Population Substructures of Castanopsis tribuloides in Northern Thailand Revealed Using Autosomal STR Variations
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. Molecular Marker for Studying Genetic Information
2.4. Statistical Analysis
3. Results
3.1. STR Locus Polymorphism
3.2. C. tribuloides Population Diversity
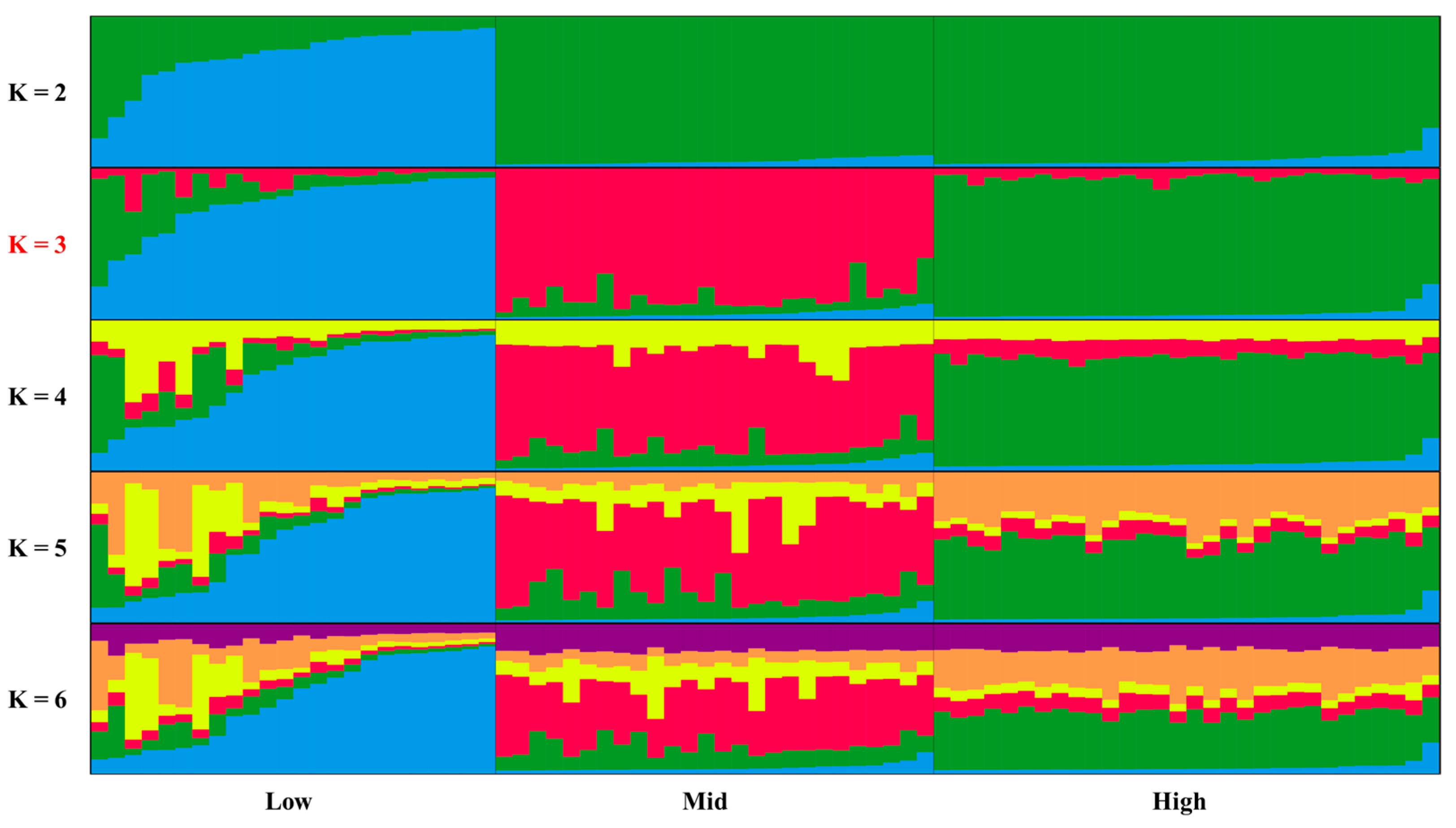
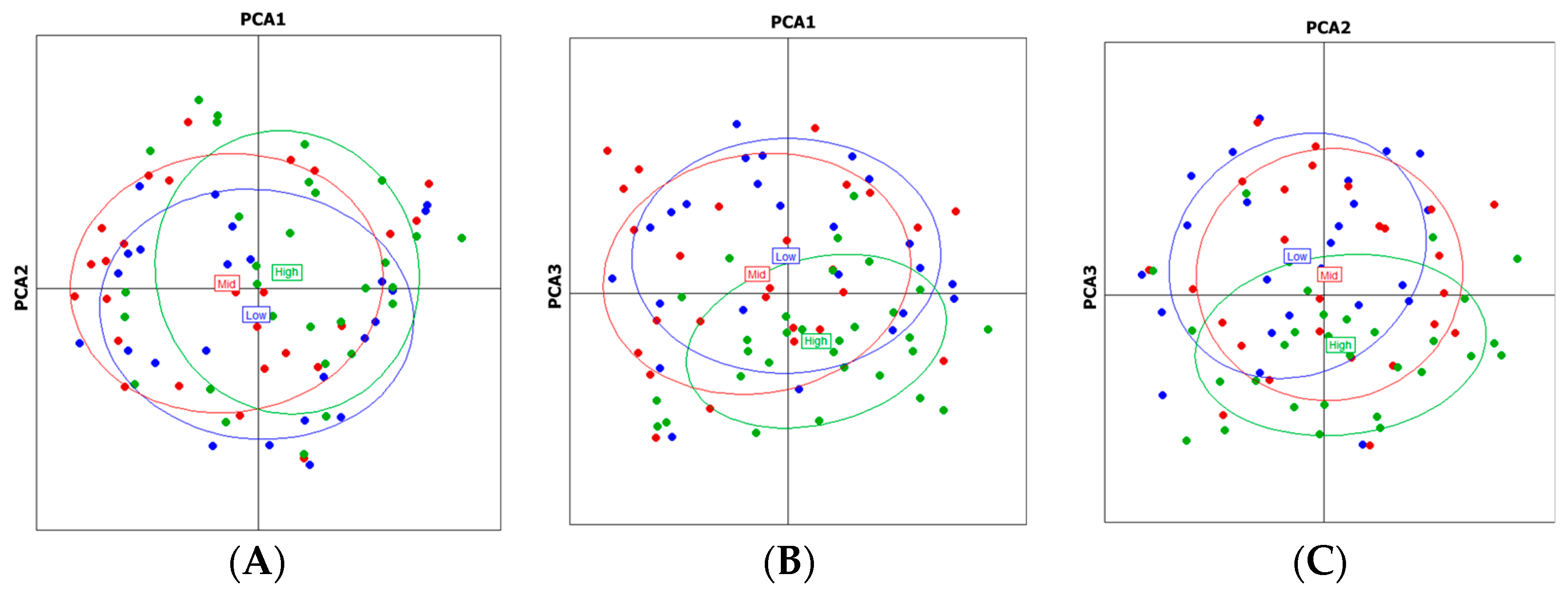
3.3. Population Structure Analysis
4. Discussion
4.1. Genetic Diversity Patterns in C. tribuloides Populations
4.2. Population Structure Across Altitudinal Gradients
4.3. Comparison with Other Castanopsis Species
4.4. Bottleneck Analysis and Historical Population Dynamics
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ratanapongsai, Y. Seedling recruitment of native tree species in active restoration forest. For. Soc. 2020, 4, 243–255. [Google Scholar] [CrossRef]
- Khamyong, N.; Wangpakapattanawong, P.; Chairuangsri, S.; Inta, A.; Tiansawat, P. Tree species composition and height-diameter allometry of three forest types in Northern Thailand. CMU J. Nat. Sci. 2018, 17, 289–308. [Google Scholar] [CrossRef]
- Marod, D.; Bootcharee, S.; Phumphuang, W.; Asanok, L.; Kamyo, T.; Thinkampaeng, S.; Thongsawi, J.; Hermhuk, S.; Khlangsap, N.; Nuipakdee, W. Diversity and spatial distribution of the Fagaceae tree species in the Doi Suthep-Pui National Park, Chiang Mai Province. J. Trop. For. Res. 2018, 2, 53–68. [Google Scholar]
- Chokchaichamnankit, P.; Anamthawat-Jónsson, K. Cytogenetics of the rare and endangered Trigonobalanus doichangensis (Fagaceae) from northern Thailand. J. Trop. For. Sci. 2015, 27, 60–68. [Google Scholar]
- Kaul, R.B. Evolution and reproductive biology of inflorescences in Lithocarpus, Castanopsis, Castanea, and Quercus (Fagaceae). Ann. Mo. Bot. Gard. 1986, 73, 284–296. [Google Scholar] [CrossRef]
- Nakanishi, A.; Yoshimaru, H.; Tomaru, N.; Miura, M.; Manabe, T.; Yamamoto, S. Patterns of pollen flow in a dense population of the insect-pollinated canopy tree species Castanopsis sieboldii. J. Hered. 2012, 103, 547–558. [Google Scholar] [CrossRef]
- Kardos, M.; Armstrong, E.E.; Fitzpatrick, S.W.; Hauser, S.; Hedrick, P.W.; Miller, J.M.; Tallmon, D.A.; Funk, W.C. The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. USA 2021, 118, e2104642118. [Google Scholar] [CrossRef]
- Bidyananda, N.; Jamir, I.; Nowakowska, K.; Varte, V.; Vendrame, W.A.; Devi, R.S.; Nongdam, P. Plant Genetic Diversity Studies: Insights from DNA Marker Analyses. Int. J. Plant Biol. 2024, 15, 607–640. [Google Scholar] [CrossRef]
- Waikham, P.; Thongkumkoon, P.; Chomdej, S.; Liu, A.; Wangpakapattanawong, P. Development of 13 microsatellite markers for Castanopsis tribuloides (Fagaceae) using next-generation sequencing. Mol. Biol. Rep. 2018, 45, 27–30. [Google Scholar] [CrossRef]
- Aoki, K.; Ueno, S.; Kamijo, T.; Setoguchi, H.; Murakami, N.; Kato, M.; Tsumura, Y. Genetic differentiation and genetic diversity of Castanopsis (Fagaceae), the dominant tree species in Japanese broadleaved evergreen forests, revealed by analysis of EST-associated microsatellites. PLoS ONE 2014, 9, e87429. [Google Scholar] [CrossRef]
- Mao, L.H.; Zhou, X.L.; Fang, Y.M. Genetic diversity and population structure of Castanopsis eyrei based on simple sequence repeat markers. Genet. Mol. Res. 2016, 15, gmr.15028256. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, R.; Bian, Y.; Qin, X.; Zhang, Z.; Sun, Y. Identification of a natural hybrid between Castanopsis sclerophylla and Castanopsis tibetana (Fagaceae) based on chloroplast and nuclear DNA sequences. Forests 2020, 11, 873. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, K.; Wen, X.; Sun, Y. Genetic Differentiation and Relationship among Castanopsis chinensis, C. qiongbeiensis, and C. glabrifolia (Fagaceae) as revealed by Nuclear SSR Markers. Plants 2024, 13, 1486. [Google Scholar] [CrossRef]
- Thongkumkoon, P.; Chomdej, S.; Kampuansai, J.; Pradit, W.; Waikham, P.; Elliott, S.; Chairuangsri, S.; Shannon, D.P.; Wangpakapattanawong, P.; Liu, A. Genetic assessment of three Fagaceae species in forest restoration trials. PeerJ 2019, 7, e6958. [Google Scholar] [CrossRef]
- Shimizu, M.; Kosaka, N.; Shimada, T.; Nagahata, T.; Iwasaki, H.; Nagai, H.; Shiba, T.; Emi, M. Universal fluorescent labeling (UFL) method for automated microsatellite analysis. DNA Res. 2002, 9, 173–178. [Google Scholar] [CrossRef]
- Hulce, D.; Li, X.; Snyder-Leiby, T.; Liu, C.S.J. GeneMarker® Genotyping software: Tools to increase the statistical power of DNA fragment analysis. J. Biomol. Tech. 2011, 22, S35–S36. [Google Scholar] [PubMed Central]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Promega. Excel PowerStats Spreadsheet; Promega Corporation: Madison, WI, USA, 1999. [Google Scholar]
- Yeh, F.C.; Yang, R.C.; Boyle, T.; Ye, Z.; Xiyan, J.M. PopGene32, Version 1.32; Molecular Biology and Biotechnology Centre: Sydney, Australia, 2000. Available online: https://sites.ualberta.ca/~fyeh/popgene.html (accessed on 1 May 2023).
- Piry, S.; Luikart, G.; Cornuet, J.-M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Li, C.; Sun, Y.; Huang, H.W.; Cannon, C.H. Footprints of divergent selection in natural populations of Castanopsis fargesii (Fagaceae). Heredity 2014, 113, 533–541. [Google Scholar] [CrossRef]
- Wang, F.; Cheng, X.; Cheng, S.; Li, W.; Huang, X. Genetic diversity of the wild ancient tea tree (Camellia taliensis) populations at different altitudes in Qianjiazhai. PLoS ONE 2023, 18, e0283189. [Google Scholar] [CrossRef]
| Primer | SSR Motif | Primer Sequence (5′–3′) | Amplification Size Range (bp) | Tm (°C) | |
|---|---|---|---|---|---|
| CT097 | (ATT)n | F: PET-CGACTTTGGGAAGGAAATAAAGG | R: TGGACTTCAACTTGCCATAGTG | 139–175 | 55 |
| CT110 | (TGT)n | F: PET-TTCTTCAGTTAGCCACATCG | R: CGCTAAGTCCATACATACAACAG | 169–205 | 55 |
| CT127 | (AGA)n | F: TAMRA- CCAGAAAACGTATGATCTTTG | R: CCATGCAACACTACCTCGTC | 207–246 | 62 |
| CT128 | (TCA)n | F: TAMRA- CCTTGGCAGACAAACTAGATA | R: GGCGCAACAACATATGAAGAAT | 160–244 | 56 |
| CT132 | (AT)n(AAG)n | F: TAMRA-TGACCCGAGCATGGTTTAT | R: GGACGTTAGGCCTGTACATT | 126–174 | 56 |
| CT135 | (TGA)n(GAA)n | F: FAM-GCCTAGCTTATGGAGTGGTT | R: GTCTTTGTGCAAGTGCTC | 124–160 | 55 |
| CT149 | (TCT)n | F: FAM-GCGCGTGACTTAGGCTCTTCAC | R: CTTCTCTGTTGGCATTTCTTGC | 144–150 | 56 |
| CT159 | (TCT)n | F: FAM- ATCCATGTCCACTTCTTCAA | R: CGTTTCCAAAACGAAGAAC | 130–193 | 55 |
| CT161 | (CAC)n | F: PET-AACGATACTAGCGACCTTGA | R: GCGAAAAACGCTCTCCAAC | 136–166 | 56 |
| CT164 | (CAC)n | F: HEX-ACAACACACCTAACATCACAAC | R: GAATGTTGCTCAGCGAAG | 124–174 | 55 |
| CT165 | (CTT)n | F: HEX-AGCGCCTTCTTAATAGAACC | R: TGGTGACCATTACTTGTTGA | 112–149 | 55 |
| Na | Ho | He | PD | MP | PIC | PE | TPI | PHWE | Null Allele | |
|---|---|---|---|---|---|---|---|---|---|---|
| CT097 | 3 | 0.4375 | 0.3768 | 0.5194 | 0.4806 | 0.3131 | 0.1384 | 0.8889 | 0.0008 | no |
| CT110 | 9 | 0.2405 | 0.3164 | 0.4509 | 0.5491 | 0.3026 | 0.0418 | 0.6583 | 0.0000 | yes |
| CT127 | 13 | 0.5417 | 0.6593 | 0.8044 | 0.1956 | 0.6313 | 0.2266 | 1.0909 | 0.0000 | yes |
| CT128 | 23 | 0.4533 | 0.9310 | 0.9614 | 0.0386 | 0.9200 | 0.1498 | 0.9146 | 0.0000 | yes |
| CT132 | 10 | 0.4359 | 0.7465 | 0.8583 | 0.1417 | 0.7067 | 0.1373 | 0.8864 | 0.0000 | yes |
| CT135 | 9 | 0.6027 | 0.7850 | 0.9022 | 0.0978 | 0.7511 | 0.2942 | 1.2586 | 0.0000 | yes |
| CT149 | 6 | 0.3924 | 0.4754 | 0.6662 | 0.3338 | 0.4467 | 0.1094 | 0.8229 | 0.0009 | yes |
| CT159 | 6 | 0.2308 | 0.2665 | 0.4270 | 0.5730 | 0.2526 | 0.0387 | 0.6500 | 0.1052 | no |
| CT161 | 6 | 0.1316 | 0.2094 | 0.3075 | 0.6925 | 0.2030 | 0.0139 | 0.5758 | 0.0006 | yes |
| CT164 | 8 | 0.2949 | 0.6099 | 0.7521 | 0.2479 | 0.5305 | 0.0615 | 0.7091 | 0.0000 | yes |
| CT165 | 15 | 0.9091 | 0.8883 | 0.9641 | 0.0359 | 0.8717 | 0.8140 | 5.5000 | 0.3610 | no |
| He | GD | GST | Fis | Nm | |
|---|---|---|---|---|---|
| Low elevation (n = 24) | 0.565 | 0.545 | 0.6207 | 0.2413 | 0.1528 |
| Mid elevation (n = 30) | 0.561 | 0.344 | 0.6523 | 0.3046 | 0.1333 |
| High elevation (n = 30) | 0.549 | 0.511 | 0.6477 | 0.2954 | 0.1360 |
| Source of Variation | Sum of Squares | Percentage of Variation | Statistics | p Value * |
|---|---|---|---|---|
| Among population | 9.889 | 2.83 | FST = 0.02826 | 0 |
| Among individual within population | 164.498 | 17 | FIS = 0.175 | 0.0059 |
| Within individuals | 120 | 80.17 | FIT = 0.19831 | 0 |
| Low Elevation | Mid Elevation | High Elevation | |
|---|---|---|---|
| Low elevation | 0 | 0.0078 | 0.0166 |
| Mid elevation | 0.0323 ** | 0 | 0.0010 |
| High elevation | 0.0256 * | 0.0395 ** | 0 |
| Population | Sign Test | Standardized | Wilcoxon Test | Allele Frequency Distribution | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IAM | TPM | SMM | IAM | TPM | SMM | IAM | TPM | SMM | ||
| Low | 0.1233 | 0.0343 | 0.0009 | 0.1228 | 0.0001 | 0.0000 | 0.8608 | 0.9919 | 0.9995 | L-shaped |
| Mid | 0.1509 | 0.0098 | 0.0107 | 0.0739 | 0.0001 | 0.0000 | 0.8799 | 0.9939 | 0.9976 | L-shaped |
| High | 0.4524 | 0.5214 | 0.0013 | 0.2731 | 0.0043 | 0.0000 | 0.5508 | 0.8174 | 0.9993 | L-shaped |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thongkumkoon, P.; Kampuansai, J.; Dansawan, M.; Tiansawat, P.; Noirungsee, N.; Punchay, K.; Khamyong, N.; Wangpakapattanawong, P. Population Substructures of Castanopsis tribuloides in Northern Thailand Revealed Using Autosomal STR Variations. Plants 2025, 14, 2306. https://doi.org/10.3390/plants14152306
Thongkumkoon P, Kampuansai J, Dansawan M, Tiansawat P, Noirungsee N, Punchay K, Khamyong N, Wangpakapattanawong P. Population Substructures of Castanopsis tribuloides in Northern Thailand Revealed Using Autosomal STR Variations. Plants. 2025; 14(15):2306. https://doi.org/10.3390/plants14152306
Chicago/Turabian StyleThongkumkoon, Patcharawadee, Jatupol Kampuansai, Maneesawan Dansawan, Pimonrat Tiansawat, Nuttapol Noirungsee, Kittiyut Punchay, Nuttaluck Khamyong, and Prasit Wangpakapattanawong. 2025. "Population Substructures of Castanopsis tribuloides in Northern Thailand Revealed Using Autosomal STR Variations" Plants 14, no. 15: 2306. https://doi.org/10.3390/plants14152306
APA StyleThongkumkoon, P., Kampuansai, J., Dansawan, M., Tiansawat, P., Noirungsee, N., Punchay, K., Khamyong, N., & Wangpakapattanawong, P. (2025). Population Substructures of Castanopsis tribuloides in Northern Thailand Revealed Using Autosomal STR Variations. Plants, 14(15), 2306. https://doi.org/10.3390/plants14152306






