Genome-Wide Analysis of KNOX Genes: Identification, Evolution, Comparative Genomics, Expression Dynamics, and Sub-Cellular Localization in Brassica napus
Abstract
1. Introduction
2. Results
2.1. Identification and Chromosome Map of KNOX Proteins and Genes from Brassica
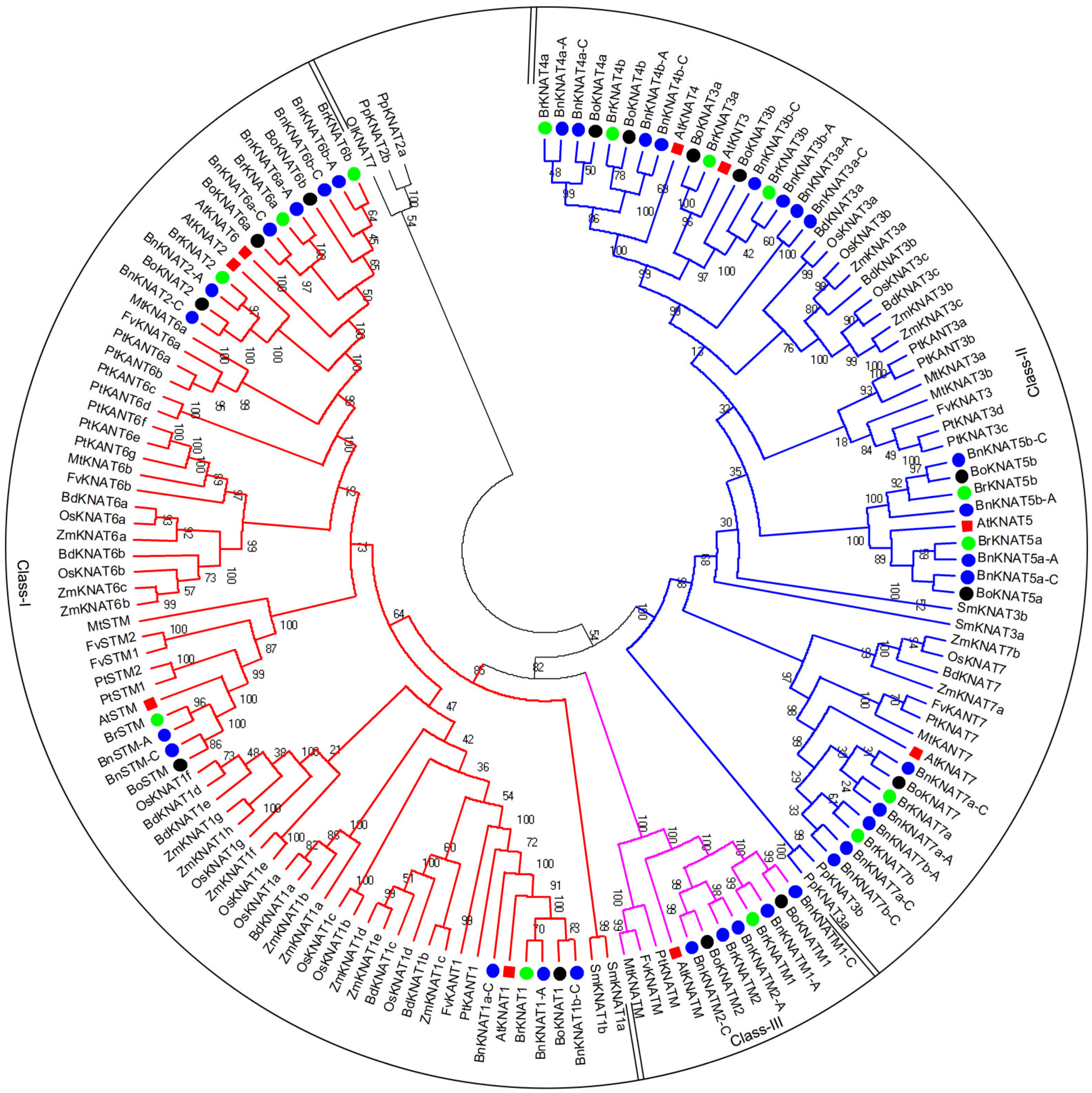
2.2. Phylogenetic and Classification Analyses of KNOX Protein Family
- To analyze the evolutionary patterns of KNOX, we constructed a comprehensive phylogenetic tree for 150 KNOX proteins, including the following: 32 from B. napus, 16 from Populus trichocarpa/Zea mays, 15 from B. rapa, 14 from B. oleracea, 13 from Oryza sativa, 11 from Brachypodium distachyon, 9 from Arabidopsis thaliana, 8 from Fragaria vesca, 7 from Medicago truncatula, 4 from Physcomitrella patens/Selaginella moellendorffii, and 1 from O. lucimarinus (Figure 2; Supplement File S1). Subsequently, excluding PpKNAT2a/2b and OlKNAT7 due to their inability to be classified into Class I or Class II despite sharing the same domain organization as others (similar results were obtained using different phylogenetic trees constructed by Neighbor-Joining or Maximum Likelihood methods), we successfully categorized the remaining set of 147 KNOX proteins into three distinct classes. Notably in Brassica species specifically, B. napus not only encompasses all KNOX proteins found in both B. rapa and B. oleracea but also exhibits an additional three unique members within its repertoire (Figure 2; Supplement File S1).
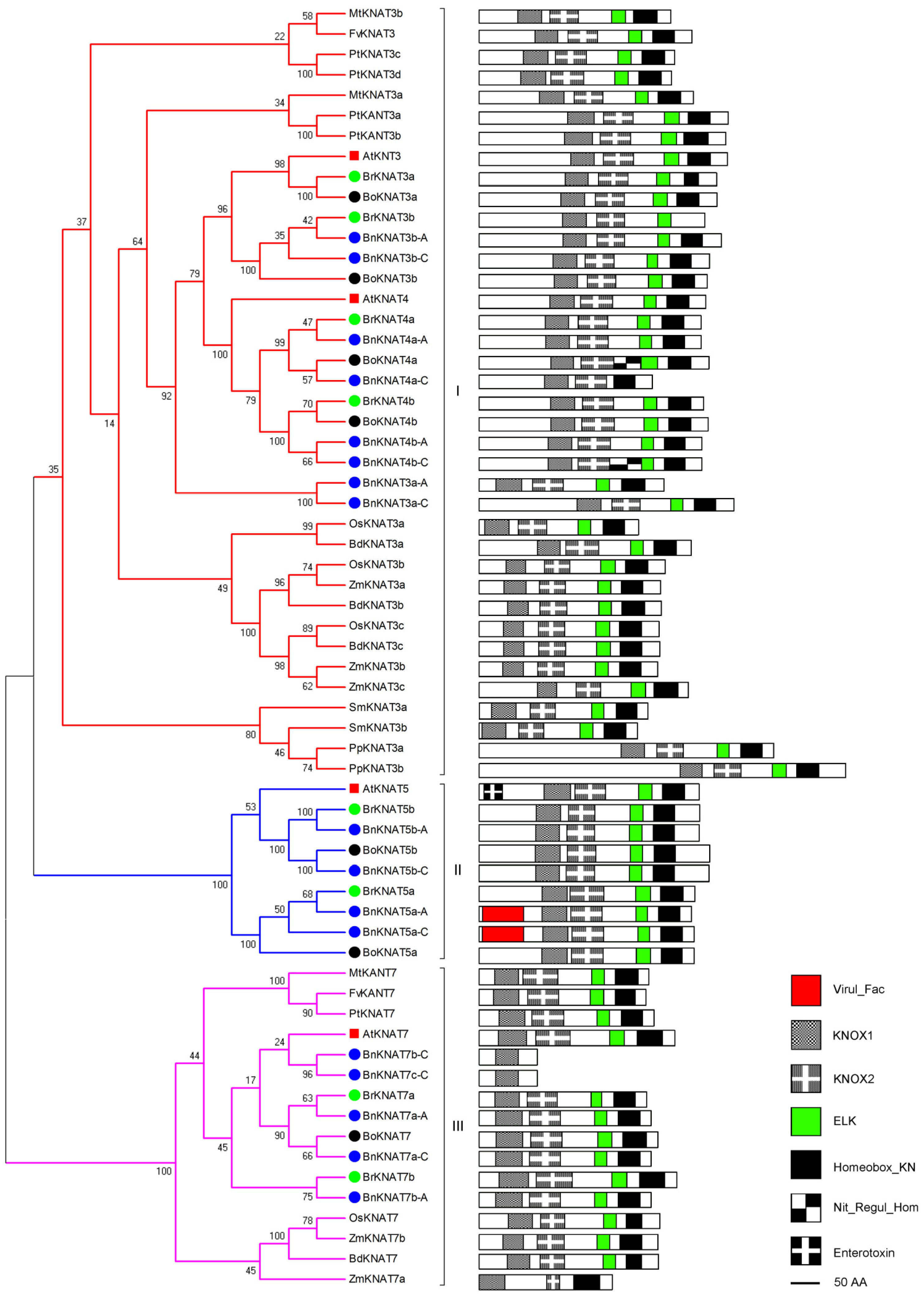
2.2.1. Phylogenetic and Domain Analyses of Class I
2.2.2. Phylogenetic and Domain Analyses of Class II
2.2.3. Phylogenetic and Domain Analyses of Class-III
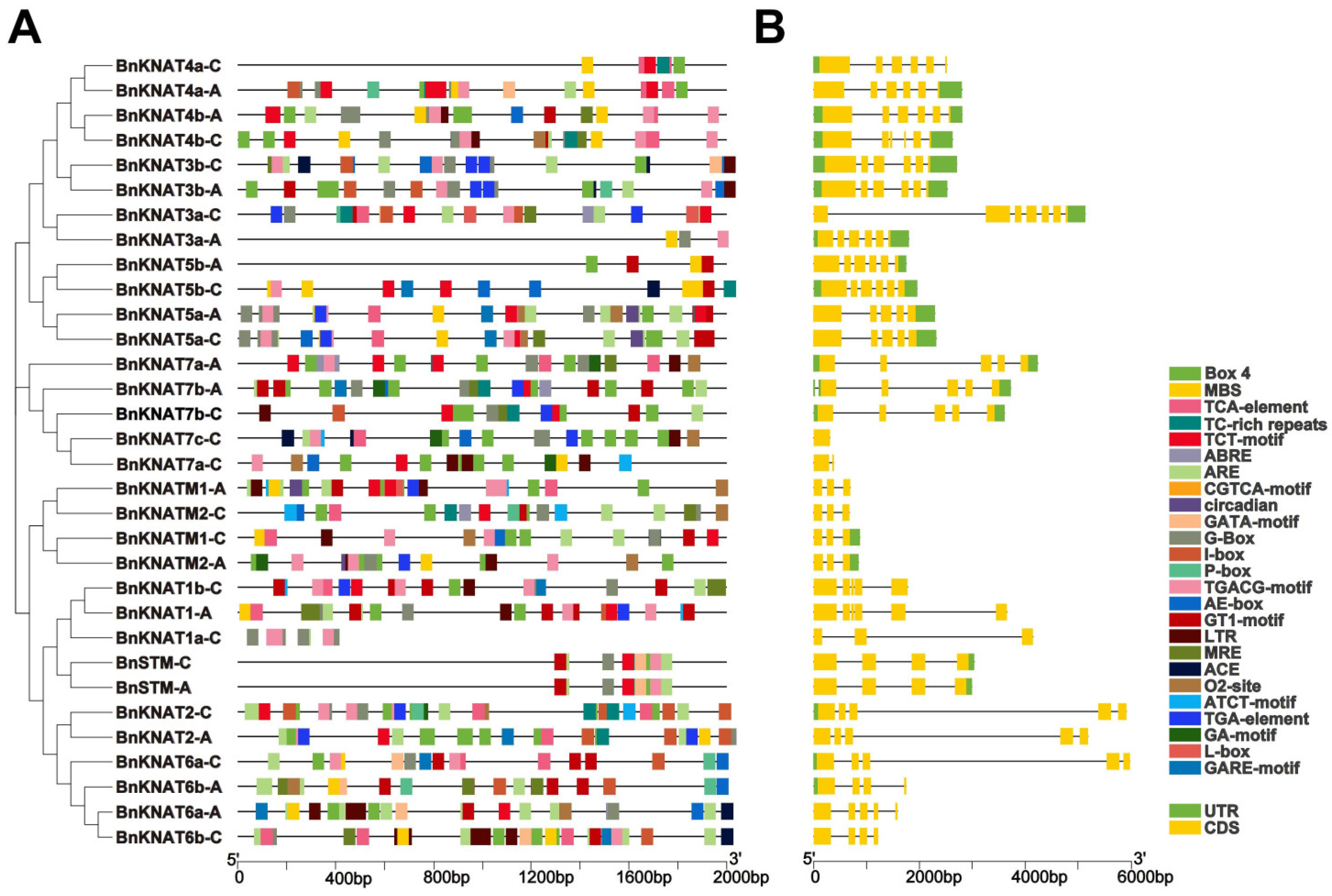
2.3. Analysis of Cis-Acting Elements of BnKNOX Gene Promoters and Gene Structure in B. napus
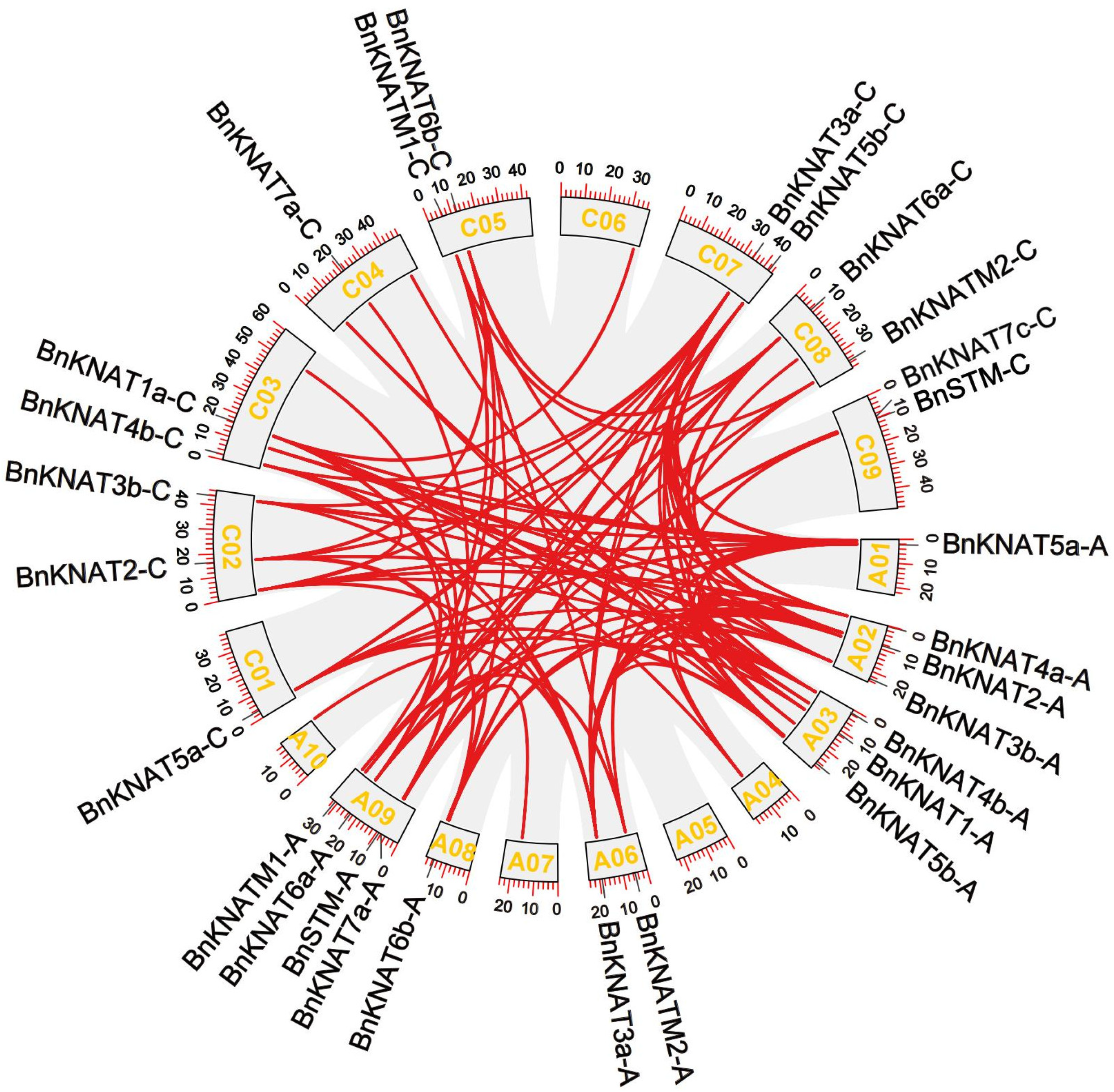
2.4. Gene Collinearity and Duplication of BnKNOXs in B. napus
2.5. Expression Patterns of BnKNOXs in Different Tissues from BrassicaEDB
2.6. BnKNOXs Expression Levels of in Reproductive Organs by qRT-PCR
2.7. Subcellular Localization Analysis
3. Discussion
3.1. Conservation and Evolution of KNOX Proteins in Plants
3.2. KNOX Orthologs Among B. napus, B. rapa and B. oleracea
3.3. KNOX Gene Duplication and Diversity in B. napus, B. rapa and B. oleracea
3.4. Potential Roles of BnKNOX Genes Related to Plant Growth and Development
4. Materials and Methods
4.1. KNOX Protein Identification and Chromosome Map Construction
4.2. Analysis of Protein Domain Organization
4.3. Phylogenetic Analysis
4.4. Analysis of Cis-Acting Elements of BnKNOXs Promoters and Gene Structures in B. napus
4.5. Collinearity Analysis
4.6. Expression Levels of BnKNOXs in Different Tissues
4.7. Validation of Expression Levels of BnKNOXs by qRT-PCR
4.8. Subcellular Localization of the BnKNOX::GFP Fusion Protein
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morata, G.; Lawrence, P. An exciting period of Drosophila developmental biology: Of imaginal discs, clones, compartments, parasegments and homeotic genes. Dev. Biol. 2022, 484, 12–21. [Google Scholar] [CrossRef]
- Jia, P.; Wang, Y.; Sharif, R.; Dong, Q.L.; Liu, Y.; Luan, H.A.; Zhang, X.M.; Guo, S.P.; Qi, G.H. KNOTTED1-like homeobox (KNOX) transcription factors—Hubs in a plethora of networks: A review. Int. J. Biol. Macromol. 2023, 253, 126878. [Google Scholar] [CrossRef]
- McGinnis, W.; Levine, M.S.; Hafen, E.; Kuroiwa, A.; Gehring, W.J. A conserved DNA sequence in homoerotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 1984, 308, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F. The evolution of floral homeotic gene function. BioEssays 2003, 25, 637–646. [Google Scholar] [CrossRef]
- Li, G.; Manzoor, M.A.; Wang, G.; Chen, C.; Song, C. Comparative analysis of KNOX genes and their expression patterns under various treatments in Dendrobium huoshanense. Front. Plant Sci. 2023, 14, 1258533. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Zheng, S.; Zhang, C.; Huang, R.; Yuan, L.; Tong, H. Identification and expression analysis of the KNOX genes during organogenesis and stress responseness in Camellia sinensis (L.) O. Kuntze. Mol. Genet. Genom. 2023, 298, 1559–1578. [Google Scholar] [CrossRef]
- Tsuda, K.; Hake, S. Diverse functions of knox transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, G.; Janssen, B.J.; Kellogg, E.A.; Sinha, N. Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Proc. Natl. Acad. Sci. USA 1997, 94, 13749–13753. [Google Scholar] [CrossRef]
- Chan, R.L.; Gago, G.M.; Palena, C.M.; Gonzalez, D.H. Homeoboxes in plant development. Biochim. Biophys. Acta 1998, 1442, 1–19. [Google Scholar] [CrossRef]
- Wen, J.; Deng, M.; Zhao, K.; Zhou, H.; Wu, R.; Li, M.; Cheng, H.; Li, P.; Zhang, R.; Lv, J. Characterization of Plant Homeodomain Transcription Factor Genes Involved in Flower Development and Multiple Abiotic Stress Response in Pepper. Genes 2023, 14, 1737. [Google Scholar] [CrossRef]
- Pick, L. Hox genes, evo-devo, and the case of the ftz gene. Chromosoma 2016, 125, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Hii, E.P.W.; Ramanathan, A.; Pandarathodiyil, A.K.; Wong, G.R.; Sekhar, E.V.S.; Binti Talib, R.; Zaini, Z.M.; Zain, R.B. Homeobox Genes in Odontogenic Lesions: A Scoping Review. Head. Neck Pathol. 2023, 17, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Bertolino, E.; Reimund, B.; Wildt-Perinic, D.; Clerc, R.G. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 1995, 270, 3178–3188. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar] [CrossRef]
- Magnani, E.; Hake, S. KNOX lost the OX: The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 2008, 20, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Chai, M.; Han, L.; Cao, X.; Zhang, J.; Kong, Y.; Fu, C.; Wang, Z.Y.; Mysore, K.S.; et al. The role of Class II KNOX family in controlling compound leaf patterning in Medicago truncatula. J. Integr. Plant Biol. 2023, 65, 2279–2291. [Google Scholar] [CrossRef]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.; Samuelsson, T. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef]
- Nagasaki, H.; Sakamoto, T.; Sato, Y.; Matsuoka, M. Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 2001, 13, 2085–2098. [Google Scholar] [CrossRef]
- Scofield, S.; Murray, J.A. KNOX gene function in plant stem cell niches. Plant Mol. Biol. 2006, 60, 929–946. [Google Scholar] [CrossRef]
- Rüscher, D.; Corra, l.J.M.; Carluccio, A.V.; Klemens, P.A.W.; Gisel, A.; Stavolone, L.; Neuhaus, H.E.; Ludewig, F.; Sonnewald, U.; Zierer, W. Auxin signaling and vascular cambium formation enable storage metabolism in cassava tuberous roots. J. Exp. Bot. 2021, 72, 3688–3703. [Google Scholar] [CrossRef]
- Yang, Q.; Cong, T.; Yao, Y.; Cheng, T.; Yuan, C.; Zhang, Q. KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium. Int. J. Mol. Sci. 2023, 24, 7081. [Google Scholar] [CrossRef]
- Hay, A.; Tsiantis, M. A KNOX family TALE. Curr. Opin. Plant Biol. 2009, 12, 593–598. [Google Scholar] [CrossRef]
- Ruiz-Estévez, M.; Bakkali, M.; Martín-Blázquez, R.; Garrido-Ramos, M. Identification and characterization of TALE Homeobox Genes in the endangered fern Vandenboschia speciosa. Genes 2017, 8, E275. [Google Scholar] [CrossRef]
- Hisanaga, T.; Fujimoto, S.; Cui, Y.; Sato, K.; Sano, R.; Yamaoka, S.; Kohchi, T.; Berger, F.; Nakajima, K. Deep evolutionary origin of gamete-directed zygote activation by KNOX/BELL transcription factors in green plants. eLife 2021, 10, e57090. [Google Scholar] [CrossRef]
- Frangedakis, E.; Saint-Marcoux, D.; Moody, L.A.; Rabbinowitsch, E.; Langdale, J.A. Nonreciprocal complementation of KNOX gene function in land plants. New Phytol. 2017, 216, 591–604. [Google Scholar] [CrossRef]
- Sakakibara, K.; Nishiyama, T.; Deguchi, H.; Hasebe, M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol. Dev. 2008, 10, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lan, S.; Yin, W.L.; Liu, Z.J. Genome-Wide Identification and Expression Pattern Analysis of KNOX Gene Family in Orchidaceae. Front. Plant Sci. 2022, 13, 901089. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Nieuwland, J.; Murray, J.A. The Arabidopsis homeobox gene SHOOT MERISTEMLESS has cellular and meristem-organisational roles with differential requirements for cytokinin and CYCD3 activity. Plant J. 2013, 75, 53–66. [Google Scholar] [CrossRef]
- Nidhi, S.; Preciado, J.; Tie, L. Knox homologs shoot meristemless (STM) and KNAT6 are epistatic to CLAVATA3 (CLV3) during shoot meristem development in Arabidopsis thaliana. Mol. Biol. Rep. 2021, 48, 6291–6302. [Google Scholar] [CrossRef]
- Gastaldi, V.; Alem, A.L.; Mansilla, N.; Ariel, F.D.; Viola, I.L.; Lucero, L.E.; Gonzalez, D.H. BREVIPEDICELLUS/KNAT1 targets TCP15 to modulate filament elongation during Arabidopsis late stamen development. Plant Physiol. 2023, 191, 29–34. [Google Scholar] [CrossRef]
- Reiser, L.; Sánchez-Baracaldo, P.; Hake, S. Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Evol. 2000, 42, 151–166. [Google Scholar] [CrossRef]
- Wang, S.; Yamaguchi, M.; Grienenberger, E.; Martone, P.T.; Samuels, A.L.; Mansfield, S.D. The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 2020, 101, 293–309. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Pei, S.; Lu, M.; Kong, Y.; Zhou, G.; Hu, R. KNAT7 regulates xylan biosynthesis in Arabidopsis seed-coat mucilage. J. Exp. Bot. 2020, 71, 4125–4139. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Gao, Y.; Yang, S. Genome-Wide Identification and Characterization of TALE Superfamily Genes in Soybean (Glycine max L.). Int. J. Mol. Sci. 2021, 22, 4117. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, Q.; Qin, W.; Gao, H.; Du, J.; Chen, J.; Li, H.; Zhou, G.; Wu, H.; Wu, A.M. The Class II KNOX family members KNAT3 and KNAT7 redundantly participate in Arabidopsis seed coat mucilage biosynthesis. J. Exp. Bot. 2022, 73, 3477–3495. [Google Scholar] [CrossRef]
- Xiong, H.; Shi, A.; Wu, D.; Weng, Y.; Qin, J.; Ravelombola, W.S.; Shu, X.; Zhou, W. Genome-wide identification, classification and evolutionary expansion of KNOX gene family in Rice (Oryza sativa) and Populus (Populus trichocarpa). Am. J. Plant Sci. 2018, 9, 1071. [Google Scholar] [CrossRef]
- Wang, T.; van Dijk, A.D.J.; Bucher, J.; Liang, J.; Wu, J.; Bonnema, G.; Wang, X. Interploidy Introgression Shaped Adaptation during the Origin and Domestication History of Brassica napus. Mol. Biol. Evol. 2023, 40, msad199. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, X.; Zhou, H.; Wei, K.; Jiang, C.; Wang, J.; Cao, Y.; Tang, F.; Zhao, S.; Lu, M.Z. KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol. 2020, 225, 1531–1544. [Google Scholar] [CrossRef]
- Wang, X.Q.; Xu, W.H.; Ma, L.G.; Fu, Z.M.; Deng, X.W.; Li, J.Y.; Wang, Y.H. Requirement of KNAT1/BP for the development of Abscission Zones in Arabidopsis thaliana. J. Integr. Plant Biol. 2006, 48, 15–26. [Google Scholar] [CrossRef]
- Yi, Z.; Sanjeev, M.; Singh, G. The Branched Nature of the Nonsense-Mediated mRNA Decay Pathway. Trends Genet. 2021, 37, 143–159. [Google Scholar] [CrossRef]
- Wang, Y.; Aïssi-Rothe, L.; Virion, J.M.; De Carvalho Bittencourt, M.; Ulas, N.; Audonnet, S.; Salmon, A.; Clement, L.; Venard, V.; Jeulin, H.; et al. Combination of Epstein-Barr virus nuclear antigen 1, 3 and lytic antigen BZLF1 peptide pools allows fast and efficient stimulation of Epstein-Barr virus-specific T cells for adoptive immunotherapy. Cytotherapy 2014, 16, 122–134. [Google Scholar] [CrossRef]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef]
- Soylev, A.; Le, T.M.; Amini, H.; Alkan, C.; Hormozdiari, F. Discovery of tandem and interspersed segmental duplications using high-throughput sequencing. Bioinformatics 2019, 35, 3923–3930. [Google Scholar] [CrossRef]
- Abdullah, M.; Sabir, I.A.; Shah, I.H.; Sajid, M.; Liu, X.; Jiu, S.; Manzoor, M.A.; Zhang, C. The role of gene duplication in the divergence of the sweet cherry. Plant Gene 2022, 32, 100379. [Google Scholar] [CrossRef]
- Yadav, S.; Chahar, N.; Lal, M.; Das, S. Phylogenetic and comparative genomics establishes origin of paralogy between homologs of AtMYB42 and AtMYB85 in last common ancestor of Brassicaceae via segmental duplication. Plant Gene 2023, 35, 100424. [Google Scholar] [CrossRef]
- Box, M.S.; Dodsworth, S.; Rudall, P.J.; Bateman, R.M.; Glover, B.J. Flower-specific KNOX phenotype in the orchid Dactylorhiza fuchsia. J. Exp. Bot. 2012, 63, 4811–4819. [Google Scholar] [CrossRef] [PubMed]
- Keren-Keiserman, A.; Shtern, A.; Levy, M.; Chalupowicz, D.; Furumizu, C.; Alvarez, J.P.; Amsalem, Z.; Arazi, T.; Alkalai-Tuvia, S.; Efroni, I.; et al. CLASS-II KNOX genes coordinate spatial and temporal ripening in tomato. Plant Physiol. 2022, 190, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Ferreira de Carvalho, J.; Stoeckel, S.; Eber, F.; Lodé-Taburel, M.; Gilet, M.M.; Trotoux, G.; Morice, J.; Falentin, C.; Chèvre, A.M.; Rousseau-Gueutin, M. Untangling structural factors driving genome stabilization in nascent Brassica napus allopolyploids. New Phytol. 2021, 230, 2072–2084. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008, 275, 2845–2861. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Yang, T.J.; Seol, Y.J.; Jin, M.; Kim, J.A.; Lim, M.H.; Kim, J.S.; Baek, S.; Choi, B.S.; et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Lukens, L.; Zou, F.; Lydiate, D.; Parkin, I.; Osborn, T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 2003, 164, 359–372. [Google Scholar] [CrossRef]
- Zeng, P.; Ge, X.; Li, Z. Transcriptional Interactions of Single B-Subgenome Chromosome with C-Subgenome in B. oleracea-nigra Additional Lines. Plants 2023, 12, 2029. [Google Scholar] [CrossRef]
- Glover, N.; Sheppard, S.; Dessimoz, C. Homoeolog Inference Methods Requiring Bidirectional Best Hits or Synteny Miss Many Pairs. Genome Biol. Evol. 2021, 13, evab077. [Google Scholar] [CrossRef]
- Bird, K.A.; Niederhuth, C.E.; Ou, S.; Gehan, M.; Pires, J.C.; Xiong, Z.; VanBuren, R.; Edger, P.P. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. New Phytol. 2021, 230, 354–371. [Google Scholar] [CrossRef]
- Dumonceaux, T.; Venglat, S.P.; Kushalappa, K.; Selvaraj, G.; Datla, R. Molecular and functional characterization of Brassica BREVIPEDICELLUS orthologs involved in inflorescence architecture. Botany 2009, 87, 604–615. [Google Scholar] [CrossRef]
- Parkin, I.A.; Gulden, S.M.; Sharpe, A.G.; Lukens, L.; Trick, M.; Osborn, T.C.; Lydiate, D.J. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 2005, 171, 2765–2781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Roth, C.; Rastogi, S.; Arvestad, L.; Dittmar, K.; Light, S.; Ekman, D.; Liberles, D.A. Evolution after gene duplication: Models, mechanisms, sequences, systems, and organisms. JEZ-B Mol. Dev. Evol. 2007, 308, 58–73. [Google Scholar] [CrossRef]
- Otto, S.P.; Yong, P. The evolution of gene duplicates. Adv. Genet. 2002, 46, 451–483. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, 384–388. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterso, F.A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Pi, B.; Pan, J.; Xiao, M.; Hu, X.; Zhang, L.; Chen, M.; Liu, B.; Ruan, Y.; Huang, Y. Systematic analysis of CCCH zinc finger family in Brassica napus showed that BnRR-TZFs are involved in stress resistance. BMC Plant Biol. 2021, 21, 555. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yu, F.; Shi, D.; Wang, Y.; Xu, F.; Zeng, S. Selection and validation of reference genes for RT-qPCR analysis in Desmodium styracifolium Merr. 3 Biotech 2021, 11, 403. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Zheng, R.; Chen, Y.; Tan, C. Genome-Wide Analysis of KNOX Genes: Identification, Evolution, Comparative Genomics, Expression Dynamics, and Sub-Cellular Localization in Brassica napus. Plants 2025, 14, 2167. https://doi.org/10.3390/plants14142167
He X, Zheng R, Chen Y, Tan C. Genome-Wide Analysis of KNOX Genes: Identification, Evolution, Comparative Genomics, Expression Dynamics, and Sub-Cellular Localization in Brassica napus. Plants. 2025; 14(14):2167. https://doi.org/10.3390/plants14142167
Chicago/Turabian StyleHe, Xiaoli, Ruiyi Zheng, Yan Chen, and Chengfang Tan. 2025. "Genome-Wide Analysis of KNOX Genes: Identification, Evolution, Comparative Genomics, Expression Dynamics, and Sub-Cellular Localization in Brassica napus" Plants 14, no. 14: 2167. https://doi.org/10.3390/plants14142167
APA StyleHe, X., Zheng, R., Chen, Y., & Tan, C. (2025). Genome-Wide Analysis of KNOX Genes: Identification, Evolution, Comparative Genomics, Expression Dynamics, and Sub-Cellular Localization in Brassica napus. Plants, 14(14), 2167. https://doi.org/10.3390/plants14142167




