Genome-Wide Insights into Streptomyces Novel Species Qhu-G9 and Its Potential for Enhancing Salt Tolerance and Growth in Avena sativa L. and Onobrychis viciifolia Scop
Abstract
1. Introduction
2. Results
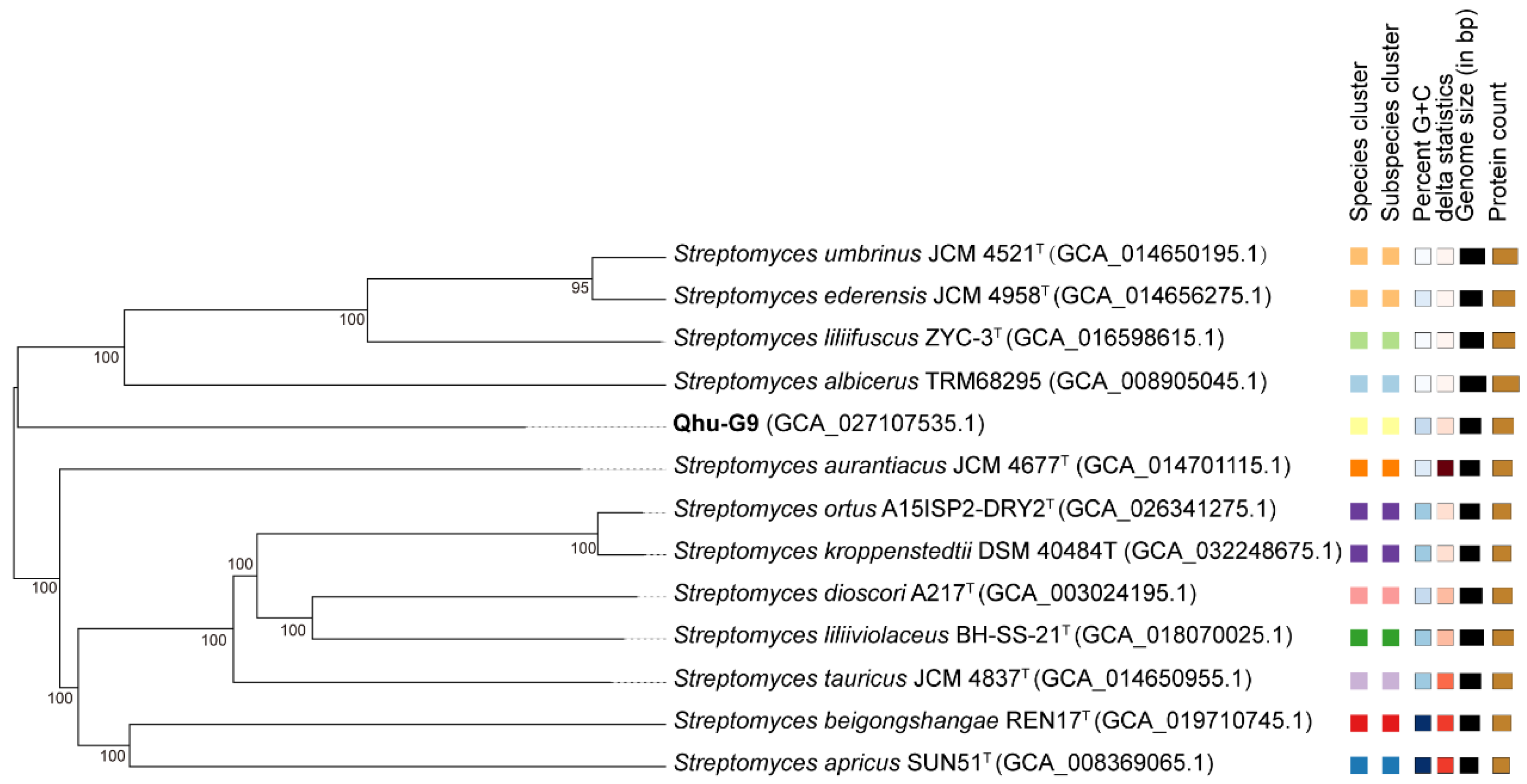
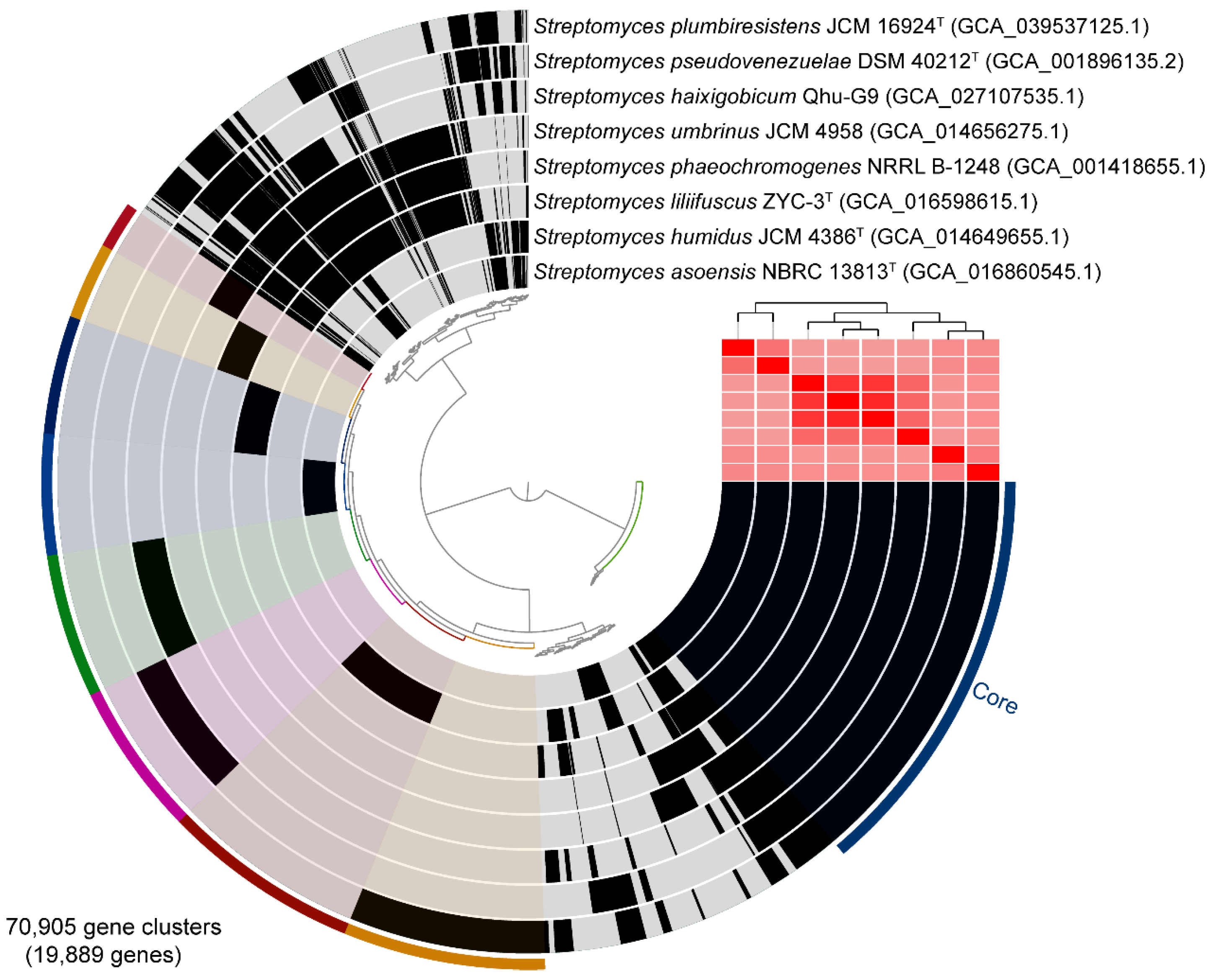
2.1. Phylogenetic Relationships and Comparative Genomics of Qhu-G9
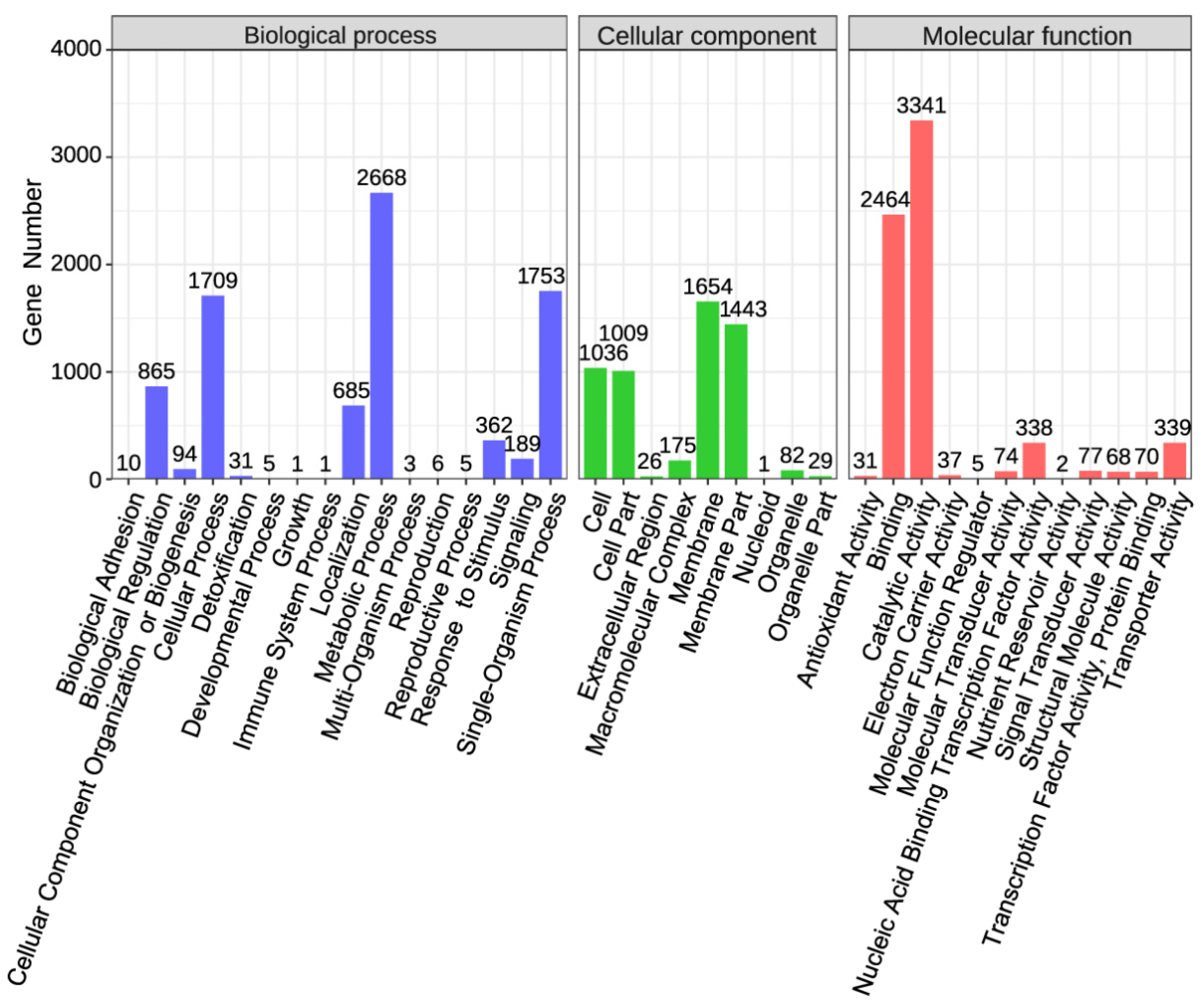
2.2. Functional Annotation of the Qhu-G9 Genome
2.3. Identification of Plant Growth-Promoting Genes in Qhu-G9
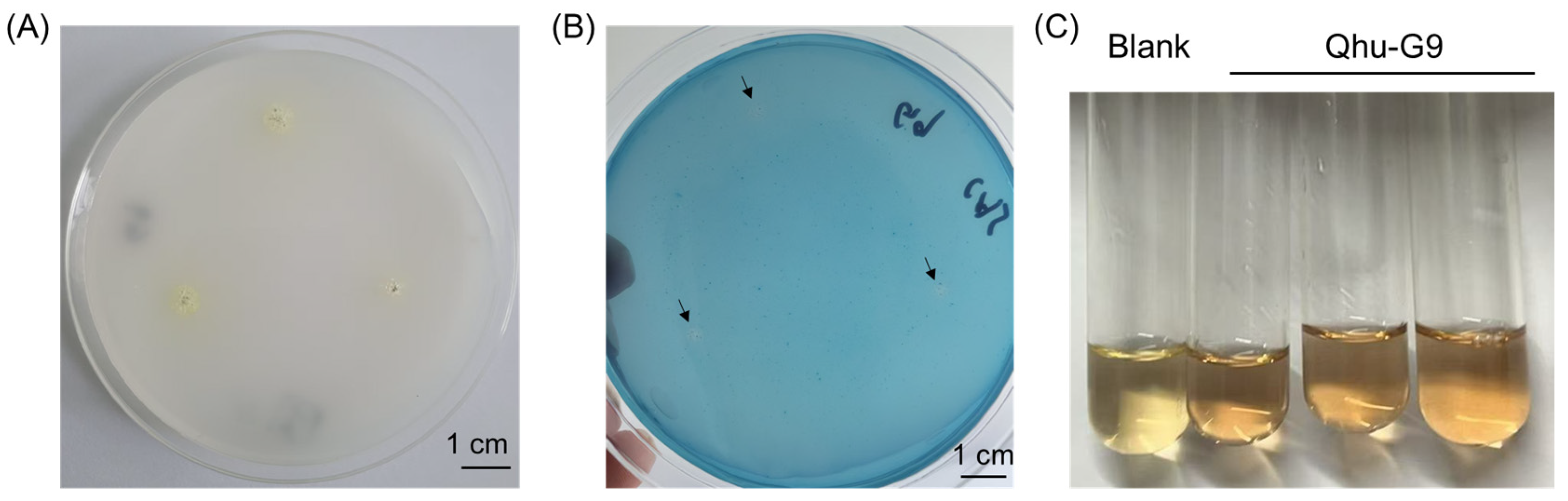
2.4. Evaluation of Plant Growth-Promoting Traits in Qhu-G9
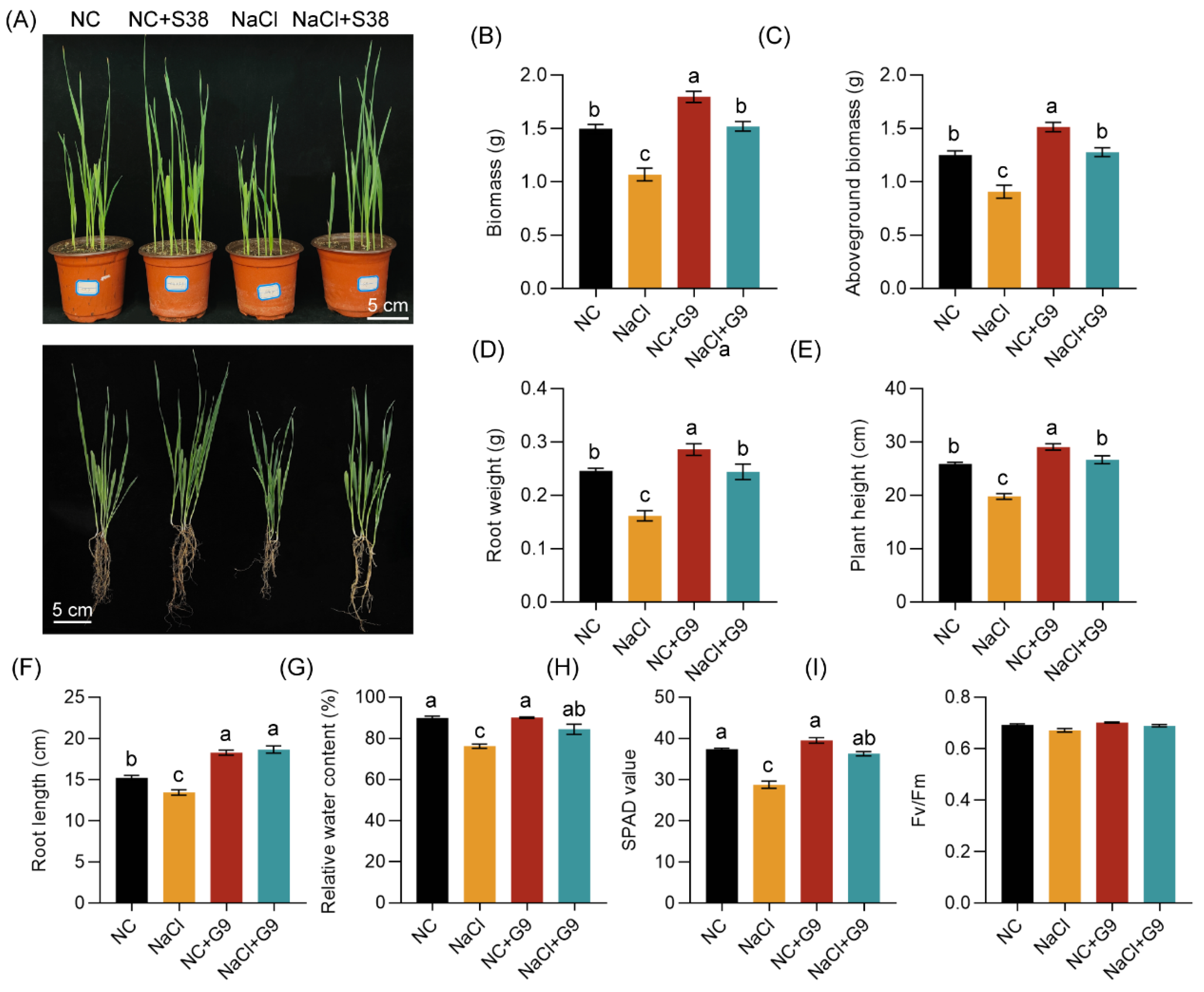
2.5. Effect of Qhu-G9 on A. sativa Growth Promotion and Salt Stress Tolerance
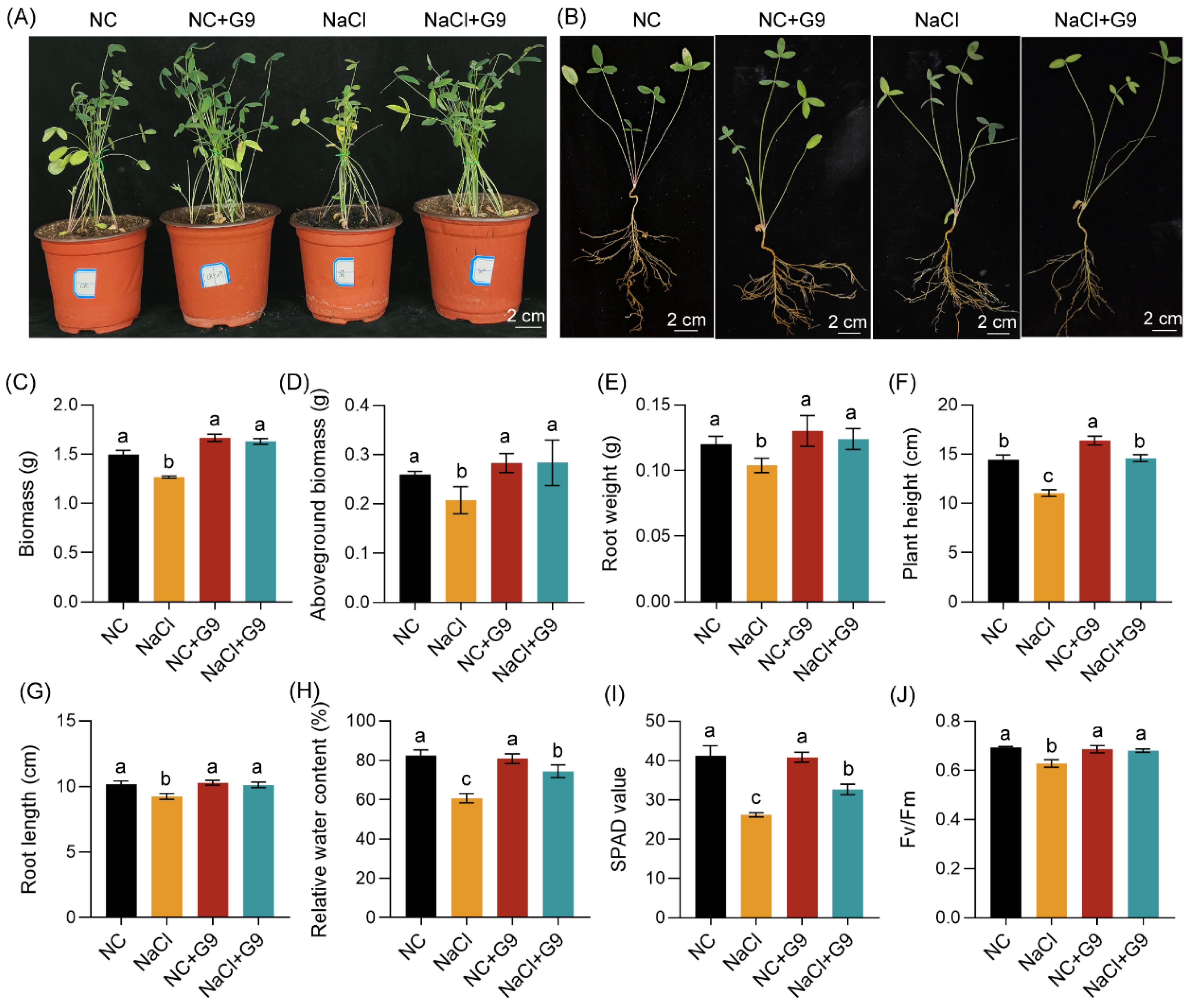
2.6. Qhu-G9 Promotes Growth and Salt Tolerance in O. viciifolia
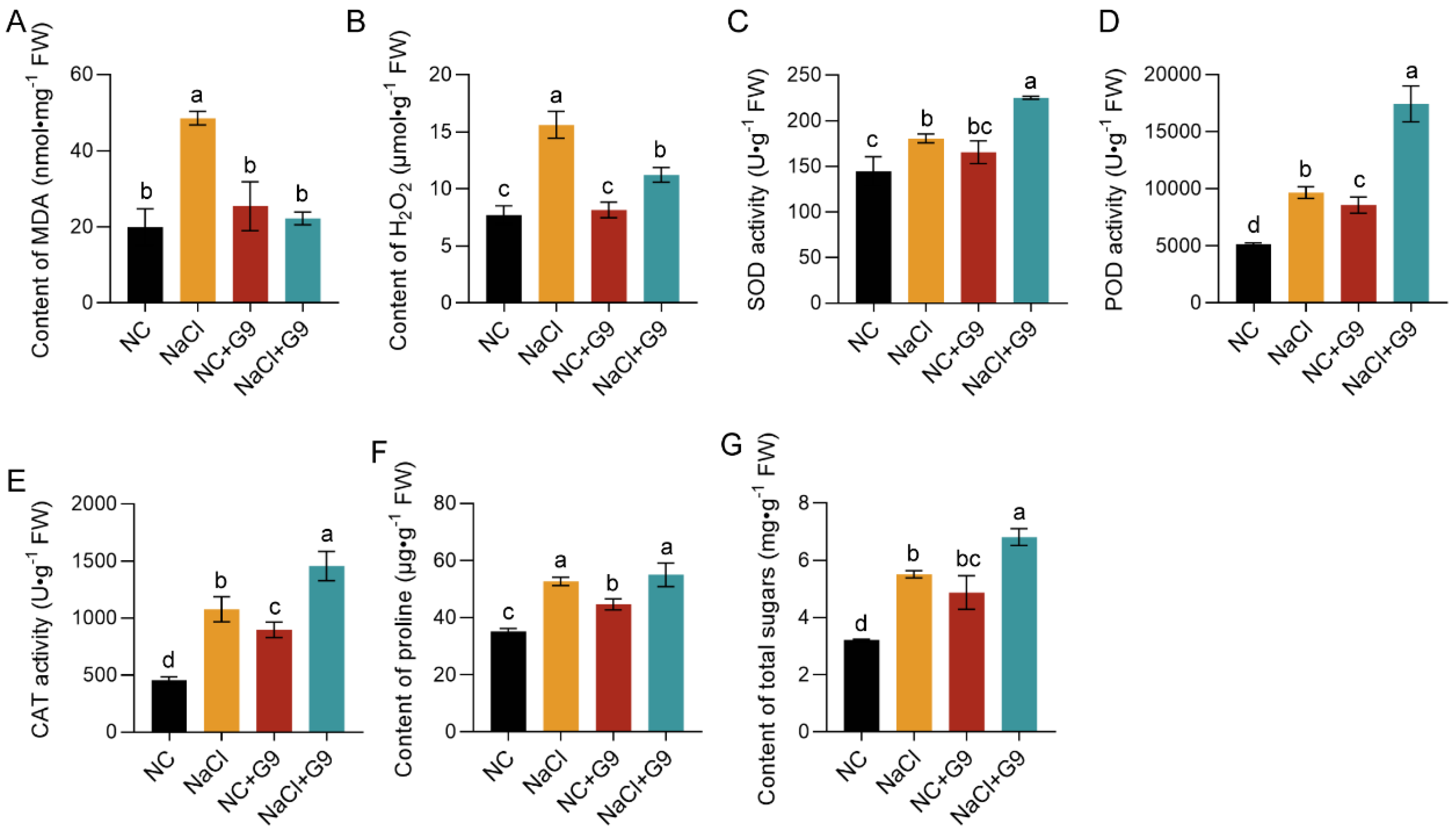
2.7. Biochemical Responses of O. viciifolia Seedlings Inoculated with Qhu-G9 Under Salt Stress
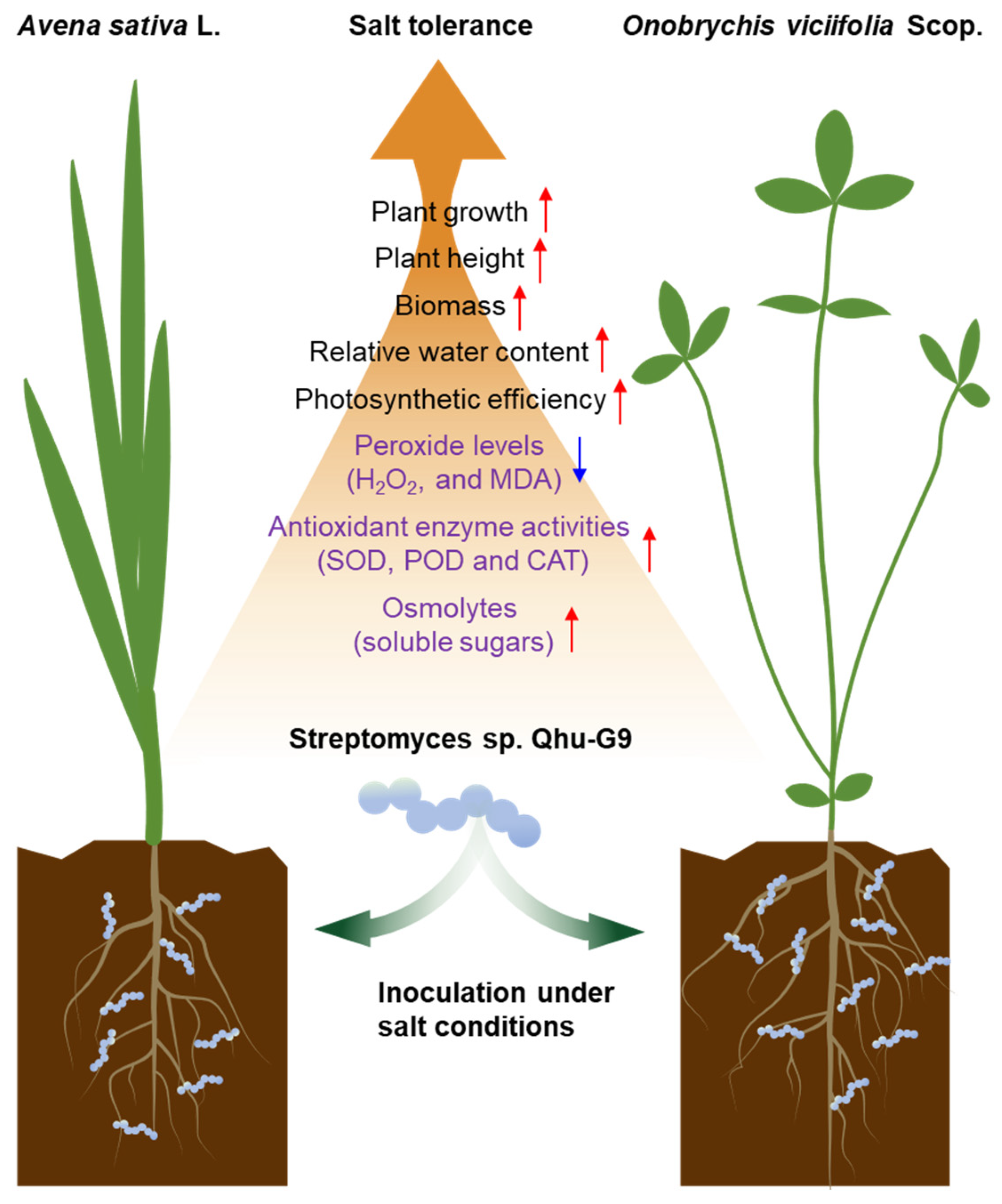
3. Discussion
4. Materials and Methods
4.1. Microbial and Plant Materials
4.2. Whole-Genome Phylogenetic Analysis and Comparative Genomic Analysis
4.3. Functional Annotation, Virulence Factor, and Genomic Islands Analysis
4.4. Assays of the Plant Growth-Promoting Properties
4.5. Preparation of Bacterial Suspension
4.6. Pot Experiments and Salt Stress Treatment
4.7. Plant Physiological and Biochemical Assays
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | 1-Aminocyclopropane-1-carboxylate |
| ANIb | ANI algorithm using BLAST |
| ANOVA | One-way analysis of variance |
| CAT | Catalase |
| EPSs | Exopolysaccharides |
| GBDP | Genome BLAST Distance Phylogeny |
| H2O2 | Hydrogen peroxide |
| IAA | Indole-3-acetic acid |
| MDA | Malondialdehyde |
| PGPR | Plant growth-promoting rhizobacterium |
| POD | Peroxidase |
| Fv/Fm | Quantum efficiency of photosystem II |
| SOD | Superoxide dismutase |
| TYGS | Type (Strain) Genome Server |
| UBCG2 | Up-to-Date Bacterial Core Gene 2 |
References
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil Salinity: Historical Perspectives and a World Overview of the Problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Zaman, M., Shahid, S.A., Heng, L., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 43–53. [Google Scholar]
- Wang, X.; Du, L.; Wang, W.; Zhang, Z.; Wu, Y.; Wang, Y. Functional identification of ZDS gene in apple (Malus halliana) and demonstration of it’s role in improving saline-alkali stress tolerance. Physiol. Mol. Biol. Plants 2023, 29, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Han, J.; Zhang, H. Rapid saline-alkali sensitivity testing using hydrogel/gold nanoparticles-modified screen-printed electrodes. Sci. Total Environ. 2023, 862, 160814. [Google Scholar] [CrossRef] [PubMed]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e914. [Google Scholar] [CrossRef]
- Tkacz, A.; Poole, P. Role of root microbiota in plant productivity. J. Exp. Bot. 2015, 66, 2167–2175. [Google Scholar] [CrossRef]
- Krishnan, K.S.; Rangasamy, A.; Arunan, Y.E.; Dananjeyan, B.; Subramanium, T.; Saminathan, V. Microbial inoculants—Fostering sustainability in groundnut production. Sci. Prog. 2025, 108, 368504251338943. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Liu, H.; Macdonald, C.A.; Singh, B.K. Microbial inoculants with higher capacity to colonize soils improved wheat drought tolerance. Microb. Biotechnol. 2023, 16, 2131–2144. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Lohmaneeratana, K.; Bunyoo, C.; Thamchaipenet, A. Actinobacteria-Plant Interactions in Alleviating Abiotic Stress. Plants 2022, 11, 2976. [Google Scholar] [CrossRef]
- Al-Turki, A.; Murali, M.; Omar, A.F.; Rehan, M.; Sayyed, R.Z. Recent advances in PGPR-mediated resilience toward interactive effects of drought and salt stress in plants. Front. Microbiol. 2023, 14, 1214845. [Google Scholar] [CrossRef]
- He, S.; Li, L.; Lv, M.; Wang, R.; Wang, L.; Yu, S.; Gao, Z.; Li, X. PGPR: Key to Enhancing Crop Productivity and Achieving Sustainable Agriculture. Curr. Microbiol. 2024, 81, 377. [Google Scholar] [CrossRef]
- Giannelli, G.; Potestio, S.; Visioli, G. The Contribution of PGPR in Salt Stress Tolerance in Crops: Unravelling the Molecular Mechanisms of Cross-Talk between Plant and Bacteria. Plants 2023, 12, 2197. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, X.; Hu, Y.; Zhang, J.; Li, H.; Cui, Y.; Zhao, D.; Dong, X.; Zhang, X.; Liu, K.; et al. Metabacillus dongyingensis sp. nov. Is Represented by the Plant Growth-Promoting Bacterium BY2G20 Isolated from Saline-Alkaline Soil and Enhances the Growth of Zea mays L. under Salt Stress. mSystems 2022, 7, e0142621. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Hui, L.; Jianlong, Z.; Hao, Z.; Cao, C.; Le, H.; Weimin, Z.; Yang, L.; Gao, Y.; Hou, X. The application of plant growth-promoting rhizobacteria enhances the tolerance of tobacco seedling to salt stress. Ecotoxicol. Environ. Saf. 2023, 265, 115512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A.; Tribedi, P. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. Int. 2017, 24, 3315–3335. [Google Scholar] [CrossRef]
- Ferreira, C.M.H.; Soares, H.; Soares, E.V. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 2019, 682, 779–799. [Google Scholar] [CrossRef]
- Li, H.; Lei, P.; Pang, X.; Li, S.; Xu, H.; Xu, Z.; Feng, X. Enhanced tolerance to salt stress in canola (Brassica napus L.) seedlings inoculated with the halotolerant Enterobacter cloacae HSNJ4. Appl. Soil Ecol. 2017, 119, 26–34. [Google Scholar] [CrossRef]
- Qurashi, A.W.; Sabri, A.N. Bacterial exopolysaccharide and biofilm formation stimulate chickpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 2012, 43, 1183–1191. [Google Scholar] [CrossRef]
- Liu, X.; Yao, T.; Chai, J.; Han, J. Adsorption of Sodium Ions by Exopolysaccharides from Pseudomonas simiae MHR6 and Its Improvement of Na+/K+ Homeostasis in Maize under Salt Stress. J. Agric. Food Chem. 2023, 71, 19949–19957. [Google Scholar] [CrossRef]
- Ayaz, M.; Ali, Q.; Jiang, Q.; Wang, R.; Wang, Z.; Mu, G.; Khan, S.A.; Khan, A.R.; Manghwar, H.; Wu, H.; et al. Salt Tolerant Bacillus Strains Improve Plant Growth Traits and Regulation of Phytohormones in Wheat under Salinity Stress. Plants 2022, 11, 2769. [Google Scholar] [CrossRef]
- Shabaan, M.; Asghar, H.N.; Zahir, Z.A.; Zhang, X.; Sardar, M.F.; Li, H. Salt-Tolerant PGPR Confer Salt Tolerance to Maize Through Enhanced Soil Biological Health, Enzymatic Activities, Nutrient Uptake and Antioxidant Defense. Front. Microbiol. 2022, 13, 901865. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Paul, S.C.; Parveen, S.; Alam, S.; Rahman, N.; Jannat, B.; Hoque, S.; Rahman, M.T.; Karim, M.M. Isolation and identification of salt-tolerant plant-growth-promoting rhizobacteria and their application for rice cultivation under salt stress. Can. J. Microbiol. 2020, 66, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Maurya, N.; Sharma, A.; Sundaram, S. The Role of PGPB-Microalgae interaction in Alleviating Salt Stress in Plants. Curr. Microbiol. 2024, 81, 270. [Google Scholar] [CrossRef]
- Pang, F.; Solanki, M.K.; Wang, Z. Streptomyces can be an excellent plant growth manager. World J. Microbiol. Biotechnol. 2022, 38, 193. [Google Scholar] [CrossRef]
- Akbari, A.; Gharanjik, S.; Koobaz, P.; Sadeghi, A. Plant growth promoting Streptomyces strains are selectively interacting with the wheat cultivars especially in saline conditions. Heliyon 2020, 6, e03445. [Google Scholar] [CrossRef]
- Lu, L.; Liu, N.; Fan, Z.; Liu, M.; Zhang, X.; Tian, J.; Yu, Y.; Lin, H.; Huang, Y.; Kong, Z. A novel PGPR strain, Streptomyces lasalocidi JCM 3373(T), alleviates salt stress and shapes root architecture in soybean by secreting indole-3-carboxaldehyde. Plant Cell Environ. 2024, 47, 1941–1956. [Google Scholar] [CrossRef]
- Gong, Y.; Chen, L.-J.; Pan, S.-Y.; Li, X.-W.; Xu, M.-J.; Zhang, C.-M.; Xing, K.; Qin, S. Antifungal potential evaluation and alleviation of salt stress in tomato seedlings by a halotolerant plant growth-promoting actinomycete Streptomyces sp. KLBMP5084. Rhizosphere 2020, 16, 100262. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, M.; Yang, L.; Wang, W.; Zhang, Y.; Liu, L.; Chai, J.; Liu, H.; Zhao, G. Comparative Physiological and Transcriptomic Analyses of Oat (Avena sativa) Seedlings under Salt Stress Reveal Salt Tolerance Mechanisms. Plants 2024, 13, 2238. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, X.; Lu, X.; Zhao, B.; Yang, Y.; Liu, J. Integrative analysis of transcriptome and metabolome reveal mechanism of tolerance to salt stress in oat (Avena sativa L.). Plant Physiol. Biochem. 2021, 160, 315–328. [Google Scholar] [CrossRef]
- Yin, H.; Zhou, H.; Wang, W.; Tran, L.P.; Zhang, B. Transcriptome Analysis Reveals Potential Roles of Abscisic Acid and Polyphenols in Adaptation of Onobrychis viciifolia to Extreme Environmental Conditions in the Qinghai-Tibetan Plateau. Biomolecules 2020, 10, 967. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.K.; Cho, Y.G. Genetic Research and Plant Breeding. Genes 2022, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Yin, H.X.; Fan, J.K.; Deng, J.W.; Zhu, Z.Y.; Qiu, Q.H.; Liu, Y.R.; Zhang, B.Y. Polyphasic identification and genome mining of secondary metabolites of a novel species Streptomyces haixigobicum sp. nov. Qhu-G9 isolated from the Gobi habitat of the Qinghai-Tibetan Plateau. Microbiol. China 2023, 50, 1872–1886. [Google Scholar]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Tambong, J.T.; Xu, R.; Fleitas, M.C.; Wang, L.; Hubbard, K.; Kutcher, R. Phylogenomic Insights on the Xanthomonas translucens Complex, and Development of a TaqMan Real-Time Assay for Specific Detection of pv. translucens on Barley. Phytopathology 2023, 113, 2091–2102. [Google Scholar] [CrossRef]
- Pishchik, V.N.; Chizhevskaya, E.P.; Chebotar, V.K.; Mirskaya, G.V.; Khomyakov, Y.V.; Vertebny, V.E.; Kononchuk, P.Y.; Kudryavtcev, D.V.; Bortsova, O.A.; Lapenko, N.G.; et al. PGPB Isolated from Drought-Tolerant Plants Help Wheat Plants to Overcome Osmotic Stress. Plants 2024, 13, 338. [Google Scholar] [CrossRef]
- Samayoa, B.E.; Shen, F.T.; Lai, W.A.; Chen, W.C. Screening and Assessment of Potential Plant Growth-promoting Bacteria Associated with Allium cepa Linn. Microbes Environ. 2020, 35, ME19147. [Google Scholar] [CrossRef]
- do Amaral, F.P.; Tuleski, T.R.; Pankievicz, V.C.S.; Melnyk, R.A.; Arkin, A.P.; Griffitts, J.; Tadra-Sfeir, M.Z.; Maltempi de Souza, E.; Deutschbauer, A.; Monteiro, R.A.; et al. Diverse Bacterial Genes Modulate Plant Root Association by Beneficial Bacteria. mBio 2020, 11, e03078-20. [Google Scholar] [CrossRef]
- Zang, Z.; Zhang, C.; Park, K.J.; Schwartz, D.A.; Podicheti, R.; Lennon, J.T.; Gerdt, J.P. Streptomyces secretes a siderophore that sensitizes competitor bacteria to phage infection. Nat. Microbiol. 2025, 10, 362–373. [Google Scholar] [CrossRef]
- Kumawat, K.C.; Sharma, P.; Singh, I.; Sirari, A.; Gill, B.S. Co-existence of Leclercia adecarboxylata (LSE-1) and Bradyrhizobium sp. (LSBR-3) in nodule niche for multifaceted effects and profitability in soybean production. World J. Microbiol. Biotechnol. 2019, 35, 172. [Google Scholar] [CrossRef]
- Nordlund, S.; Högbom, M. ADP-ribosylation, a mechanism regulating nitrogenase activity. FEBS J. 2013, 280, 3484–3490. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Ma, Y.; Shadan, A. Perspective of ACC-deaminase producing bacteria in stress agriculture. J. Biotechnol. 2022, 352, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, Z.; He, Y.; Huang, Y.; Li, X.; Ye, B.C. Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotoxicol. Environ. Saf. 2018, 164, 520–529. [Google Scholar] [CrossRef]
- Wang, W.; He, Y.; Wu, Z.; Li, T.; Xu, X.; Liu, X. De novo transcriptome sequencing of Capsicum frutescens L. and comprehensive analysis of salt stress alleviating mechanism by Bacillus atrophaeus WU-9. Physiol. Plant 2022, 174, e13728. [Google Scholar] [CrossRef]
- Nozari, R.M.; Ortolan, F.; Astarita, L.V.; Santarém, E.R. Streptomyces spp. enhance vegetative growth of maize plants under saline stress. Braz. J. Microbiol. 2021, 52, 1371–1383. [Google Scholar] [CrossRef]
- Kurniawan, A.; Chuang, H.W. Rhizobacterial Bacillus mycoides functions in stimulating the antioxidant defence system and multiple phytohormone signalling pathways to regulate plant growth and stress tolerance. J. Appl. Microbiol. 2022, 132, 1260–1274. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, L.; Zhang, S.; Li, B.; Li, J.; Wang, X.; Zhang, J.; Guan, C.; Ji, J. The combined use of a plant growth promoting Bacillus sp. strain and GABA promotes the growth of rice under salt stress by regulating antioxidant enzyme system, enhancing photosynthesis and improving soil enzyme activities. Microbiol. Res. 2023, 266, 127225. [Google Scholar] [CrossRef]
- Tian, T.; Wang, J.; Wang, H.; Cui, J.; Shi, X.; Song, J.; Li, W.; Zhong, M.; Qiu, Y.; Xu, T. Nitrogen application alleviates salt stress by enhancing osmotic balance, ROS scavenging, and photosynthesis of rapeseed seedlings (Brassica napus). Plant Signal. Behav. 2022, 17, 2081419. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Kim, J.; Na, S.I.; Kim, D.; Chun, J. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J. Microbiol. 2021, 59, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liu, B.; Zheng, D.; Chen, L.; Yang, J. VFDB 2025: An integrated resource for exploring anti-virulence compounds. Nucleic Acids Res. 2025, 53, D871–D877. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–w35. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, B.; Xiang, X.; Li, W.; Li, J.; Li, Y.; Tran, L.P.; Yin, H. Characterization of Bacillus pacificus G124 and Its Promoting Role in Plant Growth and Drought Tolerance. Plants 2024, 13, 2864. [Google Scholar] [CrossRef]

| Public Database | Annotated Gene Number | 100 bp ≤ Length < 300 bp | Length ≥ 300 bp |
|---|---|---|---|
| eggNOG_Annotation | 6529 | 2759 | 3519 |
| GO_Annotation | 5824 | 2355 | 3200 |
| KEGG_Annotation | 2849 | 1018 | 1742 |
| Nr_Annotation | 8515 | 3850 | 4034 |
| Pfam_Annotation | 6792 | 2916 | 3643 |
| Swissprot_Annotation | 3964 | 1358 | 2514 |
| TrEMBL_Annotation | 3964 | 1358 | 2514 |
| All_Annotated | 8525 | 3852 | 4035 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, X.; Ma, X.; Yin, H.; Chen, L.; Li, J.; Li, W.; Zhang, S.; Sun, C.; Zhang, B. Genome-Wide Insights into Streptomyces Novel Species Qhu-G9 and Its Potential for Enhancing Salt Tolerance and Growth in Avena sativa L. and Onobrychis viciifolia Scop. Plants 2025, 14, 2135. https://doi.org/10.3390/plants14142135
Xiang X, Ma X, Yin H, Chen L, Li J, Li W, Zhang S, Sun C, Zhang B. Genome-Wide Insights into Streptomyces Novel Species Qhu-G9 and Its Potential for Enhancing Salt Tolerance and Growth in Avena sativa L. and Onobrychis viciifolia Scop. Plants. 2025; 14(14):2135. https://doi.org/10.3390/plants14142135
Chicago/Turabian StyleXiang, Xin, Xiaolan Ma, Hengxia Yin, Liang Chen, Jiao Li, Wenjing Li, Shuhan Zhang, Chenghang Sun, and Benyin Zhang. 2025. "Genome-Wide Insights into Streptomyces Novel Species Qhu-G9 and Its Potential for Enhancing Salt Tolerance and Growth in Avena sativa L. and Onobrychis viciifolia Scop" Plants 14, no. 14: 2135. https://doi.org/10.3390/plants14142135
APA StyleXiang, X., Ma, X., Yin, H., Chen, L., Li, J., Li, W., Zhang, S., Sun, C., & Zhang, B. (2025). Genome-Wide Insights into Streptomyces Novel Species Qhu-G9 and Its Potential for Enhancing Salt Tolerance and Growth in Avena sativa L. and Onobrychis viciifolia Scop. Plants, 14(14), 2135. https://doi.org/10.3390/plants14142135






