Phylogenetic Insights from a Novel Rehubryum Species Challenge Generic Boundaries in Orthotrichaceae
Abstract
1. Introduction
2. Results
2.1. Morphological Approach
2.2. Assembly Summary Statistics
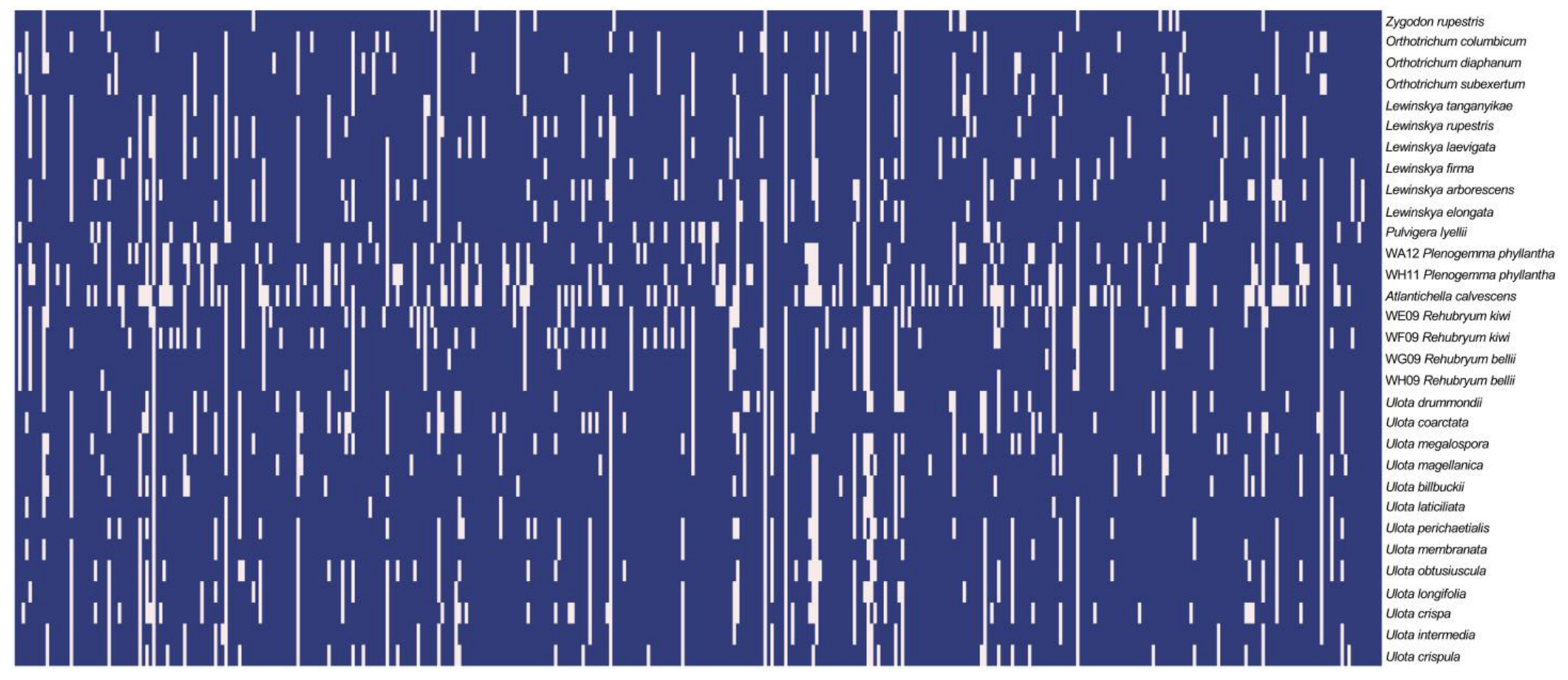
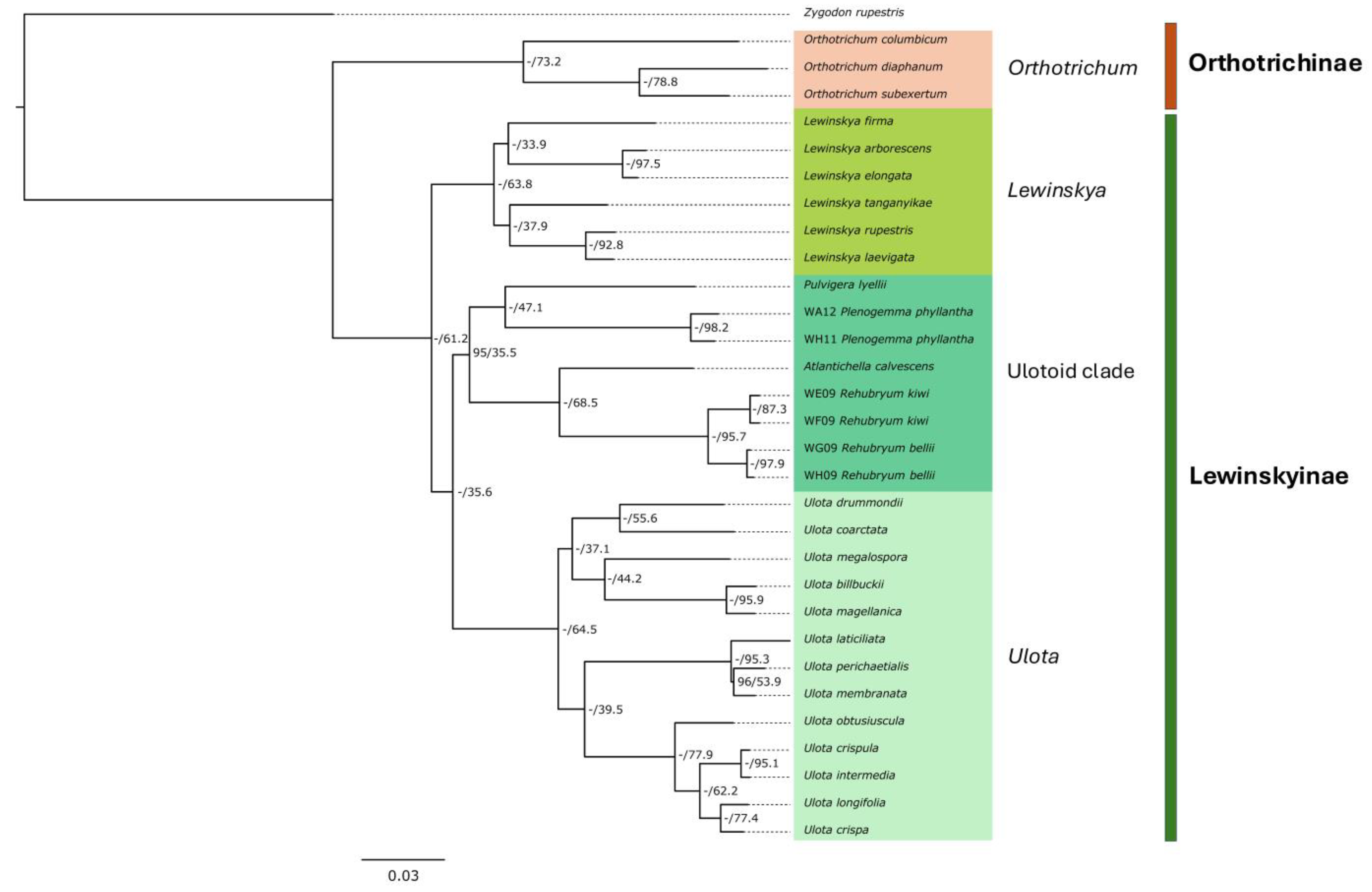
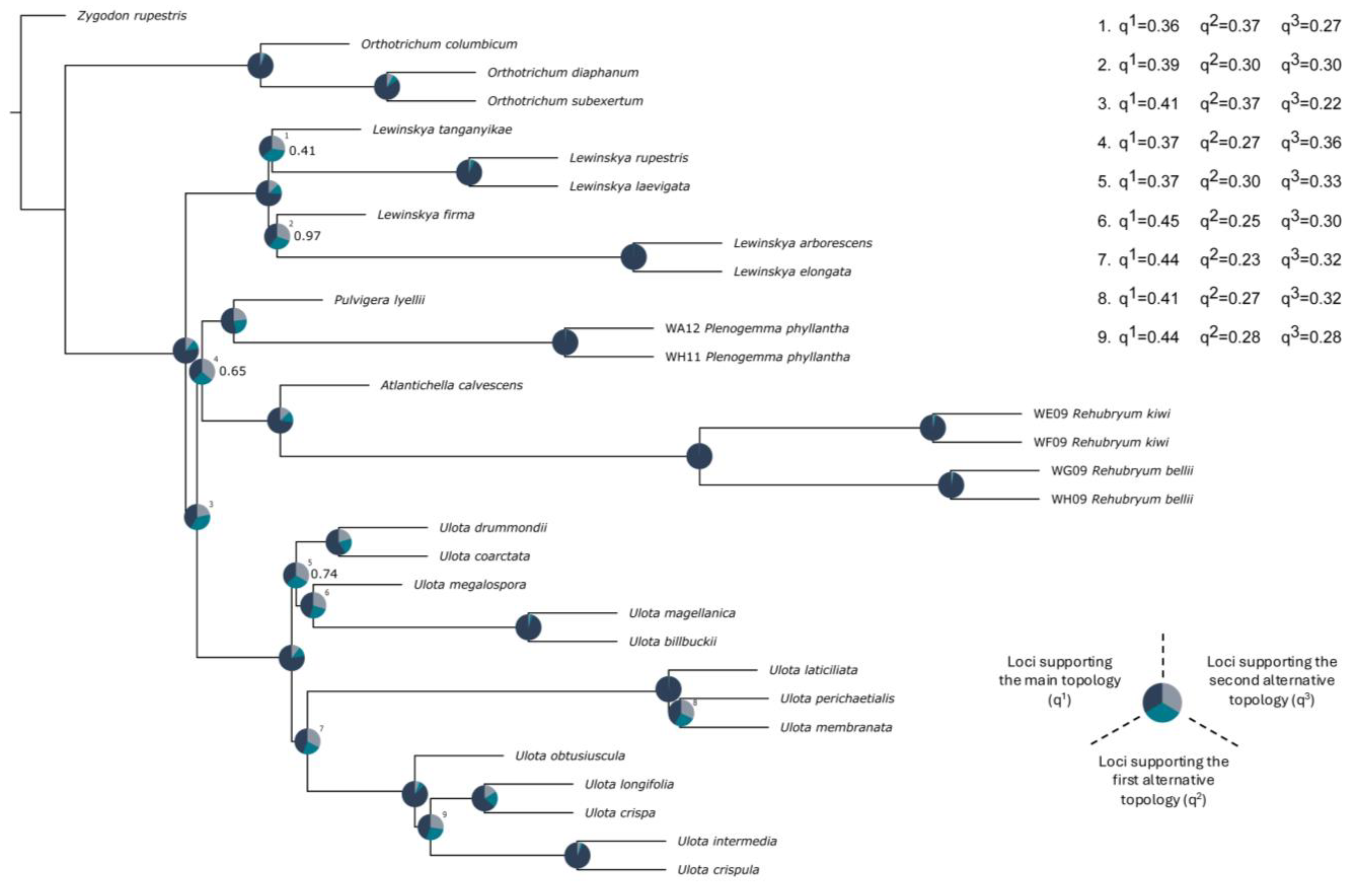
2.3. Phylogenomic Reconstructions and Support
2.4. Taxonomy
Rehubryum kiwi F.Lara, Garilleti & Matanov sp. nov. (Figure 4 and Figure 5)
3. Discussion
3.1. Systematic and Taxonomical Delimitation
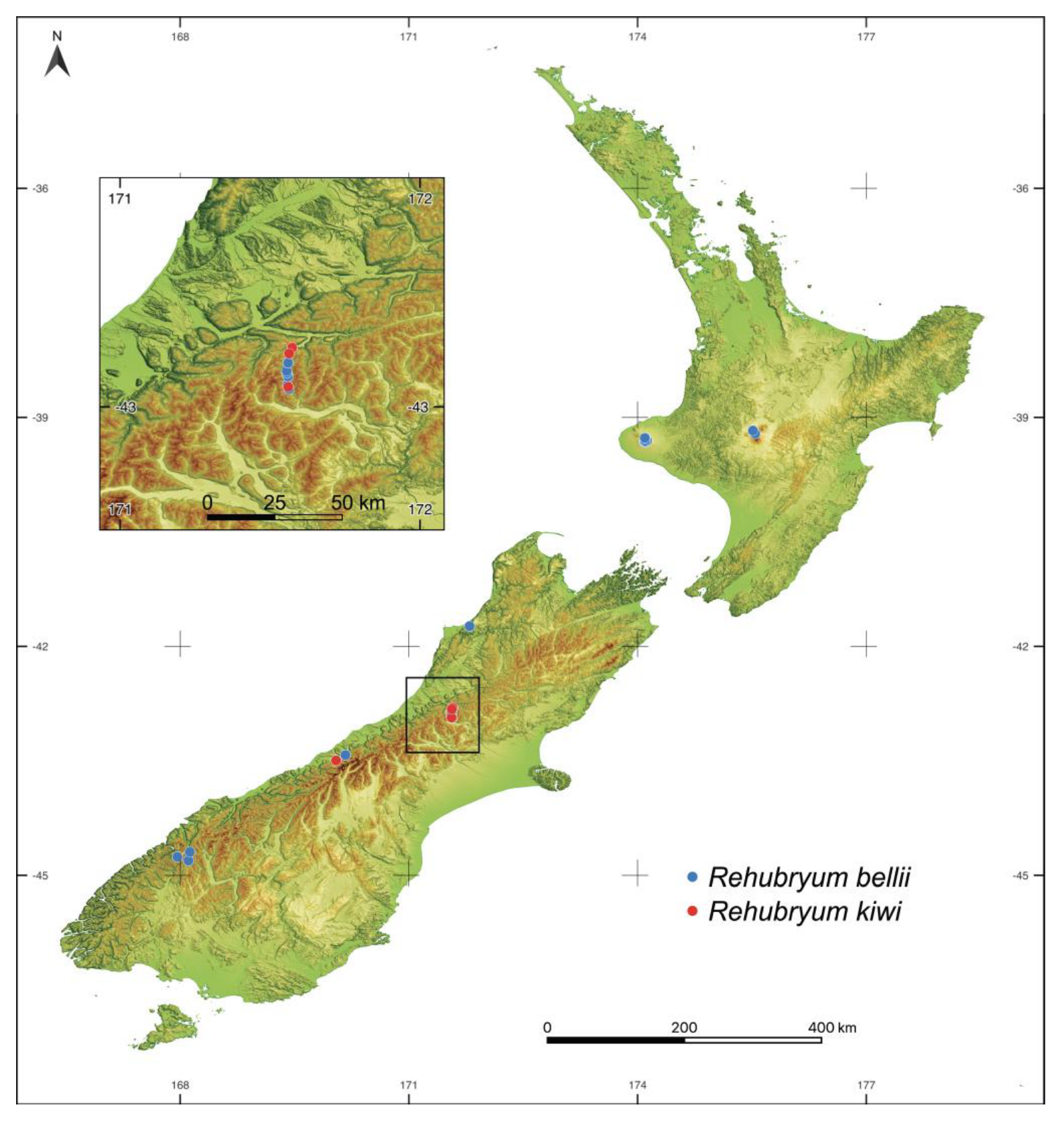
3.2. Phylogeographical Interpretation of Rehubryum Origin
4. Materials and Methods
4.1. Taxon Sampling
4.2. Morphological Analyses
4.3. DNA Extraction
4.4. Target Enrichment and Sequencing Assembly
4.5. Species Tree Inference and Evaluation of Support
4.6. Studied Specimens (Paratypes)
5. Nomenclatural Coda
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brotherus, V.F. Musci (Laubmoose) 2. Hälfte. In Die natürlichen Pflanzenfamilien, nebst ihren Gattungen und wichtigeren Arten, insbesondere den Nutzpflanzen. Zweite Auflage Band 11; Engler, A., Ed.; Wilhelm Engelmann: Leipzig, Germany, 1925. [Google Scholar]
- Schuster, R.M. The Hepaticae and Anthocerotae of North America, East of the Hundredth Meridian; Columbia University Press: New York, NY, USA, 1966–1992; Volume 1–6. [Google Scholar]
- Shaw, A.J.; Szövényi, P.; Shaw, B. Bryophyte Diversity and Evolution: Windows into the Early Evolution of Land Plants. Am. J. Bot. 2011, 98, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Bechteler, J.; Peñaloza-Bojacá, G.; Bell, D.; Gordon Burleigh, J.; McDaniel, S.F.; Christine Davis, E.; Sessa, E.B.; Bippus, A.; Christine Cargill, D.; Chantanoarrapint, S.; et al. Comprehensive Phylogenomic Time Tree of Bryophytes Reveals Deep Relationships and Uncovers Gene Incongruences in the Last 500 Million Years of Diversification. Am. J. Bot. 2023, 110, e16249. [Google Scholar] [CrossRef]
- Li, Y.; Luo, L.; Liu, Y.; He, Q.; Yu, N.; Gaowa, N.; Yi, Z.; Wang, J.; Han, W.; Peng, T.; et al. The Bryophyte Phylogeny Group: A Revised Familial Classification System Based on Plastid Phylogenomic Data. J. Syst. Evol. 2024, 62, 577–588. [Google Scholar] [CrossRef]
- Dhyani, A.; Kasana, S.; Uniyal, P.L. From Barcodes to Genomes: A New Era of Molecular Exploration in Bryophyte Research. Front. Plant Sci. 2025, 15, 1500607. [Google Scholar] [CrossRef] [PubMed]
- Plášek, V.; Sawicki, J.; Ochyra, R.; Szczecińska, M.; Kulik, T. New Taxonomical Arrangement of the Traditionally Conceived Genera Orthotrichum and Ulota (Orthotrichaceae, Bryophyta). Acta Mus. Siles. Sci. Nat. 2015, 64, 169–174. [Google Scholar] [CrossRef]
- Draper, I.; Garilleti, R.; Calleja, J.A.; Flagmeier, M.; Mazimpaka, V.; Vigalondo, B.; Lara, F. Insights into the Evolutionary History of the Subfamily Orthotrichoideae (Orthotrichaceae, Bryophyta): New and Former Supra-Specific Taxa So Far Obscured by Prevailing Homoplasy. Front. Plant Sci. 2021, 12, 629035. [Google Scholar] [CrossRef]
- Draper, I.; Villaverde, T.; Garilleti, R.; Burleigh, J.G.; McDaniel, S.F.; Mazimpaka, V.; Calleja, J.A.; Lara, F. An NGS-Based Phylogeny of Orthotricheae (Orthotrichaceae, Bryophyta) with the Proposal of the New Genus Rehubryum from Zealandia. Front. Plant Sci. 2022, 13, 882960. [Google Scholar] [CrossRef]
- Matanov, N.; Lara, F.; Draper, I.; Calleja, J.A.; Albertos, B.; Garilleti, R. On the Differentiation and Distribution of the Rare New Zealand Endemic Rehubryum bellii (Malta) F.Lara, Garilleti & Draper (Orthotrichaceae, Bryophyta). J. Bryol. 2023, 45, 85–95. [Google Scholar] [CrossRef]
- Breinholt, J.W.; Carey, S.B.; Tiley, G.P.; Davis, E.C.; Endara, L.; McDaniel, S.F.; Neves, L.G.; Sessa, E.B.; von Konrat, M.; Chantanaorrapint, S.; et al. A Target Enrichment Probe Set for Resolving the Flagellate Land Plant Tree of Life. Appl. Plant Sci. 2021, 9, e11406. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Reneau, J.; Gordon Burleigh, J.; Sigel, E.M. Target Capture Methods Offer Insight into the Evolution of Rapidly Diverged Taxa and Resolve Allopolyploid Homeologs in the Fern Genus Polypodium s.s. Syst. Bot. 2023, 48, 96–109. [Google Scholar] [CrossRef]
- Pelosi, J.A.; Zumwalde, B.A.; Testo, W.L.; Kim, E.H.; Burleigh, J.G.; Sessa, E.B. All Tangled up: Unraveling Phylogenetics and Reticulate Evolution in the Vining Ferns, Lygodium (Schizaeales). Am. J. Bot. 2024, 111, e16389. [Google Scholar] [CrossRef] [PubMed]
- Budke, J.M.; Patel, N.R.; Consortium, G.; Wienhold, M.D.; Bruggeman-Nannenga, M.A. Exploring Morphological Evolution in Relation to Habitat Moisture in the Moss Genus Fissidens Using Molecular Data Generated from Herbarium Specimens. J. Syst. Evol. 2022, 61, 868–889. [Google Scholar] [CrossRef]
- Jauregui-Lazo, J.; Brinda, J.C.; Consortium, G.; Mishler, B.D. The Phylogeny of Syntrichia: An Ecologically Diverse Clade of Mosses with an Origin in South America. Am. J. Bot. 2023, 110, e16103. [Google Scholar] [CrossRef]
- Aguado-Ramsay, P.; Villaverde, T.; Garilleti, R.; Burleigh, J.G.; McDaniel, S.F.; Flagmeier, M.; Nieuwkoop, J.; Van Der Pluijm, A.; Hans, F.; Lara, F.; et al. Seeking the Identity of an Enigmatic Moss by Embracing Phylogenomics. J. Syst. Evol. 2024, 62, 979–992. [Google Scholar] [CrossRef]
- Minh, B.Q.; Hahn, M.W.; Lanfear, R. New Methods to Calculate Concordance Factors for Phylogenomic Datasets. Mol. Biol. Evol. 2020, 37, 2727–2733. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.R.; Suárez, G.M.; Hyvönen, J. Reassessing the Role of Morphology in Bryophyte Phylogenetics: Combined Data Improves Phylogenetic Inference Despite Character Conflict. Mol. Phylogenetics Evol. 2020, 143, 106662. [Google Scholar] [CrossRef]
- Goffinet, B.; Vitt, D.H. Revised Generic Classification of the Orthotrichaceae Based on a Molecular Phylogeny and Comparative Morphology. In Bryology for the Twenty-First Century; Bates, J.W., Ashton, N.W., Duckett, J.G., Eds.; Maney Publishing and the British Bryological Society: Leeds, UK, 1998; pp. 143–160. [Google Scholar]
- Huttunen, S.; Ignatov, M.S. Phylogeny of the Brachytheciaceae (Bryophyta) Based on Morphology and Sequence Level Data. Cladistics 2004, 20, 151–183. [Google Scholar] [CrossRef]
- Liu, Y.; Budke, J.M.; Goffinet, B. Phylogenetic Inference Rejects Sporophyte Based Classification of the Funariaceae (Bryophyta): Rapid Radiation Suggests Rampant Homoplasy in Sporophyte Evolution. Mol. Phylogenetics Evol. 2012, 62, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Caparrós, R.; Garilleti, R.; Lara, F. Ulota D.Mohr. In Flora Briofítica Ibérica, vol. V. Orthotrichales: Orthotrichaceae; Hedwigiales: Hedwigiaceae; Leucodontales: Fontinalaceae, Climaciaceae, Anomodontaceae, Cryphaeaceae, Leptodontaceae, Leucodontaceae, Neckeraceae; Hookeriales: Hypopterygiaceae, Hookeriaceae, Leucomiaceae, Pilotrichaceae; Guerra, J., Cano, M.J., Brugués, M., Eds.; Universidad de Murcia & Sociedad Española de Briología: Murcia, Spain, 2014; pp. 34–50. [Google Scholar]
- Garilleti, R.; Lara, F.; Albertos, B.; Mazimpaka, V. Peristomal Ornamentation, a Precise Character for Discrimination of Ulota bruchii and U. crispa (Bryopsida, Orthotrichaceae). J. Bryol. 2000, 22, 273–278. [Google Scholar] [CrossRef]
- Garilleti, R.; Mazimpaka, V.; Lara, F. New Ulota Species with Multicellular Spores from Southern South America. Bryologist 2012, 115, 585–600. [Google Scholar] [CrossRef]
- Garilleti, R.; Albertos, B.; Draper, I.; Calleja, J.A.; Mazimpaka, V.; Lara, F. Two Complex Typifications and a New Name to Unravel Ulota germana Sensu Malta Non (Mont.) Mitt. (Orthotrichaceae). Bryologist 2020, 123, 163–178. [Google Scholar] [CrossRef]
- Caparrós, R.; Lara, F.; Draper, I.; Mazimpaka, V.; Garilleti, R. Integrative Taxonomy Sheds Light on an Old Problem: The Ulota crispa Complex (Orthotrichaceae, Musci). Bot. J. Linn. Soc. 2016, 180, 427–451. [Google Scholar] [CrossRef]
- Matanov, N.; Draper, I.; Calleja, J.A.; Flagmeier, M.; Lara, F.; Garilleti, R. One More Word on Patagonian Ulota macrodontia: Ulota brachypoda sp. nov. (Orthotrichaceae, Bryophyta). Bot. J. Linn. Soc. 2024, 206, 201–213. [Google Scholar] [CrossRef]
- Lara, F.; Draper, I.; Flagmeier, M.; Calleja, J.A.; Mazimpaka, V.; Garilleti, R. Let ’s Make Pulvigera Great Again: Re-Circumscription of a Misunderstood Group of Orthotrichaceae That Diversified in North America. Bot. J. Linn. Soc. 2020, 193, 180–206. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental Niche Equivalency Versus Conservatism: Quantitative Approaches to Niche Evolution. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef]
- Hurtado, F.; Gonçalves, J.; Hespanhol, H.; Ronquillo, C.; Estébanez, B.; Aragón, P.; Medina, N.G.; Hortal, J. Macroclimatic Niche Similarity and Species Relatedness Shift Their Influence on Species Co-Occurrence in Bryophyte Forest Communities Across Scales. J. Ecol. 2025, 113, 1546–1559. [Google Scholar] [CrossRef]
- Vigalondo, B.; Garilleti, R.; Vanderpoorten, A.; Patiño, J.; Draper, I.; Calleja, J.A.; Mazimpaka, V.; Lara, F. Do Mosses Really Exhibit So Large Distribution Ranges? Insights from the Integrative Taxonomic Study of the Lewinskya affinis Complex (Orthotrichaceae, Bryopsida). Mol. Phylogenetics Evol. 2019, 140, 106598. [Google Scholar] [CrossRef]
- Vigalondo, B.; Draper, I.; Mazimpaka, V.; Calleja, J.A.; Garilleti, R. The Lewinskya affinis Complex (Orthotrichaceae) Revisited: Species Description and Differentiation. Bryologist 2020, 123, 454–481. [Google Scholar] [CrossRef]
- Rundell, R.J.; Price, T.D. Adaptive Radiation, Nonadaptive Radiation, Ecological Speciation and Nonecological Speciation. Trends Ecol. Evol. 2009, 24, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Natcheva, R.; Cronberg, N. What Do We Know About Hybridization Among Bryophytes in Nature? Can. J. Bot. 2004, 82, 1687–1704. [Google Scholar] [CrossRef]
- Glime, J.M.; Bisang, I. Sexuality: Reproductive Barriers and Tradeoffs. In Bryophyte Ecology; Glime, J.M., Ed.; Volume 1. Physiological Ecology; Michigan Technological University and International Association of Bryologist: Houghton, MI, USA, 2017; pp. 3-4-1–3-4-33. [Google Scholar]
- Rasmussen, M.D.; Kellis, M. Unified Modeling of Gene Duplication, Loss, and Coalescence Using a Locus Tree. Genome Res. 2012, 22, 755–765. [Google Scholar] [CrossRef]
- Salichos, L.; Rokas, A. Inferring Ancient Divergences Requires Genes with Strong Phylogenetic Signals. Nature 2013, 497, 327–331. [Google Scholar] [CrossRef]
- Morales-Briones, D.F.; Kadereit, G.; Tefarikis, D.T.; Moore, M.J.; Smith, S.A.; Brockington, S.F.; Timoneda, A.; Yim, W.C.; Cushman, J.C.; Yang, Y. Disentangling Sources of Gene Tree Discordance in Phylogenomic Data Sets: Testing Ancient Hybridizations in Amaranthaceae s.l. Syst. Biol. 2021, 70, 219–235. [Google Scholar] [CrossRef]
- Mirarab, S.; Bayzid, M.S.; Warnow, T. Evaluating Summary Methods for Multilocus Species Tree Estimation in the Presence of Incomplete Lineage Sorting. Syst. Biol. 2016, 65, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Scornavacca, C.; Galtier, N. Incomplete Lineage Sorting in Mammalian Phylogenomics. Syst. Biol. 2017, 66, 112–120. [Google Scholar] [CrossRef]
- Stull, G.W.; Pham, K.K.; Soltis, P.S.; Soltis, D.E. Deep Reticulation: The Long Legacy of Hybridization in Vascular Plant Evolution. Plant J. 2023, 114, 743–766. [Google Scholar] [CrossRef]
- Hibbins, M.S.; Hahn, M.W. Phylogenomic Approaches to Detecting and Characterizing Introgression. Genetics 2022, 220, iyab173. [Google Scholar] [CrossRef] [PubMed]
- He, G.-H.; Meng, Y.; Zhang, M.-H.; Wang, D.; Meng, R.; Zhang, L.; Chu, Z.-F.; Wen, J.; Nie, Z.-L. Extensive Genome-Wide Phylogenetic Discordance Is Due to Incomplete Lineage Sorting in the Rapidly Radiated East Asian Genus Nekemias (Vitaceae). Ann. Bot. 2025, 135, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Meleshko, O.; Martin, M.D.; Korneliussen, T.S.; Schröck, C.; Lamkowski, P.; Schmutz, J.; Healey, A.; Piatkowski, B.T.; Shaw, A.J.; Weston, D.J.; et al. Extensive Genome-Wide Phylogenetic Discordance Is Due to Incomplete Lineage Sorting and Not Ongoing Introgression in a Rapidly Radiated Bryophyte Genus. Mol. Biol. Evol. 2021, 38, 2750–2766. [Google Scholar] [CrossRef]
- Lara, F.; Porley, R.D.; Draper, I.; Aleffi, M.; Garcia, C.; Garilleti, R. Ulota s.l. (Orthotrichaceae, Bryidae) at Southernmost Mediterranean Localities: Not a Simple Matter. Plant Biosyst. 2022, 156, 1448–1455. [Google Scholar] [CrossRef]
- Muñoz, J.; Felicísimo, Á.M.; Cabezas, F.; Burgaz, A.R.; Martínez, I. Wind as a Long-Distance Dispersal Vehicle in the Southern Hemisphere. Science 2004, 304, 1144–1147. [Google Scholar] [CrossRef]
- Schuster, R.M. Phytogeography of the Bryophyta. In New Manual of Bryology; The Hattori Botanical Laboratory: Nichinan, Japan, 1983; Volume 1, pp. 463–626. ISBN 4-938163-3045. [Google Scholar]
- Vanderpoorten, A.; Goffinet, B. Introduction to Bryophytes; Cambridge University Press: Cambridge, UK, 2009; ISBN 978-0-521-87712-1. [Google Scholar]
- Biersma, E.M.; Jackson, J.A.; Hyvönen, J.; Koskinen, S.; Linse, K.; Griffiths, H.; Convey, P. Global Biogeographic Patterns in Bipolar Moss Species. R. Soc. Open Sci. 2017, 4, 170147. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.R.; Biersma, E.M.; Carey, S.B.; Holsinger, K.; McDaniel, S.F.; Rozzi, R.; Goffinet, B. Resolving the Northern Hemisphere Source Region for the Long-Distance Dispersal Event That Gave Rise to the South American Endemic Dung Moss Tetraplodon Fuegianus. Am. J. Bot. 2017, 104, 1651–1659. [Google Scholar] [CrossRef]
- Schofield, W.B. Bipolar Disjunctive Mosses in the Southern Hemisphere, with Particular Reference to New Zealand. J. Hattori Bot. Lab. 1974, 38, 13–32. [Google Scholar]
- Ochyra, R.; Buck, W.R. Arctoa Fulvella, New to Tierra Del Fuego, with Notes on Trans-American Bipolar Bryogeography. Bryologist 2003, 106, 532–538. [Google Scholar] [CrossRef]
- Sanmartín, I.; Anderson, C.L.; Alarcon, M.; Ronquist, F.; Aldasoro, J.J. Bayesian Island Biogeography in a Continental Setting: The Rand Flora Case. Biol. Lett. 2010, 6, 703–707. [Google Scholar] [CrossRef]
- van Zanten, B.O. Possibilities of Long-Range Dispersal in Bryophytes with Special Reference to the Southern Hemisphere. Sonderb. Naturwiss Ver. Hambg. 1983, 7, 49–64. [Google Scholar]
- Sanmartín, I.; Wanntorp, L.; Winkworth, R.C. West Wind Drift Revisited: Testing for Directional Dispersal in the Southern Hemisphere Using Event-Based Tree Fitting. J. Biogeogr. 2007, 34, 398–416. [Google Scholar] [CrossRef]
- Wilkinson, D.M.; Koumoutsaris, S.; Mitchell, E.A.D.; Bey, I. Modelling the Effect of Size on the Aerial Dispersal of Microorganisms. J. Biogeogr. 2012, 39, 89–97. [Google Scholar] [CrossRef]
- Biersma, E.M.; Convey, P.; Wyber, R.; Robinson, S.A.; Dowton, M.; van de Vijver, B.; Linse, K.; Griffiths, H.; Jackson, J.A. Latitudinal Biogeographic Structuring in the Globally Distributed Moss Ceratodon purpureus. Front. Plant Sci. 2020, 11, 502359. [Google Scholar] [CrossRef] [PubMed]
- Wardle, P. Origin of the New Zealand Mountain Flora, with Special Reference to Trans-Tasman Relationships. N. Z. J. Bot. 1978, 16, 535–550. [Google Scholar] [CrossRef]
- Sanmartín, I.; Ronquist, F. Southern Hemisphere Biogeography Inferred by Event-Based Models: Plant versus Animal Patterns. Syst. Biol. 2004, 53, 216–243. [Google Scholar] [CrossRef]
- Axelrod, D.I.; Raven, P.H. Late Cretaceous and Tertiary Vegetation History of Africa. In Biogeography and Ecology of Southern Africa; Werger, M.J.A., Ed.; Springer: Dordrecht, The Netherlands, 1978; pp. 77–130. ISBN 978-94-009-9951-0. [Google Scholar]
- Plana, V. Mechanisms and Tempo of Evolution in the African Guineo–Congolian Rainforest. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1585–1594. [Google Scholar] [CrossRef]
- Wallis, G.P.; Trewick, S.A. New Zealand Phylogeography: Evolution on a Small Continent. Mol. Ecol. 2009, 18, 3548–3580. [Google Scholar] [CrossRef]
- McGlone, M.S.; Duncan, R.P.; Heenan, P.B. Endemism, Species Selection and the Origin and Distribution of the Vascular Plant Flora of New Zealand. J. Biogeogr. 2001, 28, 199–216. [Google Scholar] [CrossRef]
- McGlone, M.S. Goodbye Gondwana. J. Biogeogr. 2005, 32, 739–740. [Google Scholar] [CrossRef]
- McCarthy, P.M. (Ed.) Flora of Australia Volume 51 (Mosses 1); ABRS and CSIRO Publishing: Canberra, Australia; Melbourne, Australia, 2006; ISBN 0-643-09240-4. [Google Scholar]
- Lara, F.; Draper, I.; Garilleti, R. New Insights into the Genus Stoneobryum (Bryophyta: Orthotrichaceae) Based on Recent Collections of the Two Known Species. Bryophyt. Divers. Evol. 2023, 46, 92–108. [Google Scholar] [CrossRef]
- Lara, F.; Garilleti, R.; Mazimpaka, V.; Guerra, J. (Eds.) Orthotrichaceae. In Flora Briofítica Ibérica, vol. V. Orthotrichales: Orthotrichaceae; Hedwigiales: Hedwigiaceae; Leucodontales: Fontinalaceae, Climaciaceae, Anomodontaceae, Cryphaeaceae, Leptodontaceae, Leucodontaceae, Neckeraceae; Hookeriales: Hypopterygiaceae, Hookeriaceae, Leucomiaceae, Pilotrichaceae; Universidad de Murcia & Sociedad Española de Briología: Murcia, Spain, 2014; pp. 15–135. [Google Scholar]
- Vigalondo, B.; Lara, F.; Draper, I.; Valcárcel, V.; Garilleti, R.; Mazimpaka, V. Is It Really You, Orthotrichum acuminatum? Ascertaining a New Case of Intercontinental Disjunction in Mosses. Bot. J. Linn. Soc. 2016, 180, 30–49. [Google Scholar] [CrossRef]
- ORTHOTREE Orthotrichaceae. List of Used Taxonomical Characters. Available online: https://www.orthotree.net/orthotrichaceae/ (accessed on 14 February 2024).
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Krueger, F. Trim Galore! version 0.6.10; Computer Software; Babraham Bioinformatics: Cambridge, UK, 2019. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 20 July 2022).
- Breinholt, J.W.; Earl, C.; Lemmon, A.R.; Lemmon, E.M.; Xiao, L.; Kawahara, A.Y. Resolving Relationships Among the Megadiverse Butterflies and Moths with a Novel Pipeline for Anchored Phylogenomics. Syst. Biol. 2018, 67, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Chang, Z.; Li, G.; Liu, J.; Zhang, Y.; Ashby, C.; Liu, D.; Cramer, C.L.; Huang, X. Bridger: A New Framework for de Novo Transcriptome Assembly Using RNA-Seq Data. Genome Biol. 2015, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Borowiec, M.L. AMAS: A Fast Tool for Alignment Manipulation and Computing of Summary Statistics. PeerJ 2016, 4, e1660. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Package Version 1.0.13; Comprehensive R Archive Network (CRAN): Vienna, Austria, 2025; Available online: https://github.com/raivokolde/pheatmap (accessed on 22 March 2025).
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial Time Species Tree Reconstruction from Partially Resolved Gene Trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef]
- Sayyari, E.; Mirarab, S. Fast Coalescent-Based Computation of Local Branch Support from Quartet Frequencies. Mol. Biol. Evol. 2016, 33, 1654–1668. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]

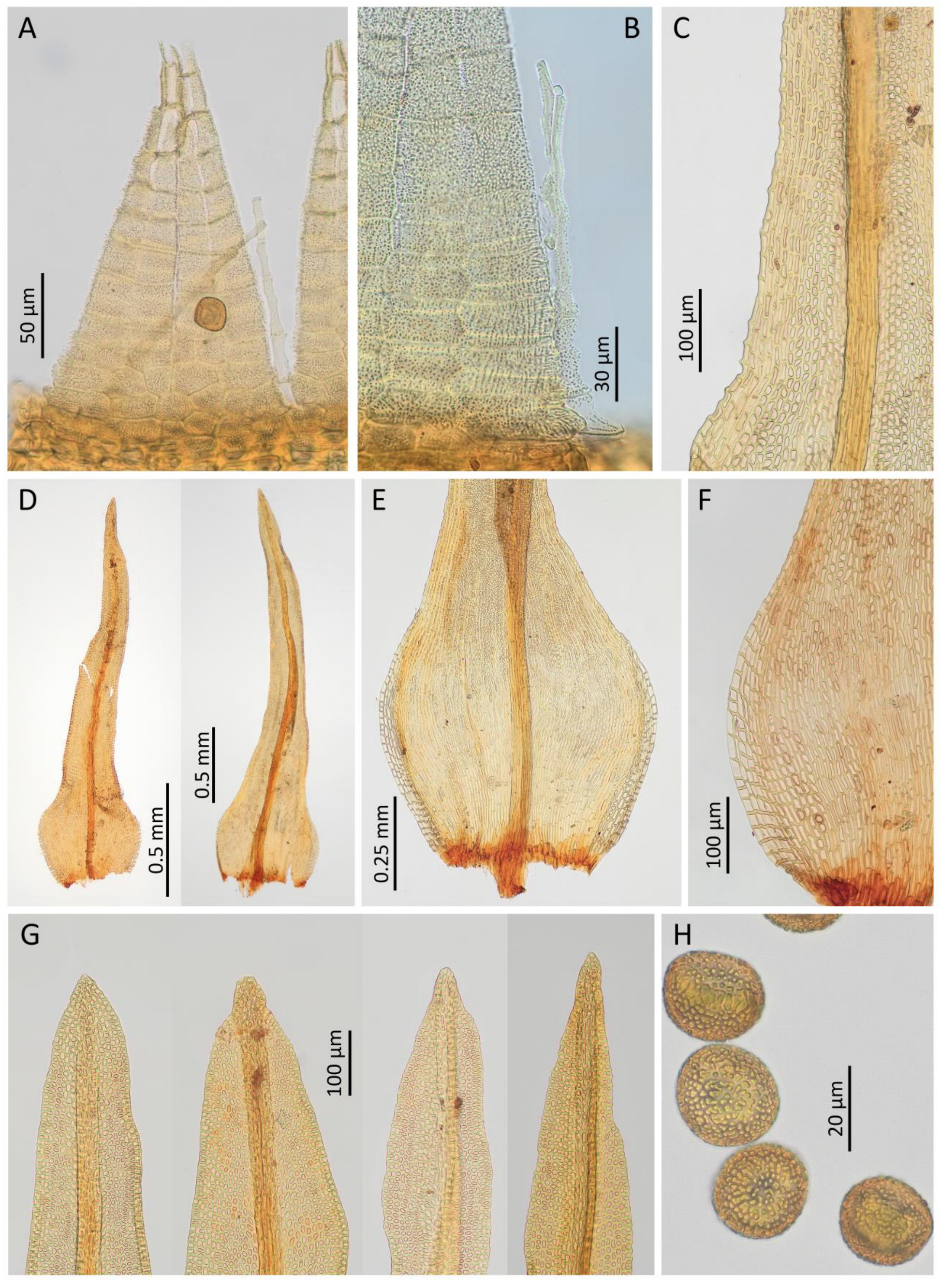

| Rehubryum bellii | Rehubryum kiwi | Atlantichella calvescens | |
|---|---|---|---|
| Capsule shape when dry | Short cylindrical to obovoid, not strongly furrowed | Short cylindrical to obovoid, not strongly furrowed | Cylindrical, constricted below mouth, and strongly furrowed |
| Exothecial bands | Mostly 4 cell rows, reaching the capsule mouth, almost concolor with the exothecial cells | Mostly 2 cell rows, reaching 3–4 rows of suboral cells, darker than the exothecial cells | Mostly 4 cell rows, reaching the capsule mouth, darker than the exothecial cells |
| Operculum basal rim | Orange to red, not undulate | Crimson, broad, not undulate | Undifferentiated or orangish, undulate |
| Exostome | Eight pairs of teeth, almost completely divided into sixteen in mature capsules | Eight pairs of teeth remaining unsplit | Eight pairs of teeth, with tendency to completely split into sixteen |
| Endostome segments | Sixteen, filiform and fragile, shorter than the exostome teeth | Eight, filiform and fragile, shorter than the exostome teeth | Eight, filiform with widened base, as long as the exostome teeth |
| Endostome connecting membrane | High and protruding mouth | Low, hardly noticeable | Low, frequently incomplete, hardly noticeable |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matanov, N.; Lara, F.; Calleja, J.A.; Draper, I.; Aguado-Ramsay, P.; Garilleti, R. Phylogenetic Insights from a Novel Rehubryum Species Challenge Generic Boundaries in Orthotrichaceae. Plants 2025, 14, 2373. https://doi.org/10.3390/plants14152373
Matanov N, Lara F, Calleja JA, Draper I, Aguado-Ramsay P, Garilleti R. Phylogenetic Insights from a Novel Rehubryum Species Challenge Generic Boundaries in Orthotrichaceae. Plants. 2025; 14(15):2373. https://doi.org/10.3390/plants14152373
Chicago/Turabian StyleMatanov, Nikolay, Francisco Lara, Juan Antonio Calleja, Isabel Draper, Pablo Aguado-Ramsay, and Ricardo Garilleti. 2025. "Phylogenetic Insights from a Novel Rehubryum Species Challenge Generic Boundaries in Orthotrichaceae" Plants 14, no. 15: 2373. https://doi.org/10.3390/plants14152373
APA StyleMatanov, N., Lara, F., Calleja, J. A., Draper, I., Aguado-Ramsay, P., & Garilleti, R. (2025). Phylogenetic Insights from a Novel Rehubryum Species Challenge Generic Boundaries in Orthotrichaceae. Plants, 14(15), 2373. https://doi.org/10.3390/plants14152373








