Transcription Factor LjWRKY50 Affects Jasmonate-Regulated Floral Bud Duration in Lonicera japonica
Abstract
1. Introduction
2. Results
2.1. Exogenous MeJA Accelerates Floral Bud Opening in L. japonica
2.2. Prolonged Floral Bud Duration Trait Is Preferred in Artificial Breeding
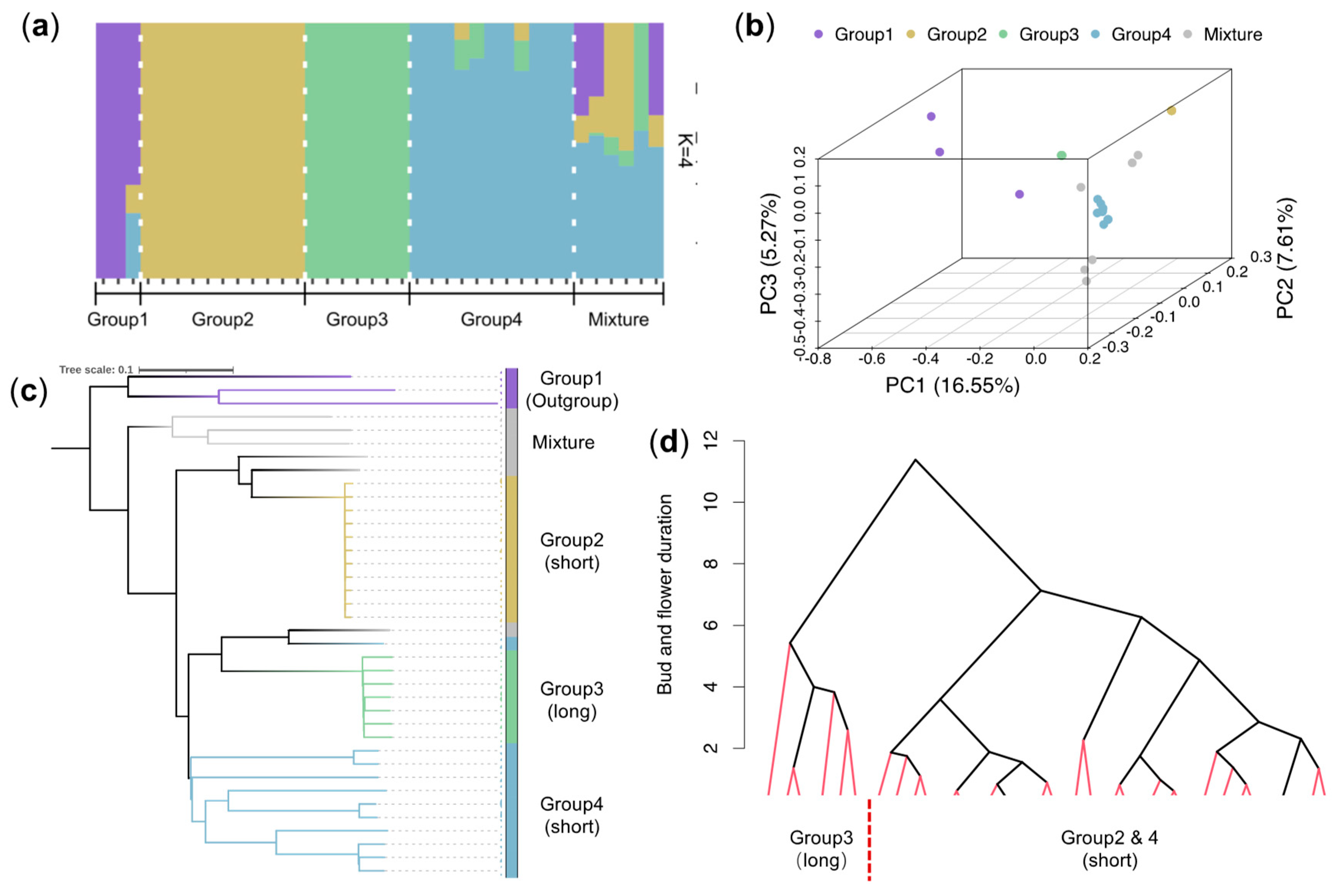
2.3. LjWRKY50 Is Associated with the Prolonged Duration of Floral Buds
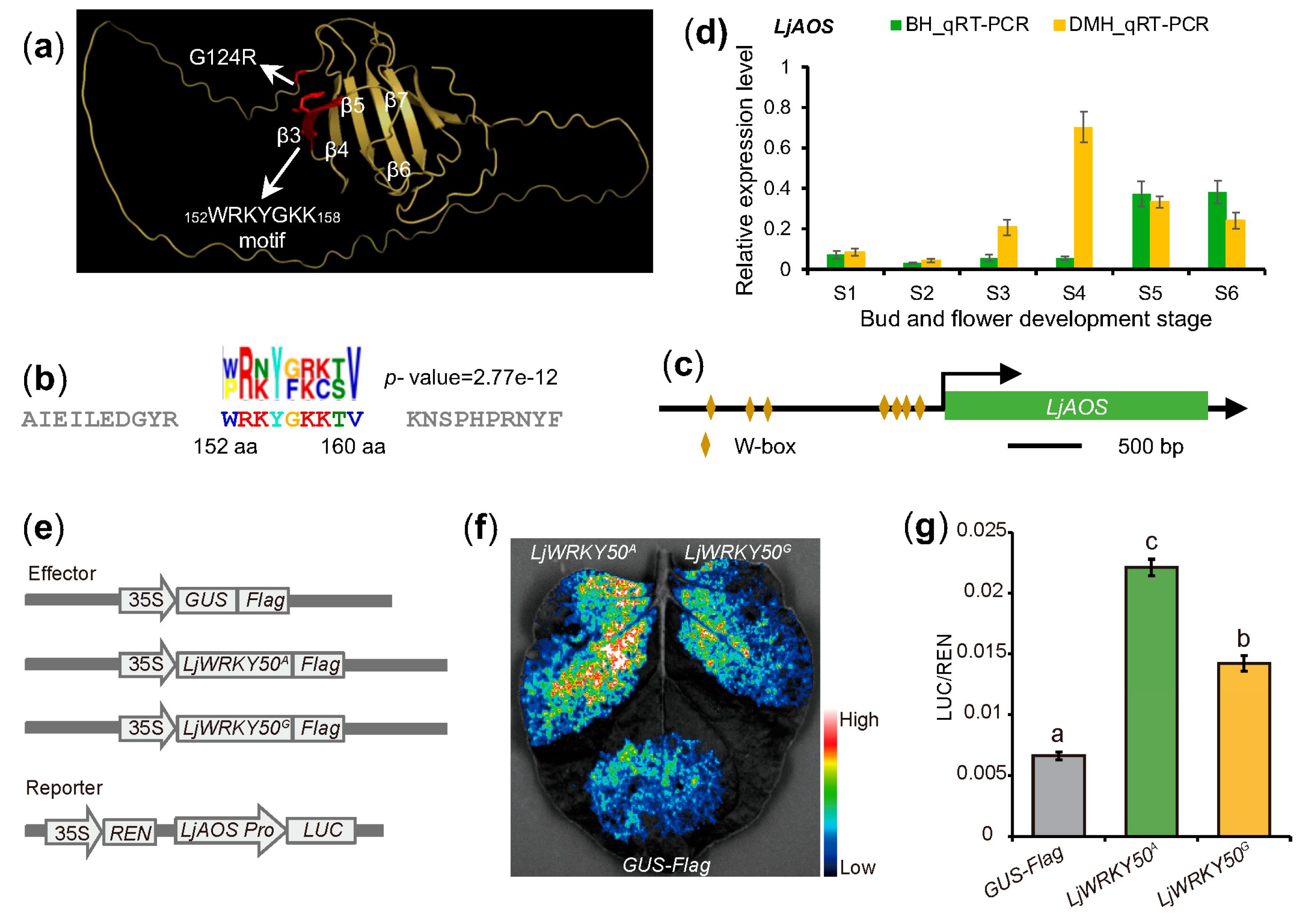
2.4. LjWRKY50 Influences Jasmonate Biosynthesis in L. japonica
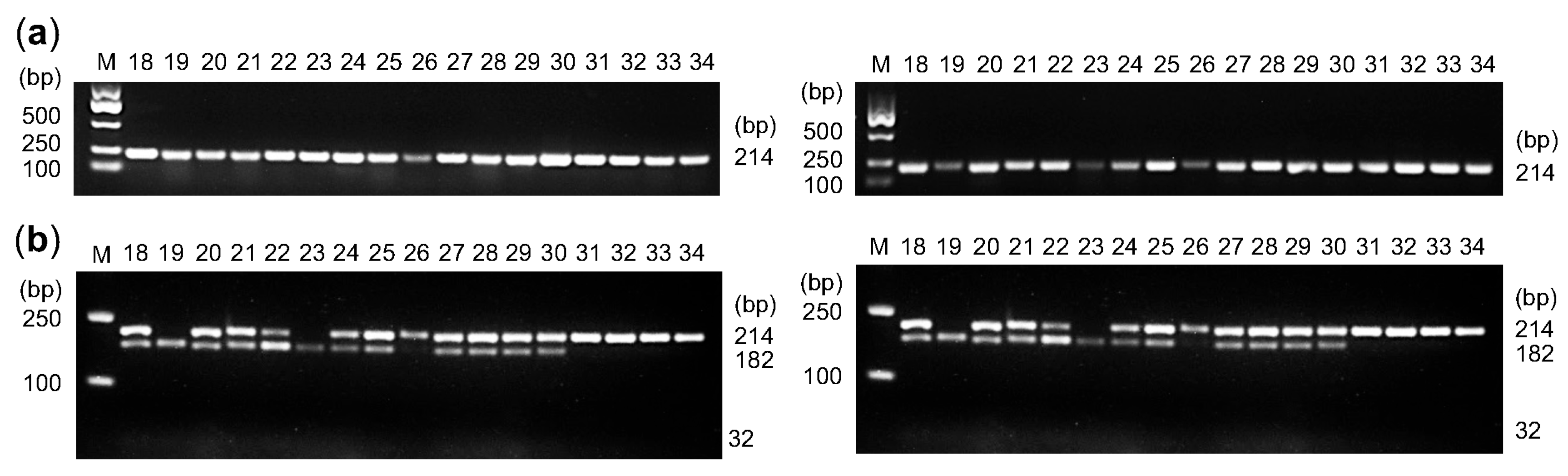
2.5. A dCAPS Marker Is Developed to Identify Germplasms with Prolonged Floral Bud Duration
3. Discussion
4. Materials and Methods
4.1. Phenotype Investigation and Analyses
4.2. Exogenous MeJA Treatment
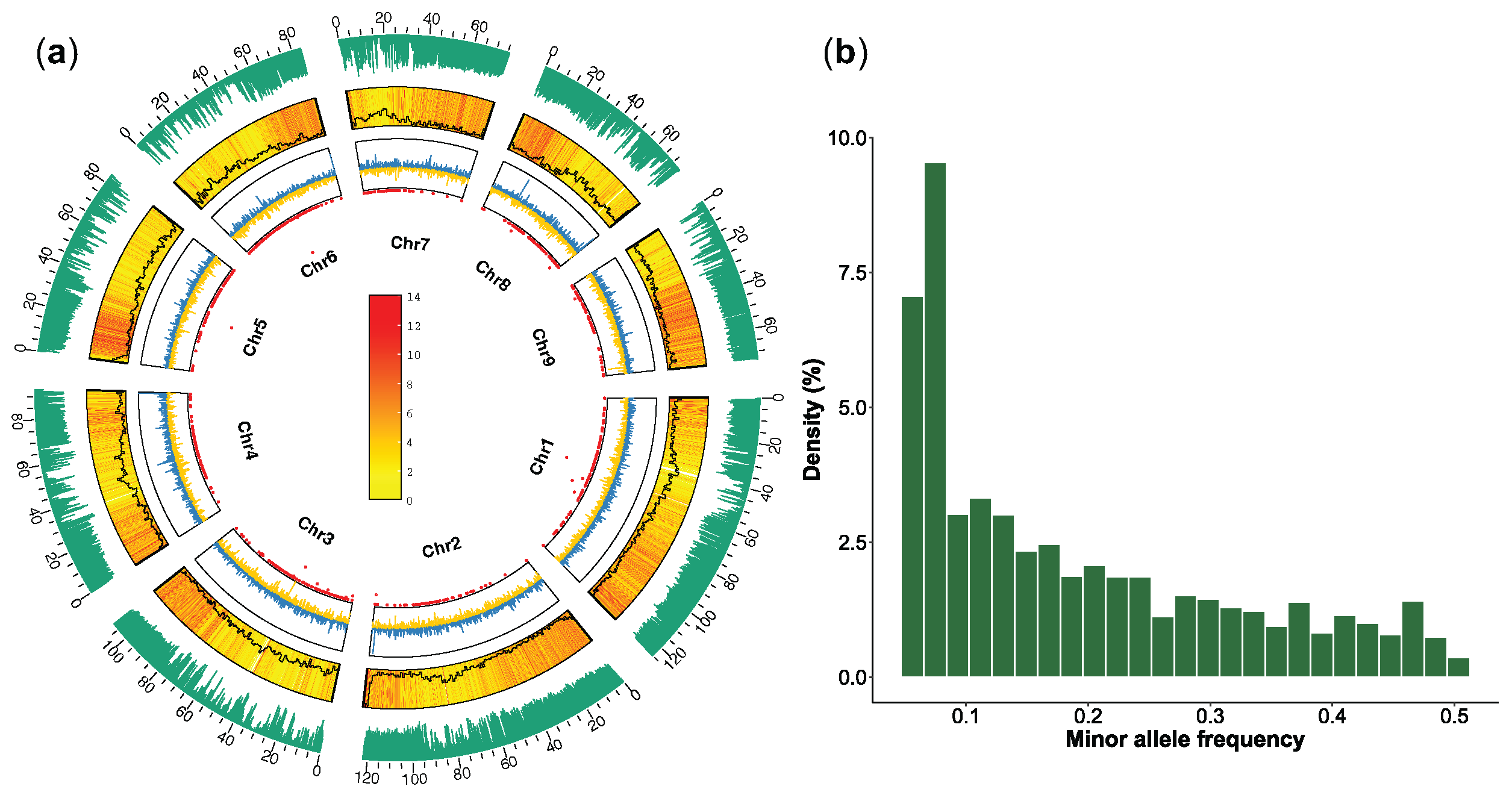
4.3. Whole Genome Resequencing, Read Alignment, and Variant Calling
4.4. Population Structure Analysis
4.5. Nucleotide Diversity and Selective Sweep Detection
4.6. Quantitative Real-Time PCR
4.7. Candidate Gene Analyses
4.8. Dual-Luciferase Reporter Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| L. japonica | Lonicera japonica Thunb. |
| S1 | The juvenile floral bud stage |
| S2 | The three-green floral bud stage |
| S3 | The two-white floral bud stage |
| S4 | The complete white floral bud stage |
| S5 | The silver flower stage |
| S6 | The gold flower stage |
| S7 | The withering flower stage |
| XP-CLR | Cross-population composite likelihood ratio |
| PCA | Principal component analysis |
| MeJA | Methyl jasmonate |
| LUC | Firefly luciferase |
| dCAPS | Derived cleaved amplified polymorphic sequence |
| MAS | Marker-assisted selection |
References
- Schierenbeck, K.A. Japanese Honeysuckle (Lonicera japonica) as an Invasive Species; History, Ecology, and Context. Crit. Rev. Plant Sci. 2004, 23, 391–400. [Google Scholar] [CrossRef]
- Pu, X.; Li, Z.; Tian, Y.; Gao, R.; Hao, L.; Hu, Y.; He, C.; Sun, W.; Xu, M.; Peters, R.J.; et al. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol. 2020, 227, 930–943. [Google Scholar] [CrossRef]
- Shang, X.; Pan, H.; Li, M.; Miao, X.; Ding, H. Lonicera japonica Thunb.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2011, 138, 1–21. [Google Scholar] [CrossRef]
- Tang, X.; Liu, X.; Zhong, J.; Fang, R. Potential Application of Lonicera japonica Extracts in Animal Production: From the Perspective of Intestinal Health. Front. Microbiol. 2021, 12, 719877. [Google Scholar] [CrossRef]
- Ge, L.; Xie, Q.; Jiang, Y.; Xiao, L.; Wan, H.; Zhou, B.; Wu, S.; Tian, J.; Zeng, X. Genus Lonicera: New drug discovery from traditional usage to modern chemical and pharmacological research. Phytomedicine 2022, 96, 153889. [Google Scholar] [CrossRef]
- Zhu, J.; Jia, Y.; Wang, C.; Zhou, W.; Shu, Y.; Zhang, K.; Zeng, X.; Guo, R. Lonicera japonica polysaccharides improve longevity and fitness of Caenorhabditis elegans by activating DAF-16. Int. J. Biol. Macromol. 2023, 229, 81–91. [Google Scholar] [CrossRef]
- Li, J.; Ye, C.; Chang, C. Comparative transcriptomics analysis revealing flower trichome development during flower development in two Lonicera japonica Thunb. cultivars using RNA-seq. BMC Plant Biol. 2020, 20, 341. [Google Scholar] [CrossRef]
- Yang, B.; Zhong, Z.; Wang, T.; Ou, Y.; Tian, J.; Komatsu, S.; Zhang, L. Integrative omics of Lonicera japonica Thunb. Flower development unravels molecular changes regulating secondary metabolites. J. Proteomics 2019, 208, 103470. [Google Scholar] [CrossRef]
- Guo, X.; Ma, M.; Ma, X.; Zhao, J.; Zhang, Y.; Wang, X.; Li, S.; Yu, Y. Quality assessment for the flower of Lonicera japonica Thunb. during flowering period by integrating GC-MS, UHPLC-HRMS, and chemometrics. Ind. Crops Prod. 2023, 191, 115938. [Google Scholar] [CrossRef]
- Kou, Y.-N.; Zhang, X.; Kong, F.-K.; Dong, H.-J.; Yang, J.; Wang, W.-H.; Kong, D.-Z.; Yang, B. Content of secondary metabolites and antioxidant activities of Lonicera japonica flowers at different developmental stages from Shandong province. China J. Chin. Mater. Medica 2024, 49, 2654–2665. [Google Scholar]
- Yu, X.; Yu, H.; Lu, Y.; Zhang, C.; Wang, H. Genetic and epigenetic variations underlying flavonoid divergence in Beihua and Sijihua honeysuckles. Epigenet. Insights 2024, 17, e002. [Google Scholar] [CrossRef]
- van Doorn, W.G.; Van Meeteren, U. Flower opening and closure: A review. J. Exp. Bot. 2003, 54, 1801–1812. [Google Scholar] [CrossRef]
- Ochiai, M.; Matsumoto, S.; Yamada, K. Methyl jasmonate treatment promotes flower opening of cut Eustoma by inducing cell wall loosening proteins in petals. Postharvest Biol. Technol. 2013, 82, 1–5. [Google Scholar] [CrossRef]
- Niwa, T.; Suzuki, T.; Takebayashi, Y.; Ishiguro, R.; Higashiyama, T.; Sakakibara, H.; Ishiguro, S. Jasmonic acid facilitates flower opening and floral organ development through the upregulated expression of SlMYB21 transcription factor in tomato. Biosci. Biotechnol. Biochem. 2018, 82, 292–303. [Google Scholar] [CrossRef]
- Ding, W.; Gou, Y.; Li, Y.; Li, J.; Fang, Y.; Liu, X.; Zhu, X.; Ye, R.; Heng, Y.; Wang, H.; et al. A jasmonate-mediated regulatory network modulates diurnal floret opening time in rice. New Phytol. 2024, 244, 176–191. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, X.; Huang, Z.; Chen, M.; Xu, P.; Liao, S.; Zhao, Y.; Gao, Y.; He, J.; Luo, Y.; et al. Controlling diurnal flower-opening time by manipulating the jasmonate pathway accelerates development of Indica-japonica hybrid rice breeding. Plant Biotechnol. J. 2024, 22, 2267–2281. [Google Scholar] [CrossRef]
- Cebrian, G.; Segura, M.; Martinez, J.; Iglesias-Moya, J.; Martinez, C.; Garrido, D.; Jamilena, M. Jasmonate-deficient mutant lox3a reveals crosstalk between jasmonate and ethylene in the differential regulation of male and female flower opening and early fruit development in Cucurbita pepo. J. Exp. Bot. 2023, 74, 1258–1274. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Lyu, X.; Li, Y.H.; Li, Y.; Li, D.; Han, C.; Hong, H.; Tian, Y.; Han, L.; Liu, B.; Qiu, L.J. The domestication-associated L1 gene encodes a eucomic acid synthase pleiotropically modulating pod pigmentation and shattering in soybean. Mol. Plant 2023, 16, 1178–1191. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, M.; Liu, Y.; Liu, J.; Li, W.; Chen, G.; Peng, Y.; Jin, M.; Wei, W.; Jian, L.; et al. Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol. 2022, 234, 513–526. [Google Scholar] [CrossRef]
- Wang, L.; Chen, H.; Chen, G.; Luo, G.; Shen, X.; Ouyang, B.; Bie, Z. Transcription factor SlWRKY50 enhances cold tolerance in tomato by activating the jasmonic acid signaling. Plant Physiol. 2024, 194, 1075–1090. [Google Scholar] [CrossRef]
- Gao, Z.; Yang, X.; Chen, J.; Rausher, M.D.; Shi, T. Expression inheritance and constraints on cis- and trans-regulatory mutations underlying lotus color variation. Plant Physiol. 2022, 191, 1662–1683. [Google Scholar] [CrossRef]
- Yvert, G.; Brem, R.B.; Whittle, J.; Akey, J.M.; Foss, E.; Smith, E.N.; Mackelprang, R.; Kruglyak, L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 2003, 35, 57–64. [Google Scholar] [CrossRef]
- Gerke, J.; Lorenz, K.; Cohen, B. Genetic Interactions Between Transcription Factors Cause Natural Variation in Yeast. Science 2009, 323, 498–501. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low Oleic Acid-Derived Repression of Jasmonic Acid-Inducible Defense Responses Requires the WRKY50 and WRKY51 Proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechn. 2021, 19, 128. [Google Scholar] [CrossRef]
- Guo, S.; Li, Y.; Qiu, H.; Hu, G.; Zhao, C.; Wang, R.; Zhang, H.; Tian, Y.; Li, X.; Liu, B.; et al. GmAP1d regulates flowering time under long-day photoperiods in soybean. Crop J. 2024, 12, 845–855. [Google Scholar] [CrossRef]
- Singh, J.; Saeedan, A.S.; Kaithwas, G.; Ansari, M.N. Small interfering RNA: From designing to therapeutic in cancer. J. Genet. Eng. Biotechn. 2025, 23, 100484. [Google Scholar] [CrossRef]
- Ma, J.; Wang, X.-q.; Ni, H.-f.; Huang, T.-y.; Yang, B.-x. GRAS transcription factors mediate flowering through signaling pathways of gibberellin and circadian rhythm in Lonicera japonica Thunb. Plant Gene 2021, 28, 100340. [Google Scholar] [CrossRef]
- Wang, X.; Kong, L.; Cheng, H.; Li, Q.; Wu, L.; Xu, Z.; Mi, Y.; Xiang, L.; Gao, R. Characterization and expression analysis of the MADS-box gene family in Lonicera japonica reveals the role of LjMADS36 in flower coloration. Ind. Crops Prod. 2024, 219, 119122. [Google Scholar] [CrossRef]
- Kroc, M.; Czepiel, K.; Wilczura, P.; Mokrzycka, M.; Swiecicki, W. Development and validation of a gene-targeted dCAPS marker for marker-assisted selection of low-alkaloid content in seeds of narrow-leafed lupin (Lupinus angustifolius L.). Genes 2019, 10, 428. [Google Scholar] [CrossRef]
- Zhang, T.; Jing, W.; Qinwei, G.; Wenqi, C.; Xueyan, W.; Chaosen, L.; Xiaomin, X.; Dongfeng, Z.; Huiqin, L.; Fang, P. Development and validation of a functional molecular marker for a CaAPRR2-like gene that controls green fruit colour in pepper. Biotechnol. Biotechnol. Equip. 2024, 38, 2356860. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Y.; Liu, Z.; He, J.; Jiang, X.; He, F.; Lu, Z.; Yang, S.; Chen, P.; Yu, H.; et al. Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants 2021, 7, 954–965. [Google Scholar] [CrossRef]
- Wang, N.; Chen, P.; Xu, Y.; Guo, L.; Li, X.; Yi, H.; Larkin, R.M.; Zhou, Y.; Deng, X.; Xu, Q. Phased genomics reveals hidden somatic mutations and provides insight into fruit development in sweet orange. Hortic. Res. 2023, 11, uhad268. [Google Scholar] [CrossRef]
- Du, P.; Wang, Q.; Yuan, D.Y.; Chen, S.S.; Su, Y.N.; Li, L.; Chen, S.; He, X.J. WRKY transcription factors and OBERON histone-binding proteins form complexes to balance plant growth and stress tolerance. EMBO J. 2023, 42, e113639. [Google Scholar]
- Xin, T.; Zhang, Y.; Pu, X.; Gao, R.; Xu, Z.; Song, J. Trends in herbgenomics. Sci. China Life Sci. 2019, 62, 288–308. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Chen, H.; Patterson, N.; Reich, D. Population differentiation as a test for selective sweeps. Genome Res. 2010, 20, 393–402. [Google Scholar] [CrossRef]
- Li, Y.F.; Li, Y.H.; Su, S.S.; Reif, J.C.; Qi, Z.M.; Wang, X.B.; Wang, X.; Tian, Y.; Li, D.L.; Sun, R.J.; et al. SoySNP618K array: A high-resolution single nucleotide polymorphism platform as a valuable genomic resource for soybean genetics and breeding. J. Integr. Plant Biol. 2022, 64, 632–648. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhang, L.; Wang, J.; Wang, X.; Guo, S.; Xu, Z.J.; Li, D.; Liu, Z.; Li, Y.H.; Liu, B.; et al. Flowering time regulator qFT13-3 involved in soybean adaptation to high latitudes. Plant Biotechnol. J. 2024, 22, 1164–1176. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef]
- DeLano, W.L. An open-source molecular graphics tool. CCP4 Newslett. Protein Cryst. 2002, 40, 82. [Google Scholar]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Gan, Y.; Qi, G.; Xu, W.; Xin, T.; Huang, Y.; Fu, L.; Hao, L.; Lou, Q.; Fu, X.; et al. Transcription Factor LjWRKY50 Affects Jasmonate-Regulated Floral Bud Duration in Lonicera japonica. Plants 2025, 14, 2328. https://doi.org/10.3390/plants14152328
Li Y, Gan Y, Qi G, Xu W, Xin T, Huang Y, Fu L, Hao L, Lou Q, Fu X, et al. Transcription Factor LjWRKY50 Affects Jasmonate-Regulated Floral Bud Duration in Lonicera japonica. Plants. 2025; 14(15):2328. https://doi.org/10.3390/plants14152328
Chicago/Turabian StyleLi, Yanfei, Yutong Gan, Guihong Qi, Wenjie Xu, Tianyi Xin, Yuanhao Huang, Lianguo Fu, Lijun Hao, Qian Lou, Xiao Fu, and et al. 2025. "Transcription Factor LjWRKY50 Affects Jasmonate-Regulated Floral Bud Duration in Lonicera japonica" Plants 14, no. 15: 2328. https://doi.org/10.3390/plants14152328
APA StyleLi, Y., Gan, Y., Qi, G., Xu, W., Xin, T., Huang, Y., Fu, L., Hao, L., Lou, Q., Fu, X., Wei, X., Liu, L., Liu, C., & Song, J. (2025). Transcription Factor LjWRKY50 Affects Jasmonate-Regulated Floral Bud Duration in Lonicera japonica. Plants, 14(15), 2328. https://doi.org/10.3390/plants14152328





