Identification of the MADS-Box Gene Family and Development of Simple Sequence Repeat Markers in Chimonanthus praecox
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Characterization of CpMADS Genes
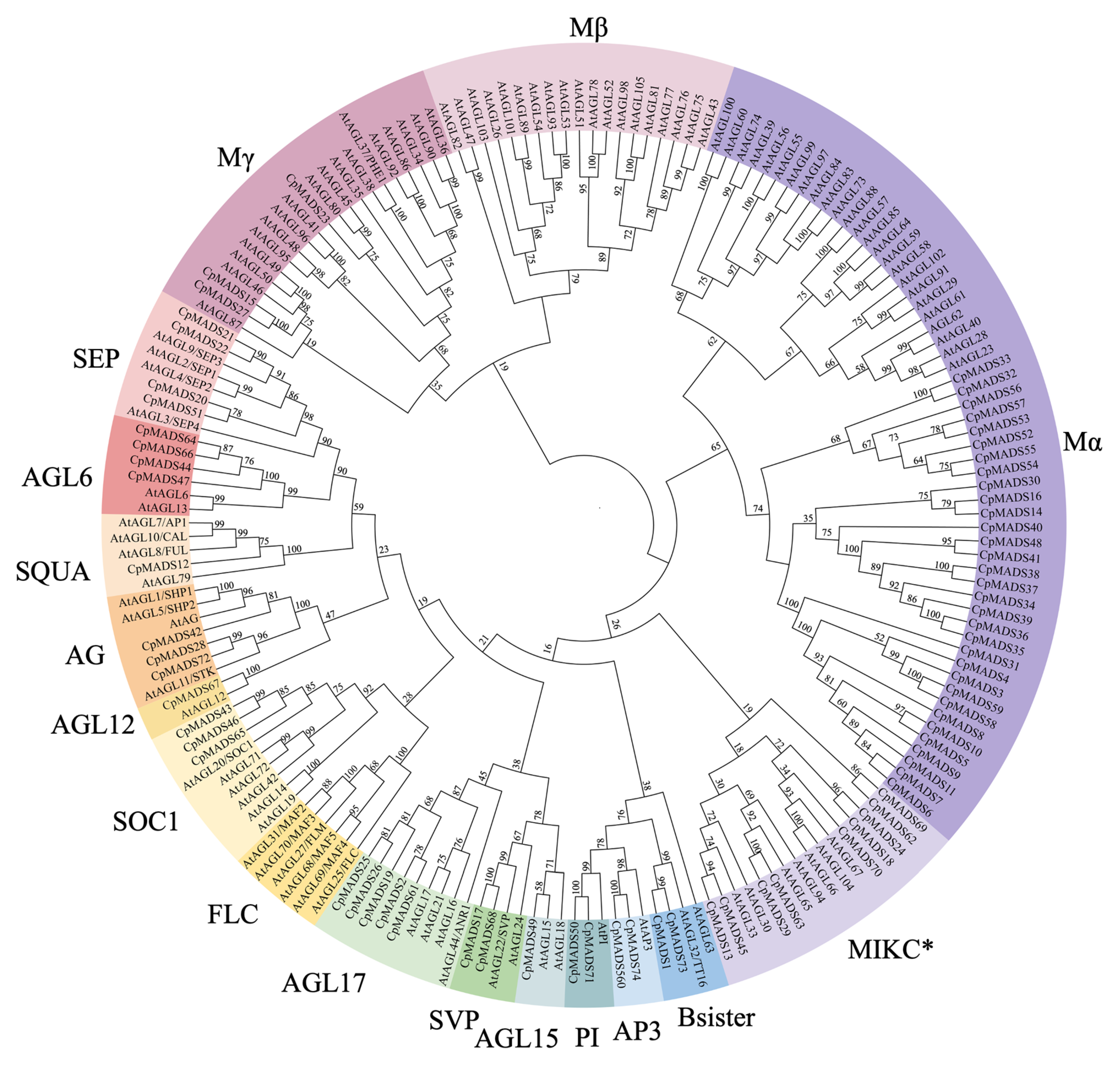
2.2. Phylogenetic Tree Analysis of the Wintersweet MADS Protein Family
2.3. Analysis of CpMADS Gene Structure, Domains, and Conserved Motifs
2.4. Identification and Characterization of SSR Loci in the CpMADS Gene Family
2.5. Chromosomal Localization of CpMADS and SSR Loci
2.6. Polymorphism and Universality of SSR Markers
2.7. Unweighted Pair Group Method with Arithmetic (UPGMA) Cluster Analysis of Different Varieties of C. praecox Based on CpMADS Markers
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. Identification and Characterization of the CpMADS Gene Family
4.3. Gene Structure, Protein Motif, and Conserved Domain Analyses
4.4. Chromosomal Distribution of MADS-Box Genes and SSR Loci
4.5. Microsatellite Marker Identification, PCR Amplification, and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- de Folter, S.; Angenent, G.C. Trans meets cis in MADS science. Trends Plant Sci. 2006, 11, 224–231. [Google Scholar] [CrossRef]
- Nam, J.; dePamphilis, C.W.; Ma, H.; Nei, M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003, 20, 1435–1447. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.H.; Ma, H.; Nei, M.S. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martinez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhu, G.P.; Li, F.D.; Wang, L.; Chen, C.; Zhao, H. MIKC-Type MADS-Box Gene Family Discovery and Evolutionary Investigation in Rosaceae Plants. Agronomy 2023, 13, 1695. [Google Scholar] [CrossRef]
- Kwantes, M.; Liebsch, D.; Verelst, W. How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 2012, 29, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Kofuji, R.; Sumikawa, N.; Yamasaki, M.; Kondo, K.; Ueda, K.; Ito, M.; Hasebe, M. Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 2003, 20, 1963–1977. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Liljegren, S.J.; Pelaz, S.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Vergara-Silva, F.; Yanofsky, M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000, 24, 457–466. [Google Scholar] [CrossRef]
- Grimplet, J.; Martinez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Qiu, Y.C.; Köhler, C. Endosperm Evolution by Duplicated and Neofunctionalized Type I MADS-Box Transcription Factors. Mol. Biol. Evol. 2022, 39, msab355. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Qiu, Z.J.; Ghazy, A.; Wang, Q.; Fiaz, S.; Al-Doss, A.A.; Attia, K.A.; Ul Haq, I.; Iqbal, R.; Hou, W.H. Identification of the complete MADS-box gene family in pea (Pisum sativum L.) and its expression pattern in development and adversity. Genet. Resour. Crop. Evol. 2025, 72, 6521–6540. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Leseberg, C.H.; Li, A.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef]
- Zhang, X.; Fatima, M.; Zhou, P.; Ma, Q.; Ming, R. Analysis of MADS-box genes revealed modified flowering gene network and diurnal expression in pineapple. BMC Genom. 2020, 21, 8. [Google Scholar] [CrossRef]
- Ruelens, P.; Zhang, Z.; van Mourik, H.; Maere, S.; Kaufmann, K.; Geuten, K. The Origin of Floral Organ Identity Quartets. Plant Cell 2017, 29, 229–242. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, X.T.; Liu, X.; Zhang, L.S. Evolutionary Analysis of MIKC-Type MADS-Box Genes in Gymnosperms and Angiosperms. Front. Plant Sci. 2017, 8, 895. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant biology. Flor. Quartets Nat. 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Denboer, B.G.W.; Vanmontagu, M.; Okamuro, J.K. Control of Arabidopsis Flower and Seed Development by the Homeotic Gene Apetala2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [CrossRef]
- Akash, M.; Shiyab, S.; Saleh, M.; Hasan, S.M.; Abuhussein, M.; Al-Awaida, W. Development and Validation of Gene-Based SSR Markers in the Genus. Scientifica 2023, 2023, 6624354. [Google Scholar] [CrossRef]
- Hou, X.J.; Liu, S.R.; Khan, M.R.G.; Hu, C.G.; Zhang, J.Z. Genome-Wide Identification, Classification, Expression Profiling, and SSR Marker Development of the MADS-Box Gene Family in Citrus. Plant Mol. Biol. Rep. 2014, 32, 28–41. [Google Scholar] [CrossRef]
- Yang, W.; Bai, Z.; Wang, F.; Zou, M.; Wang, X.; Xie, J.; Zhang, F. Analysis of the genetic diversity and population structure of Monochasma savatieri Franch. ex Maxim using novel EST-SSR markers. BMC Genom. 2022, 23, 597. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhao, L.; Chen, C.; Wang, J.; Chen, Y.; Lu, D. Identification of Quantitative Trait Loci and Analysis of Novel Candidate Genes for Resistance to False Smut of Rice Based on SSR Molecular Markers. Biomolecules 2025, 15, 186. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Y.; Wang, Y.; Han, R.; Liang, Z.; He, Q.; Jia, Q. SSR Loci Analysis in Transcriptome and Molecular Marker Development in Polygonatum sibiricum. BioMed Res. Int. 2022, 2022, 4237913. [Google Scholar] [CrossRef]
- Fu, X.M.; Yang, N.; Du, Y.Q.; Kamran, H.M.; Wang, H.B.; Chen, S.Y.; Chen, L.Q. Development of SSR Molecular Markers and Genetic Diversity Analysis of TPS Gene Family in Chimonanthus praecox. Agriculture 2023, 13, 893. [Google Scholar] [CrossRef]
- Liu, B.; Wu, H.F.; Cao, Y.Z.; Yang, X.M.; Sui, S.Z. Establishment of Novel Simple Sequence Repeat (SSR) Markers from Chimonanthus praecox Transcriptome Data and Their Application in the Identification of Varieties. Plants 2024, 13, 2131. [Google Scholar] [CrossRef]
- Zhou, P.; Lei, S.R.; Zhang, X.D.; Wang, Y.H.; Guo, R.; Yan, S.B.; Jin, G.; Zhang, X.T. Genome sequencing revealed the red-flower trait candidate gene of a peach landrace. Hortic. Res. 2023, 10, uhad210. [Google Scholar] [CrossRef]
- Yang, N. Functional Characterization of Anthocyanin Synthase CpANS1 and Transcription Factor CpMYB2 in Chimonanthus praecox Tepals. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2019. [Google Scholar]
- Zhu, T.; Feng, Y.Y.; Dong, X.X.; Yang, X.M.; Liu, B.; Yuan, P.Y.; Song, X.R.; Chen, S.X.; Sui, S.Z. Optimizing DUS testing for Chimonanthus praecox using feature selection based on a genetic algorithm. Front. Plant Sci. 2024, 14, 1328603. [Google Scholar] [CrossRef]
- Li, Z.N.; Liu, N.; Zhang, W.; Wu, C.Y.; Jiang, Y.J.; Ma, J.; Li, M.Y.; Sui, S.Z. Integrated transcriptome and proteome analysis provides insight into chilling-induced dormancy breaking in Chimonanthus praecox. Hortic. Res. 2020, 7, 198. [Google Scholar] [CrossRef]
- Shen, Z.G.; Li, W.Y.; Li, Y.L.; Liu, M.L.; Cao, H.P.; Provart, N.; Ding, X.; Sun, M.; Tang, Z.H.; Yue, C.P.; et al. The red flower wintersweet genome provides insights into the evolution of magnoliids and the molecular mechanism for tepal color development. Plant J. 2021, 108, 1662–1678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.G.; Zhou, M.Q.; Chen, L.Q.; Zhang, D.L.; Robert, G.W. Genetic diversity and discrimination of Chimonanthus praecox (L.) Link germplasm using ISSR and RAPD markers. Hortscience 2007, 42, 1144–1148. [Google Scholar] [CrossRef]
- Huang, R.W.; Liu, D.F.; Huang, M.; Ma, J.; Li, Z.N.; Li, M.Y.; Sui, S.Z. CpWRKY71, a WRKY transcription factor gene of Wintersweet (Chimonanthus praecox), promotes flowering and leaf senescence in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 5325. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Yang, N.; Chen, L.Q. Paraffin section observation of flower bud differentiation of Chimonanthus praecox in Kunming and comparison of the differentiation processes in different regions, China. Hortic. Plant J. 2022, 8, 221–229. [Google Scholar] [CrossRef]
- Tian, M.K.; Li, Q.; Liu, N.; Li, J.C.; Huo, J.T.; Sui, S.Z.; Li, Z.N. Ca2+-induced CpCBL8-CpCIPK9 module negatively regulates dormancy breaking and cold tolerance in winter-flowering wintersweet. Hortic. Plant J. 2025, 11, 877–890. [Google Scholar] [CrossRef]
- Kitagawa, N.; Ninomiya, K.; Okugawa, S.; Motai, C.; Nakanishi, Y.; Yoshikawa, M.; Muraoka, O.; Morikawa, T. Quantitative determination of principal alkaloid and flavonoid constituents in wintersweet, the flower buds of Chimonanthus praecox. Nat. Prod. Commun. 2016, 11, 953–956. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, J.M.; Xing, B.C.; Yang, B.X.; Cheng, K.J. Research progress on the molecular biology of genus Chimonanthus medicinal plants. Chin. J. Mod. Appl. Pharm. 2022, 39, 1646–1654. [Google Scholar] [CrossRef]
- Luo, J.; Wang, J. Research review on cultivar classification of Chimonanthus praecox in China. J. Jiangsu For. Sci. Technol. 2012, 39, 42–46. [Google Scholar]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef]
- Hou, H.F.; Tian, M.K.; Liu, N.; Huo, J.T.; Sui, S.Z.; Li, Z.N. Genome-wide analysis of MIKCC-type MADS-box genes and roles of CpFUL/SEP/AGL6 superclade in dormancy breaking and bud formation of Chimonanthus praecox. Plant Physiol. Biochem. 2023, 196, 893–902. [Google Scholar] [CrossRef]
- Hou, H.F.; Wu, C.Y.; Huo, J.T.; Liu, N.; Jiang, Y.J.; Sui, S.Z.; Li, Z.E. Integrated transcriptome and proteome analysis provides insights into CpFPA1 for floral induction in Chimonanthus praecox (Magnoliidae) without FLC in genome. Plant Cell Rep. 2024, 43, 66. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, B.G.; Duan, K.; Wang, L.G.; Wang, M.; Tang, X.M.; Pan, A.H.; Sui, S.Z.; Wang, G.D. The paleoAP3-type gene CpAP3, an ancestral B-class gene from the basal angiosperm Chimonanthus praecox, can affect stamen and petal development in higher eudicots. Dev. Genes Evol. 2011, 221, 83–93. [Google Scholar] [CrossRef]
- Luo, P. Cloning and Functional Analysis of CpAGL2 in Chimonanthus praecox. Master’s Thesis, Southwest University, Chongqing, China, 2014. [Google Scholar]
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.; Jie, C.; Jitang, Z.; Jingbo, L.; Xinzhu, L.; Qingwei, L. Research on Postharvest Preservation Technology of Wintersweet Cut Flowers. North. Hortic. 2023, 95–102. [Google Scholar]
- Fu, X.; Wang, H.; Tao, X.; Liu, Y.; Chen, L.; Yang, N. Integrated Multiomics Analysis Sheds Light on the Mechanisms of Color and Fragrance Biosynthesis in Wintersweet Flowers. Int. J. Mol. Sci. 2025, 26, 1684. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.Z.; Tian, J.P.; Cheng, H.H.; Yan, Q.M.; Li, L.; Jamal, A.; Xu, Z.P.; Xiang, L.; Saski, C.A.; Jin, S.X.; et al. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020, 21, 200. [Google Scholar] [CrossRef]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Wei, X.; Wang, L.; Yu, J.; Zhang, Y.; Li, D.; Zhang, X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 2015, 569, 66–76. [Google Scholar] [CrossRef]
- Liu, W.; Ma, Y.; Ying, Z.; Yang, L.; Xie, M.; Ma, H. Genome-wide identification and characterization of MADS-box gene family of Rhododendron griersonianum Balf. f. et Forrest. Plant Sci. J. 2024, 42, 624–633. [Google Scholar]
- Liang, M.; Du, Z.; Yang, Z.; Luo, T.; Ji, C.; Cui, H.; Li, R. Genome-wide characterization and expression analysis of MADS-box transcription factor gene family in Perilla frutescens. Front. Plant Sci. 2023, 14, 1299902. [Google Scholar] [CrossRef]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.H.; Wang, Z.M.; Li, S.L.; Hou, M.L.; Zhou, Y.; Zhao, Y.Q.; Li, G.J.; Zhao, H.; Ma, H.L. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, N.; Zhao, K.; Chen, Y.; Tang, R.; Chen, L. Development and primer selection of EST-SSR molecular markers based on transcriptome sequencing of Chimonanthus praecox. J. Beijing For. Univ. 2013, 35, 25–32. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Wang, J.; LI, Z.; Zhang, J.; Li, Q. Genetic diversity analysis and fingerprinting of 175 Chimonanthus praecox germplasm based on SSR molecular marker. Chin. J. Biotechnol. 2024, 40, 252–268. [Google Scholar] [CrossRef]
- Li, X.; Qiao, L.; Chen, B.; Zheng, Y.; Zhi, C.; Zhang, S.; Pan, Y.; Cheng, Z. SSR markers development and their application in genetic diversity evaluation of garlic (Allium sativum) germplasm. Plant Divers. 2022, 44, 481–491. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakse, J.; Javornik, B. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica 2004, 136, 93–102. [Google Scholar] [CrossRef]
- Liu, B.; Wu, H.F.; Cao, Y.Z.; Ma, G.P.; Zheng, X.W.; Zhu, H.X.; Song, X.R.; Sui, S.Z. Reducing costs and shortening the cetyltrimethylammonium bromide (CTAB) method to improve DNA extraction efficiency from wintersweet and some other plants. Sci. Rep. 2025, 15, 13441. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Applied Biostatistics; Exeter Publishing: Exeter, UK, 1992. [Google Scholar]




| Item | Number |
|---|---|
| Total sequences analyzed | 74 |
| Total length of sequences analyzed (bp) | 1,209,009 |
| Total SSRs identified | 80 |
| Number of sequences containing SSRs | 41 |
| Number of sequences with multiple SSRs | 21 |
| Number of compound SSRs | 3 |
| Number of SSR primers designed | 295 |
| Total repeat motifs detected | 29 |
| Number of Repetitions | Mononucleotide (Mono) | Dinucleotide (Di) | Trinucleotide (Tri) | Tetranucleotide (Tetra) | Pentanucleotide (Pentra) | Total | Percentage (%) |
|---|---|---|---|---|---|---|---|
| 5 | 0 | 0 | 0 | 5 | 1 | 6 | 7.5 |
| 6 | 0 | 0 | 9 | 3 | 0 | 12 | 15 |
| 7 | 0 | 0 | 3 | 1 | 0 | 4 | 5 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 1 | 0 | 0 | 1 | 1.25 |
| 10 | 0 | 6 | 0 | 0 | 0 | 6 | 7.5 |
| 11 | 0 | 8 | 1 | 0 | 0 | 9 | 11.25 |
| 12 | 0 | 6 | 0 | 0 | 0 | 6 | 7.5 |
| 13 | 0 | 3 | 0 | 0 | 0 | 3 | 3.75 |
| 14 | 0 | 5 | 0 | 0 | 0 | 5 | 6.25 |
| 15 | 0 | 3 | 0 | 0 | 0 | 3 | 3.75 |
| 16 | 0 | 2 | 0 | 0 | 0 | 2 | 2.5 |
| 17 | 0 | 3 | 0 | 0 | 0 | 3 | 3.75 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 1 | 0 | 0 | 0 | 1 | 1.25 |
| 20 | 5 | 1 | 0 | 0 | 0 | 6 | 7.5 |
| >20 | 4 | 9 | 0 | 0 | 0 | 13 | 16.25 |
| Total | 9 | 47 | 14 | 9 | 1 | 80 | |
| Percentage (%) | 11.25 | 58.75 | 17.5 | 11.25 | 1.25 |
| Locus | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|---|---|
| CpMADS41-31 | CCGATCAGAGCTGAATCCCC | GGAACGTCCCTGATAACGCA | 7 | 4.60 | 1.71 | 0.70 | 0.78 | 0.76 |
| CpMADS60-50 | GGTTTTGAGTTCGATTTCTCCCT | GGTTTTGAGTTCGATTTCTCCCT | 9 | 5.90 | 1.94 | 0.45 | 0.83 | 0.82 |
| CpMADS61-53 | CAGTCAAGCCCCAACCTGAT | CCCCCAAACCCACCACTATC | 7 | 5.60 | 1.80 | 0.74 | 0.82 | 0.80 |
| CpMADS23-16 | AGCAATATTGGCCATGTGGG | AGCAGAGGTGAAAGTATCCGC | 7 | 5.44 | 1.78 | 0.91 | 0.82 | 0.81 |
| CpMADS66-69 | AGGTGAGCACATGTGAGTGA | TTCTCCCGACTTTGGCTGAA | 8 | 4.79 | 1.76 | 0.78 | 0.79 | 0.76 |
| CpMADS19-9 | TCACATGTGTATCCTTAAACCGT | TGTCGATTTCAAGTGCATCAAA | 8 | 5.39 | 1.84 | 0.21 | 0.81 | 0.82 |
| CpMADS28-20 | CGGGAAAAGCATTTCGACCG | AGTCCGTAATCTCAGGCGAG | 6 | 3.74 | 1.47 | 0.81 | 0.73 | 0.74 |
| CpMADS46-41 | TCATTTTCGGGCCTTGTCCA | CAATGCGTGGAAACAGCACA | 6 | 5.31 | 1.72 | 0.89 | 0.81 | 0.82 |
| CpMADS50-46 | GCAACAAAACACAAGGCTGG | TGTGTTTGTGAGGGAGGCAA | 7 | 3.87 | 1.60 | 0.86 | 0.74 | 0.74 |
| CpMADS51-47 | GCCCTAAGGTTTGCTAGAGGA | AACGGCAACACCCAAATTGG | 5 | 3.50 | 1.38 | 0.82 | 0.71 | 0.75 |
| Mean | 7 | 4.81 | 1.70 | 0.72 | 0.79 | 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Liu, B.; Cao, Y.; Ma, G.; Zheng, X.; Yang, X.; Dai, Q.; Zhu, H.; Zhu, H.; Song, X.; et al. Identification of the MADS-Box Gene Family and Development of Simple Sequence Repeat Markers in Chimonanthus praecox. Plants 2025, 14, 2450. https://doi.org/10.3390/plants14152450
Wu H, Liu B, Cao Y, Ma G, Zheng X, Yang X, Dai Q, Zhu H, Zhu H, Song X, et al. Identification of the MADS-Box Gene Family and Development of Simple Sequence Repeat Markers in Chimonanthus praecox. Plants. 2025; 14(15):2450. https://doi.org/10.3390/plants14152450
Chicago/Turabian StyleWu, Huafeng, Bin Liu, Yinzhu Cao, Guanpeng Ma, Xiaowen Zheng, Ximeng Yang, Qianli Dai, Hengxing Zhu, Haoxiang Zhu, Xingrong Song, and et al. 2025. "Identification of the MADS-Box Gene Family and Development of Simple Sequence Repeat Markers in Chimonanthus praecox" Plants 14, no. 15: 2450. https://doi.org/10.3390/plants14152450
APA StyleWu, H., Liu, B., Cao, Y., Ma, G., Zheng, X., Yang, X., Dai, Q., Zhu, H., Zhu, H., Song, X., & Sui, S. (2025). Identification of the MADS-Box Gene Family and Development of Simple Sequence Repeat Markers in Chimonanthus praecox. Plants, 14(15), 2450. https://doi.org/10.3390/plants14152450






