeconomy. J. Exp. Bot. 2022, 73, 646–664. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Siddique, A.B.; Shabala, S.; Li, C.; Chen, Z.-H.; Varshney, R.K.; Zhao, C.; Zhou, M. Reducing Heat Stress Damage in Cereal Crops through Agronomic Management and Breeding Strategies. Plant Stress 2025, 16, 100888. [Google Scholar] [CrossRef]
- Sargent, D.; Conaty, W.C.; Tissue, D.T.; Sharwood, R.E. Synthetic Biology and Opportunities within Agricultural Crops. J. Sustain. Agric. Environ. 2022, 1, 89–107. [Google Scholar] [CrossRef]
- Liu, S.; Zenda, T.; Tian, Z.; Huang, Z. Metabolic Pathways Engineering for Drought or/and Heat Tolerance in Cereals. Front. Plant Sci. 2023, 14, 1111875. [Google Scholar] [CrossRef]
- Yaschenko, A.E.; Alonso, J.M.; Stepanova, A.N. Arabidopsis as a Model for Translational Research. Plant Cell 2025, 37, koae065. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Nelissen, H.; Chan, R.L.; Napier, J.A.; Seung, D.; Liu, L.; McKim, S.M. Challenges of Translating Arabidopsis Insights into Crops. Plant Cell 2025, 37, koaf059. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R. Plant Responses to Multifactorial Stress Combination. New Phytol. 2022, 234, 1161–1167. [Google Scholar] [CrossRef]
- Horváth, Á.; Berki, Z.; Balla, K.; Bányai, J.; Mayer, M.; Cseh, A.; Kiss, T.; Karsai, I. Field versus Controlled Environmental Experiments to Evaluate the Heat Stress Response of Barley (Hordeum vulgare L.). Environ. Exp. Bot. 2024, 228, 106038. [Google Scholar] [CrossRef]
- de Souza, L.P.; Borghi, M.; Fernie, A. Plant Single-Cell Metabolomics-Challenges and Perspectives. Int. J. Mol. Sci. 2020, 21, 8987. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Jiao, Y. Advances and Applications of Single-Cell Omics Technologies in Plant Research. Plant J. Cell Mol. Biol. 2022, 110, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Scolnick, J. Complex Biological Questions Being Addressed Using Single Cell Sequencing Technologies. SLAS Technol. 2022, 27, 143–149. [Google Scholar] [CrossRef]
- Zhao, D.; Zhu, X.; Zhou, H.; Sun, N.; Wang, T.; Bi, C.; Zhang, X. CRISPR-Based Metabolic Pathway Engineering. Metab. Eng. 2021, 63, 148–159. [Google Scholar] [CrossRef]
- Guo, Y.; Hao, D.; Wang, X.; Wang, H.; Wu, Z.; Yang, P.; Zhang, B. Comparative Transcriptomics Reveals Key Genes Contributing to the Differences in Drought Tolerance among Three Cultivars of Foxtail Millet (Setaria italica). Plant Growth Regul. 2023, 99, 45–64. [Google Scholar] [CrossRef]
- Kaur, S.; Seem, K.; Duhan, N.; Kumar, S.; Kaundal, R.; Mohapatra, T. Transcriptome and Physio-Biochemical Profiling Reveals Differential Responses of Rice Cultivars at Reproductive-Stage Drought Stress. Int. J. Mol. Sci. 2023, 24, 1002. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Wu, X.; Wang, J.; Li, H.; Zhang, R. Transcriptomic and Physiological Responses of Contrasting Maize Genotypes to Drought Stress. Front. Plant Sci. 2022, 13, 928897. [Google Scholar] [CrossRef]
- Wu, G.; Sun, X.; Sun, Q.; Kang, X.; Wang, J.; He, X.; Liu, W.; Xu, D.; Dai, X.; Ma, W. Genetic Variation in Wheat Root Transcriptome Responses to Salinity: A Comparative Study of Tolerant and Sensitive Genotypes. Int. J. Mol. Sci. 2025, 26, 331. [Google Scholar] [CrossRef]
- Wang, W.; Zou, J.; Zhang, Y.; Niu, L.; Yu, L.; Wang, Z.; Wang, F.; Zhang, S.; Yang, X. Salinity Stress Phenotyping of Wheat Germplasm under Multiple Growth Conditions and Transcriptomic Analysis of Two Wheat Varieties Contrasting in Their Salinity Stress Tolerance. Plant Growth Regul. 2025, 105, 739–757. [Google Scholar] [CrossRef]
- Alyahya, N.; Taybi, T. Comparative Transcriptomic Profiling Reveals Differentially Expressed Genes and Important Related Metabolic Pathways in Shoots and Roots of a Saudi Wheat Cultivar (Najran) under Salinity Stress. Front. Plant Sci. 2023, 14, 1225541. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, Z.; Chen, J.; Kuang, L.; Yan, T.; Wu, D.; Gao, F. Comparative Transcriptomics of Indica and Japonica Rice Roots under Heat Stress Reveals the Crucial Role of OsMAPK3 in Heat Response. Plant Physiol. Biochem. 2025, 221, 109668. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Wang, H.; Zhou, L.; Li, B.; Zhang, S.; He, Y.; Guo, Y.; You, A.; Jiao, C.; Xu, Y. Time-Series Transcriptomic Analysis of Contrasting Rice Materials under Heat Stress Reveals a Faster Response in the Tolerant Cultivar. Int. J. Mol. Sci. 2023, 24, 9408. [Google Scholar] [CrossRef]
- Azameti, M.K.; Ranjan, A.; Singh, P.; Gaikwad, K.; Singh, A.K.; Dalal, M.; Arora, A.; Rai, V.; Padaria, J.C. Transcriptome Profiling Reveals the Genes and Pathways Involved in Thermo-Tolerance in Wheat (Triticum aestivum L.) Genotype Raj 3765. Sci. Rep. 2022, 12, 14831. [Google Scholar] [CrossRef]
- Konstantinov, D.K.; Zubairova, U.S.; Ermakov, A.A.; Doroshkov, A.V. Comparative Transcriptome Profiling of a Resistant vs Susceptible Bread Wheat (Triticum aestivum L.) Cultivar in Response to Water Deficit and Cold Stress. PeerJ 2021, 9, e11428. [Google Scholar] [CrossRef]
- Thapa, R.; Tabien, R.E.; Johnson, C.D.; Septiningsih, E.M. Comparative Transcriptomic Analysis of Germinating Rice Seedlings to Individual and Combined Anaerobic and Cold Stress. BMC Genom. 2023, 24, 185. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, X.; Chen, Y.; Ji, G.; Ma, X.; Zhang, Y.; Xiang, J.; Wang, Y.; Wang, Z.; Li, L. Comparative Transcriptome Combined with Morphophysiological Analyses Revealed Carotenoid Biosynthesis for Differential Chilling Tolerance in Two Contrasting Rice (Oryza sativa L.) Genotypes. Rice 2023, 16, 52. [Google Scholar] [CrossRef]
- Zhang, T.; Xiao, J.; Zhao, Y.; Zhang, Y.; Jie, Y.; Shen, D.; Yue, C.; Huang, J.; Hua, Y.; Zhou, T. Comparative Physiological and Transcriptomic Analyses Reveal Ascorbate and Glutathione Coregulation of Cadmium Toxicity Resistance in Wheat Genotypes. BMC Plant Biol. 2021, 21, 459. [Google Scholar] [CrossRef]
- Luo, D.; Li, Q.; Pang, F.; Zhang, W.; Li, Y.; Xing, Y.; Dong, D. Commonalities and Specificities in Wheat (Triticum aestivum L.) Responses to Aluminum Toxicity and Low Phosphorus Revealed by Transcriptomics and Targeted Metabolomics. Int. J. Mol. Sci. 2024, 25, 9273. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Zhou, L.; Guo, G.; Li, Y.; Chen, Z.; Lu, R.; Liu, C.; Chen, J. Comparative Analysis of Root Transcriptome of High-NUE Mutant and Wild-Type Barley under Low-Nitrogen Conditions. Agronomy 2023, 13, 806. [Google Scholar] [CrossRef]
- Li, J.; Zenda, T.; Liu, S.; Dong, A.; Wang, Y.; Liu, X.; Wang, N.; Duan, H. Integrated Transcriptomic and Proteomic Analyses of Low-Nitrogen-Stress Tolerance and Function Analysis of ZmGST42 Gene in Maize. Antioxidants 2023, 12, 1831. [Google Scholar] [CrossRef]
- Wang, G.; Wang, J.; Yao, L.; Li, B.; Ma, X.; Si, E.; Yang, K.; Li, C.; Shang, X.; Meng, Y. Transcriptome and Metabolome Reveal the Molecular Mechanism of Barley Genotypes Underlying the Response to Low Nitrogen and Resupply. Int. J. Mol. Sci. 2023, 24, 4706. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Yang, Z.; Lu, J.; Yang, J.; Li, N.; Shi, H. Transcriptomics Uncovers Pathways Mediating Low-Nitrogen Stress Tolerance in Two Foxtail Millet Varieties. Agriculture 2025, 15, 628. [Google Scholar] [CrossRef]
- Guo, H.; Tao, W.; Gao, H.; Chen, L.; Zhong, X.; Tang, M.; Gao, G.; Liang, T.; Zhang, X. Physiological Traits, Gene Expression Responses, and Proteomics of Rice Varieties Varying in Heat Stress Tolerance at the Flowering Stage. Front. Plant Sci. 2024, 15, 1489331. [Google Scholar] [CrossRef]
- Kumar, R.; Ghatak, A.; Goyal, I.; Sarkar, N.K.; Weckwerth, W.; Grover, A.; Chaturvedi, P. Heat-Induced Proteomic Changes in Anthers of Contrasting Rice Genotypes under Variable Stress Regimes. Front. Plant Sci. 2023, 13, 1083971. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Peng, Y.; Zhao, X.; Wu, B.; Chen, F.; Ren, B.; Zhuang, Z.; Gao, Q.; Ding, Y. Comparative Proteomics Analysis of the Seedling Root Response of Drought-Sensitive and Drought-Tolerant Maize Varieties to Drought Stress. Int. J. Mol. Sci. 2019, 20, 2793. [Google Scholar] [CrossRef]
- Jha, S.; Maity, S.; Singh, J.; Chouhan, C.; Tak, N.; Ambatipudi, K. Integrated Physiological and Comparative Proteomics Analysis of Contrasting Genotypes of Pearl Millet Reveals Underlying Salt-responsive Mechanisms. Physiol. Plant. 2022, 174, e13605. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, M.; Zhao, C.; Wang, Y.; Zhang, R. Physiological and Proteomic Analyses Revealed the Response Mechanisms of Two Different Drought-Resistant Maize Varieties. BMC Plant Biol. 2021, 21, 513. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Shi, Z.; Zhou, M.; Zhao, B.; Li, H.; Wang, J.; Liu, Y.; Zhao, J. iTRAQ-Based Quantitative Proteomic Analysis Provides Insight into the Drought-Stress Response in Maize Seedlings. Sci. Rep. 2022, 12, 9520. [Google Scholar] [CrossRef] [PubMed]
- Prathap, V.; Kumar, S.; Tyagi, A. Comparative Proteome Analysis of Phosphorus-Responsive Genotypes Reveals the Proteins Differentially Expressed under Phosphorous Starvation Stress in Rice. Int. J. Biol. Macromol. 2023, 234, 123760. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Liu, S.; Dong, A.; Zenda, T.; Liu, X.; Li, J.; Duan, H. Comparative Proteomic Analysis of Two Contrasting Maize Hybrids’ Responses to Low Nitrogen Stress at the Twelve Leaf Stage and Function Verification of ZmTGA Gene. Genes 2022, 13, 670. [Google Scholar] [CrossRef]
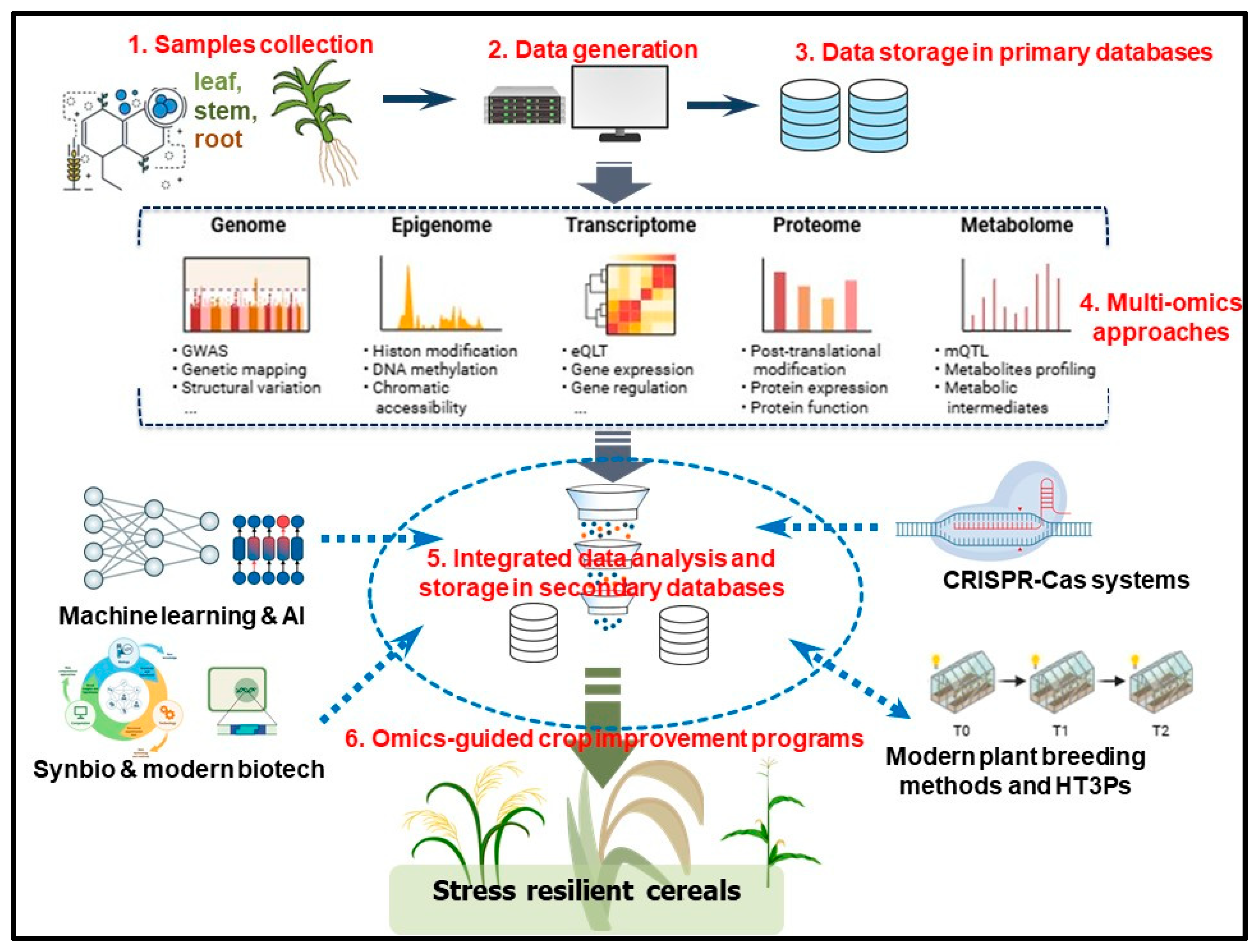
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goche, T.; Mavindidze, P.; Zenda, T. Advances in Functional Genomics for Exploring Abiotic Stress Tolerance Mechanisms in Cereals. Plants 2025, 14, 2459. https://doi.org/10.3390/plants14162459
Goche T, Mavindidze P, Zenda T. Advances in Functional Genomics for Exploring Abiotic Stress Tolerance Mechanisms in Cereals. Plants. 2025; 14(16):2459. https://doi.org/10.3390/plants14162459
Chicago/Turabian StyleGoche, Tatenda, Peter Mavindidze, and Tinashe Zenda. 2025. "Advances in Functional Genomics for Exploring Abiotic Stress Tolerance Mechanisms in Cereals" Plants 14, no. 16: 2459. https://doi.org/10.3390/plants14162459
APA StyleGoche, T., Mavindidze, P., & Zenda, T. (2025). Advances in Functional Genomics for Exploring Abiotic Stress Tolerance Mechanisms in Cereals. Plants, 14(16), 2459. https://doi.org/10.3390/plants14162459






