Quinoa–Peanut Relay Intercropping Promotes Peanut Productivity Through the Temporal Optimization of Soil Physicochemical Properties and Microbial Community Composition in Saline Soil
Abstract
1. Introduction
2. Results
2.1. Quinoa–Peanut Relay Intercropping Alters Peanut Agronomic Characteristics and Improves Peanut Pod Yield
2.2. Quinoa–Peanut Relay Intercropping Changes Soil Properties
2.3. A Close Relationship Among Peanut Pod Yield, Pod Dry Weight, Biomass, and Soil Properties Was Observed
2.4. Quinoa–Peanut Relay Intercropping Alters Soil Bacterial Diversity in the Rhizosphere of Peanut Plants
2.5. Quinoa–Peanut Relay Intercropping Alters Soil Bacterial Community Composition in the Rhizosphere of Peanut Plants
2.6. Correlation Analysis of Soil Bacterial Community Composition and Soil Properties of Peanut
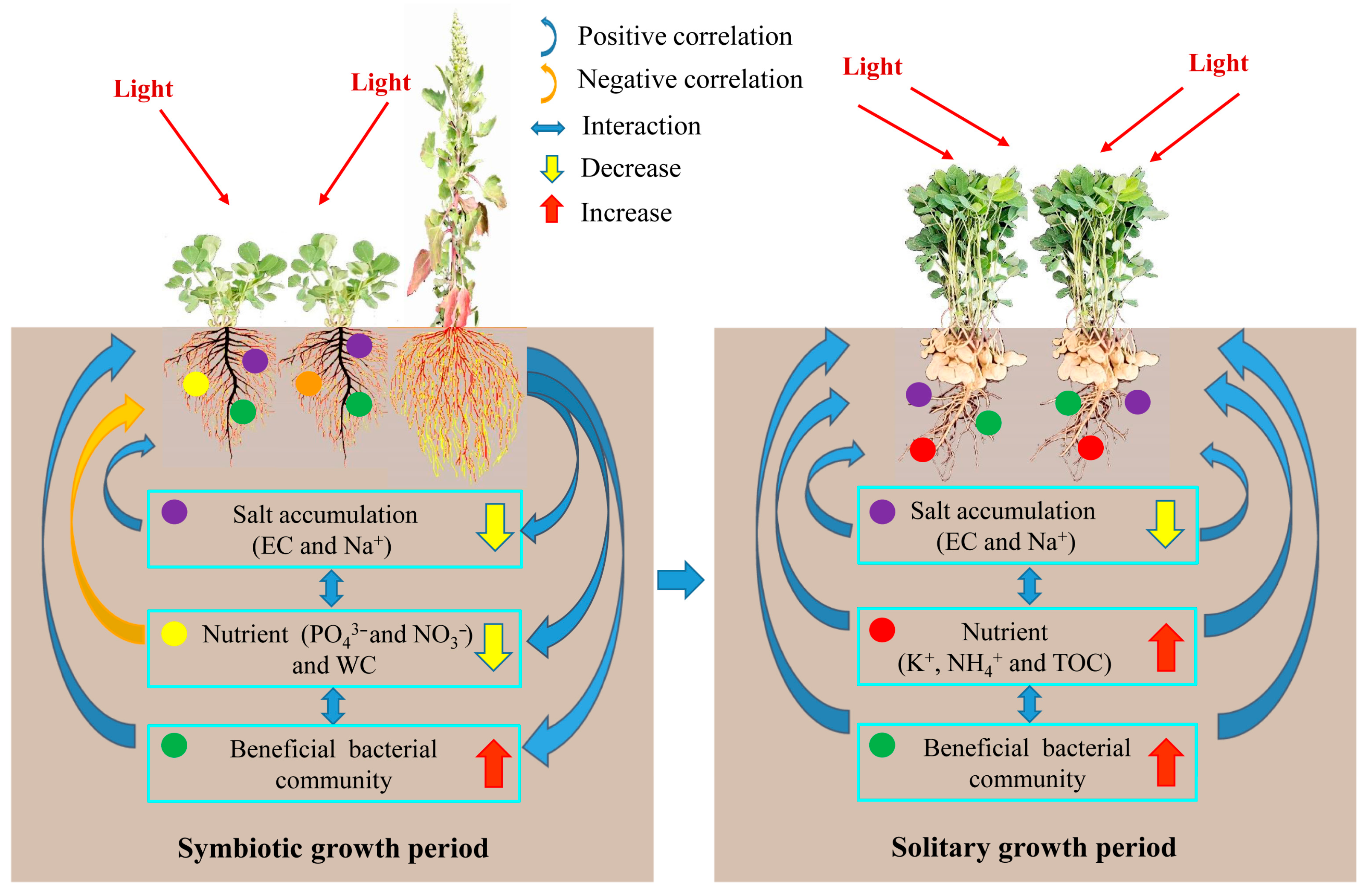
3. Discussion
3.1. Growth Dynamics and Yield Performance of Peanut
3.2. Soil Physicochemical Properties and Their Relationship with the Pod Yield of Peanut
3.3. Soil Microbial Community Characteristics and Their Relationships with Soil Properties
4. Materials and Methods
4.1. Field Experimental Site
4.2. Experimental Design and Field Management
4.3. Plant Sampling and Pod Yield
4.4. Soil Sampling
4.5. Soil Physicochemical Property Analyses
4.6. Soil DNA Extraction, PCR Amplification, and Sequencing Analysis
4.7. Llumina MiSeq and Processing of Sequencing Data
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Meena, H.N.; Meena, M.; Yadav, R.S. Comparative performance of seed types on yield potential of peanut arachis hypogaea l. under saline irrigation. Field Crop Res. 2016, 196, 305–310. [Google Scholar] [CrossRef]
- Xie, W.; Zhang, K.; Wang, X.Y.; Zou, X.X.; Zhang, X.J.; Yu, X.N.; Wang, Y.F.; Si, T. Peanut and cotton intercropping increases productivity and economic returns through regulating plant nutrient accumulation and soil microbial communities. BMC Plant Biol. 2022, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.P.; Bharadwaj, R.; Nayak, H.; Mahto, R.; Singh, R.K.; Prasad, S.K. Impact of salt stress on growth; productivity and physicochemical properties of plants, a Review. Int. J. Chem. Stud. 2019, 7, 1793–1798. [Google Scholar]
- Zhao, X.L.; Meng, T.Z.; Jin, S.H.; Ren, K.X.; Cai, Z.; Cai, B.; Li, S.B. The salinity survival strategy of Chenopodium quinoa, investigating microbial community shifts and nitrogen cycling in saline soils. Microorganisms 2023, 1112, 2829. [Google Scholar] [CrossRef]
- Liu, S.H.; Wang, N.; Li, D.Z.; Tian, C.Y.; Zhang, K.; Hu, M.F.; Niu, H.S.; Zhao, Z.Y. Effects of halophyte Suaeda salsa continuous cropping on physical and chemical properties of saline soil under drip irrigation in arid regions. Agric. Ecosyst. Environ. 2024, 371, 109076. [Google Scholar] [CrossRef]
- Jurado, C.; Díaz-Vivancos, P.; Gregorio, B.E.; Acosta-Motos, J.R.; Hernández, J.A. Effect of halophyte-based management in physiological and biochemical responses of tomato plants under moderately saline greenhouse conditions. Plant Physiol. Biochem. 2024, 206, 108228. [Google Scholar] [CrossRef]
- Wang, J.; Song, M.; Yao, L.; Li, P.; Si, E.; Li, B.; Meng, Y.; Ma, X.; Yang, K.; Zhang, H.; et al. Metagenomic analysis reveal the phytoremediation effects of monocropping and intercropping of halophytes Halogeton glomeratus and Suaeda glauca in saline soil of Northwestern China. BMC Plant Biol. 2025, 25, 213. [Google Scholar] [CrossRef]
- Liang, J.P.; Shi, W.J. Cotton/halophytes intercropping decreases salt accumulation and improves soil physicochemical properties and crop productivity in saline-alkali soils under mulched drip irrigation: A three-year field experiment-ScienceDirect. Field Crop Res. 2021, 2621, 108027. [Google Scholar] [CrossRef]
- Shen, L.; Wang, X.Y.; Liu, T.T.; Wei, W.W.; Zhang, S.; Keyhani, A.B.; Li, L.H.; Zhang, W. Border row effects on the distribution of root and soil resources in maize–soybean strip intercropping systems. Soil Tillage Res. 2023, 233, 105812. [Google Scholar] [CrossRef]
- Brooker, R.W.; Bennett, A.E.; Cong, W.F.; Daniell, T.J.; George, T.S.; Hallett, P.D.; Hawes, C.; Iannetta, P.P.M.; Jones, H.G.; Karley, A.J. Improving intercropping, a synthesis of research in agronomy; plant physiology and ecology. New Phytol. 2015, 206, 107–117. [Google Scholar] [CrossRef]
- Tanveer, M.; Anjum, S.A.; Hussain, S.; Cerdà, A.; Ashraf, U. Relay cropping as a sustainable approach: Problems and opportunities for sustainable crop production. Environ. Sci. Pollut. Res. 2017, 248, 6973–6988. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Hoffland, E.; Kuyper, T.W.; Yu, Y.; Li, H.G.; Zhang, C.C.; Zhang, F.S.; van der Werf, W. Yield gain, complementarity and competitive dominance in intercropping in China: A meta-analysis of drivers of yield gain using additive partitioning. Eur. J. Agron. 2020, 113, 125987. [Google Scholar] [CrossRef]
- Lin, P.; Wang, J.; Chen, P.; Fu, Z.; Luo, K.; Li, Y.; Pu, T.; Wang, X.; Yong, T.; Yang, W. Relay intercropped soybean promotes nodules development and nitrogen fixation by root exudates deposition. Front. Plant Sci. 2024, 15, 1447447. [Google Scholar] [CrossRef]
- Zhao, J.H.; Bedoussac, L.; Sun, J.H.; Chen, W.; Li, W.Q.; Bao, X.G.; van der Werf, W. Competition-recovery and overyielding of maize in intercropping depend on species temporal complementarity and nitrogen supply. Field Crop Res. 2023, 292, 108820. [Google Scholar] [CrossRef]
- Raza, M.A.; Bin Khalid, M.H.; Zhang, X.; Feng, L.Y.; Khan, I.; Hassan, M.J.; Ahmed, M.; Ansar, M.; Chen, Y.K.; Fan, Y.F.; et al. Effect of planting patterns on yield, nutrient accumulation and distribution in maize and soybean under relay intercropping systems. Sci. Rep. 2019, 91, 4947. [Google Scholar] [CrossRef]
- Te, X.; Hassan, M.J.; Cui, K.; Xiao, J.; Aslam, M.N.; Saeed, A.; Yang, W.; Ali, S. Effect of different planting pattern arrangements on soil organic matter and soil nitrogen content under a maize/soybean strip relay intercropping system. Front Plant Sci. 2022, 13, 95750. [Google Scholar] [CrossRef]
- Li, Q.S.; Chen, J.; Wu, L.K.; Luo, X.M.; Li, N.; Arafat, Y.; Lin, S.; Lin, W.X. Belowground interactions impact the soil bacterial community, soil fertility, and crop yield in maize/peanut intercropping systems. Int. J. Mol. Sci. 2018, 19, 622. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Keel, C. Seed-borne bacteria drive wheat rhizosphere microbiome assembly via niche partitioning and facilitation. Nat. Microbiol. 2025, 10, 1130–1144. [Google Scholar] [CrossRef]
- Shi, X.; Zhao, X.; Ren, J.; Dong, J.; Zhang, H.; Dong, Q.; Jiang, C.; Zhong, C.; Zhou, Y.; Yu, H. Influence of peanut, sorghum, and soil salinity on microbial community composition in interspecific interaction zone. Front. Microbiol. 2021, 12, 678250. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, N.; Beßler, H.; Engels, C.; Gleixner, G.; Habekost, M.; Milcu, A.; Partsch, S.; Sabais, A.C.; Scherber, C.; Steinbeiss, S.; et al. Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology 2010, 92, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.R.; Zhang, R.F. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 2014, 91, e85301. [Google Scholar] [CrossRef]
- Chang, X.; Wei, D.; Zeng, Y.; Zhao, X.; Hu, Y.; Wu, X.; Song, C.; Gong, G.; Chen, H.; Yang, C.; et al. Maize-soybean relay strip intercropping reshapes the rhizosphere bacterial community and recruits beneficial bacteria to suppress Fusarium root rot of soybean. Front. Microbiol. 2022, 13, 1009689. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Zhang, H.; Xu, N.; Sun, G. Use of mulberry-soybean intercropping in salt-alkali soil impacts the diversity of the soil bacterial community. Microb. Biotechnol. 2016, 9, 293–304. [Google Scholar] [CrossRef]
- He, C.; Zhou, B.; Wang, H.; Wei, Y.; Huang, J. A first-year maize/cassava relay intercropping system improves soil nutrients and changes the soil microbial community in the symbiotic period. Front. Microbiol. 2023, 14, 1087202. [Google Scholar] [CrossRef]
- Adolf, V.I.; Jacobsen, S.E.; Shabala, S. Salt tolerance mechanisms in quinoa (Chenopodium quinoa Willd). Environ. Exp. Bot. 2013, 92, 43–54. [Google Scholar] [CrossRef]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the nutritional composition of quinoa Chenopodium quinoa Willd. Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef]
- Ci, D.W.; Zhang, Z.M.; Ding, H.; Song, W.W.; Fu, F.P.; Kang, T.; Dai, L.X. Evaluation and selection indices of salinity tolerance in peanut seedling. Acta Ecol. Sin. 2015, 35, 805–814. [Google Scholar]
- Xie, H.; Wang, Q.C.; Zhang, P. Transcriptomic and metabolomic analysis of the response of quinoa seedlings to low temperatures. Biomolecules 2022, 127, 977. [Google Scholar] [CrossRef]
- Ranjan, A.; Kumari, A.; Pandey, D.M. Annotation of stress responsive candidate genes in peanut ESTs. Interdiscip. Sci. 2015, 72, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Khan, M.U.; Lin, X.Q.; Lin, Z.M.; Lin, S.; Lin, W.X. Evaluation of maize/peanut intercropping effects on microbial assembly, root exudates and peanut nitrogen uptake. Plant Physiol. Biochem. 2022, 171, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; He, Y.; Zhang, Z.; Wu, H.; He, L.; Jiang, J.; Meng, W.; Huang, Z.; Xiong, F.; Liu, J.; et al. Beneficial shift of rhizosphere soil nutrients and metabolites under a sugarcane/peanut intercropping system. Front. Plant Sci. 2022, 13, 1018727. [Google Scholar] [CrossRef]
- Boote, K. Growth Stages of Peanut (Arachis hypogaea L.). Peanut Sci. 1982, 9, 35–40. [Google Scholar] [CrossRef]
- Chi, B.J.; Zhang, Y.J.; Zhang, D.M.; Zhang, X.J.; Dai, J.L.; Dong, H.Z. Wide-strip intercropping of cotton and peanut combined with strip rotation increases crop productivity and economic returns. Field Crops Res. 2019, 243, 107617. [Google Scholar] [CrossRef]
- Wang, R.N.; Sun, Z.X.; Zhang, L.Z.; Yang, N.; Werf, W.V.D. Border-row proportion determines strength of interspecific interactions and crop yields in maize/peanut strip intercropping. Field Crop Res. 2020, 253, 107819. [Google Scholar] [CrossRef]
- Secco, D.; Bassegio, D.; Marins, A.C.D.; Chang, P.; Savioli, M.R.; Castro, M.B.S.; Mesa, V.R.; Silva, E.L.; Wendt, E.J. Short-term impacts of different intercropping times of maize and ruzigrass on soil physical properties in subtropical Brazil. Soil Tillage Res. 2023, 234, 105838. [Google Scholar] [CrossRef]
- Jacobsen, S.E. Adaptation of quinoa (Chenopodium quinoa) to Northern European agriculture: Studies on developmental pattern. Euphytica 1997, 96, 41–48. [Google Scholar] [CrossRef]
- Sudini, H.; Upadhyaya, H.D.; Reddy, S.V.; Mangala, U.N.; Rathore, A.; Kumar, K.V.K. Resistance to late leaf spot and rust diseases in ICRISAT’s mini core collection of peanut Arachis hypogaea L. Australas. Plant Pathol. 2015, 44, 557–566. [Google Scholar]
- Anco, D.; Thomas, J.; Jordan, D.; Shew, B.; Monfort, W.; Mehl, H.; Small, I.; Wright, D.; Tillman, B.; Dufault, N. Peanut yield loss in the presence of defoliation caused by late or early leaf spot. Plant Dis. 2020, 104, 1390–1399. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, G.; Ding, H.; Ci, D.; Dai, L.; Zhang, Z. Influence of salt stress on the rhizosphere soil bacterial community structure and growth performance of groundnut Arachis hypogaea L. Int. Microbiol. 2020, 233, 453–465. [Google Scholar]
- Shabala, S.; Hariadi, Y.; Jacobsen, S.E. Genotypic difference in salinity tolerance in quinoa is determined by differential control of xylem Na+ loading and stomatal density. J. Plant Physiol. 2013, 170, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Hauggaard-Nielsen, H.; Gooding, M.; Ambus, P.; Corre-Hellou, G.; Crozat, Y.; Dahlmann, C.; Dibet, A.; von Fragstein, P.; Pristeri, A.; Monti, M.; et al. Pea-barley intercropping for efficient symbiotic N2-fixation, soil N acquisition and use of other nutrients in European organic cropping systems. Field Crops Res. 2009, 113, 64–71. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, X.; Tang, Y.; Xiong, S.; Han, Y.; Wang, Z.; Feng, L.; Wang, G.; Yang, B.; Lei, Y. Directly linking plant N, P and K nutrition to biomass production in cotton-based intercropping systems. Eur. J. Agron. 2023, 151, 126960. [Google Scholar] [CrossRef]
- Zheng, B.C.; Zhou, Y.; Chen, P.; Zhang, X.N.; Du, Q.; Yang, H.; Wang, X.C.; Yang, F.; Xiao, T.; Li, L.; et al. Maize–legume intercropping promote N uptake through changing the root spatial distribution, legume nodulation capacity, and soil N availability. J. Integr. Agric. 2022, 216, 1755–1771. [Google Scholar]
- Bybee-Finley, K.A.; Bybee-Finley, U.D.; Pelzer, C.J.; Ruhl, L.; Lounsbury, N.P.; Warren, N.D.; Cherney, J.H.; Darby, H.M.; Smith, R.G.; Ryan, M.R. Quantifying the roles of intraspecific and interspecific diversification strategies in forage cropping systems. Field Crops Res. 2023, 302, 109036. [Google Scholar] [CrossRef]
- Han, Y.; Dong, Q.; Zhang, K.; Sha, D.; Jiang, C.; Yang, X.; Liu, X.; Zhang, H.; Wang, X.; Guo, F.; et al. Maize-peanut rotational strip intercropping improves peanut growth and soil properties by optimizing microbial community diversity. Peer J. 2022, 10, e13777. [Google Scholar] [CrossRef]
- Yang, Z.P.; Yang, W.P.; Li, S.C.; Hao, J.M.; Su, Z.F.; Sun, M.; Gao, Z.Q.; Zhang, C.L. Variation of bacterial community diversity in rhizosphere soil of sole-Cropped versus intercropped wheat field after harvest. PLoS ONE 2016, 113, e0150618. [Google Scholar] [CrossRef]
- Cavalieri, A.; Bak, F.; Garcia-Lemos, A.M.; Weiner, J.; Nicolaisen, M.H.; Nybroe, O. Effects of intra- and interspecific plant density on rhizosphere bacterial communities. Front. Microbiol. 2020, 11, 1045. [Google Scholar] [CrossRef]
- Qiao, Y.J.; Li, Z.Z.; Wang, X.; Zhu, B.; Hu, Y.G.; Zeng, Z.H. Effect of legume-cereal mixtures on the diversity of bacterial communities in the rhizosphere. Plant Soil Environ. 2012, 58, 174–180. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Janzen, H.H.; Bremer, E.; Smith, E.G.; Kanashiro, D.A.; Eastman, A.H.; Petri, R.M. The core soil bacterial genera and enzyme activities in incubated soils from century-old wheat rotations. Geoderma 2021, 404, 115275. [Google Scholar] [CrossRef]
- Yuan, M.; Yu, T.; Shi, Q.; Han, D.; Yu, K.; Wang, L.; Wang, S.; Xiang, H.; Wen, R.; Nian, H.; et al. Rhizosphere soil bacterial communities of continuous cropping-tolerant and sensitive soybean genotypes respond differently to long-term continuous cropping in Mollisols. Front. Microbiol. 2021, 12, 729047. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Wang, S.; Wang, J.; Qi, X.; Long, Q.; Huang, M. The shift of soil bacterial community after afforestation influence soil organic carbon and aggregate stability in Karst region. Front. Microbiol. 2022, 13, 901126. [Google Scholar] [CrossRef]
- Schmitz, E.V.; Just, C.L.; Schilling, K.; Streeter, M.; Mattes, T.E. Reconnaissance of oxygenic denitrifiers in agriculturally impacted soils. mSphere 2023, 83, e0057122. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria, Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Dai, L.X.; Zhang, G.K.; Yu, Z.P.; Ding, H.; Xu, Y.; Zhang, Z.M. Effect of drought stress and developmental stages on microbial community structure and diversity in peanut rhizosphere soil. Int. J. Mol. Sci. 2019, 20, 2265–2281. [Google Scholar] [CrossRef]
- Zhang, G.; Bai, J.; Tebbe, C.C.; Zhao, Q.; Jia, J.; Wang, W.; Wang, X.; Yu, L. Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ. Microbiol. 2021, 232, 1020–1037. [Google Scholar] [CrossRef]
- Wu, X.; Rensing, C.; Han, D.; Xiao, K.Q.; Dai, Y.; Tang, Z.; Liesack, W.; Peng, J.; Cui, Z.; Zhang, F. Genome-resolved metagenomics reveals distinct phosphorus acquisition strategies between soil microbiomes. mSystems 2022, 71, e0110721. [Google Scholar] [CrossRef]
- Xia, M.M.; Ma, X.L.; Liu, J.; Wu, M.; Li, Z.P.; Liu, M. Potential effect of key soil bacterial taxa on the increase of rice yield under milk vetch rotation. Front. Microbiol. 2023, 14, 1150505. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 34, 470–480. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, P.; Zhang, X.; Du, Q.; Zheng, B.; Yang, H.; Luo, K.; Lin, P.; Li, Y.; Pu, T.; et al. Maize-legume intercropping achieves yield advantages by improving leaf functions and dry matter partition. BMC Plant Biol. 2023, 231, 438. [Google Scholar] [CrossRef] [PubMed]
- Dick, W.A.; Cheng, L.; Wang, P. Soil acid and alkaline phosphtase activity as PH ajustment indicators. Soil Biol. Biochem. 2000, 32, 1915–1919. [Google Scholar] [CrossRef]
- Walkley, A. An Examination of methods for determining organic carbon and nitrogen in soils. J. Agric. Sci. 1935, 25, 598–609. [Google Scholar] [CrossRef]
- Sun, R.; Zheng, H.; Yin, S.; Zhang, X.; You, X.; Wu, H.; Suo, F.; Han, K.; Cheng, Y.; Zhang, C.; et al. Comparative study of pyrochar and hydrochar on peanut seedling growth in a coastal salt-affected soil of Yellow River Delta, China. Sci. Total Environ. 2022, 10, 833, 155183. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 1010, 996–998. [Google Scholar] [CrossRef]
- Song, Y.J.; Ma, L.; Zhang, H.Y.; Fu, R.; Liang, X.Y.; Li, J.L.; Li, J.J.; Li, M.; Shan, Y.; Cheng, J.S.; et al. The diversity and structure of diazotrophic communities in the rhizosphere of coastal saline plants is mainly affected by soil physicochemical factors but not host plant species. Front. Mar. Sci. 2022, 9, 1100289. [Google Scholar] [CrossRef]
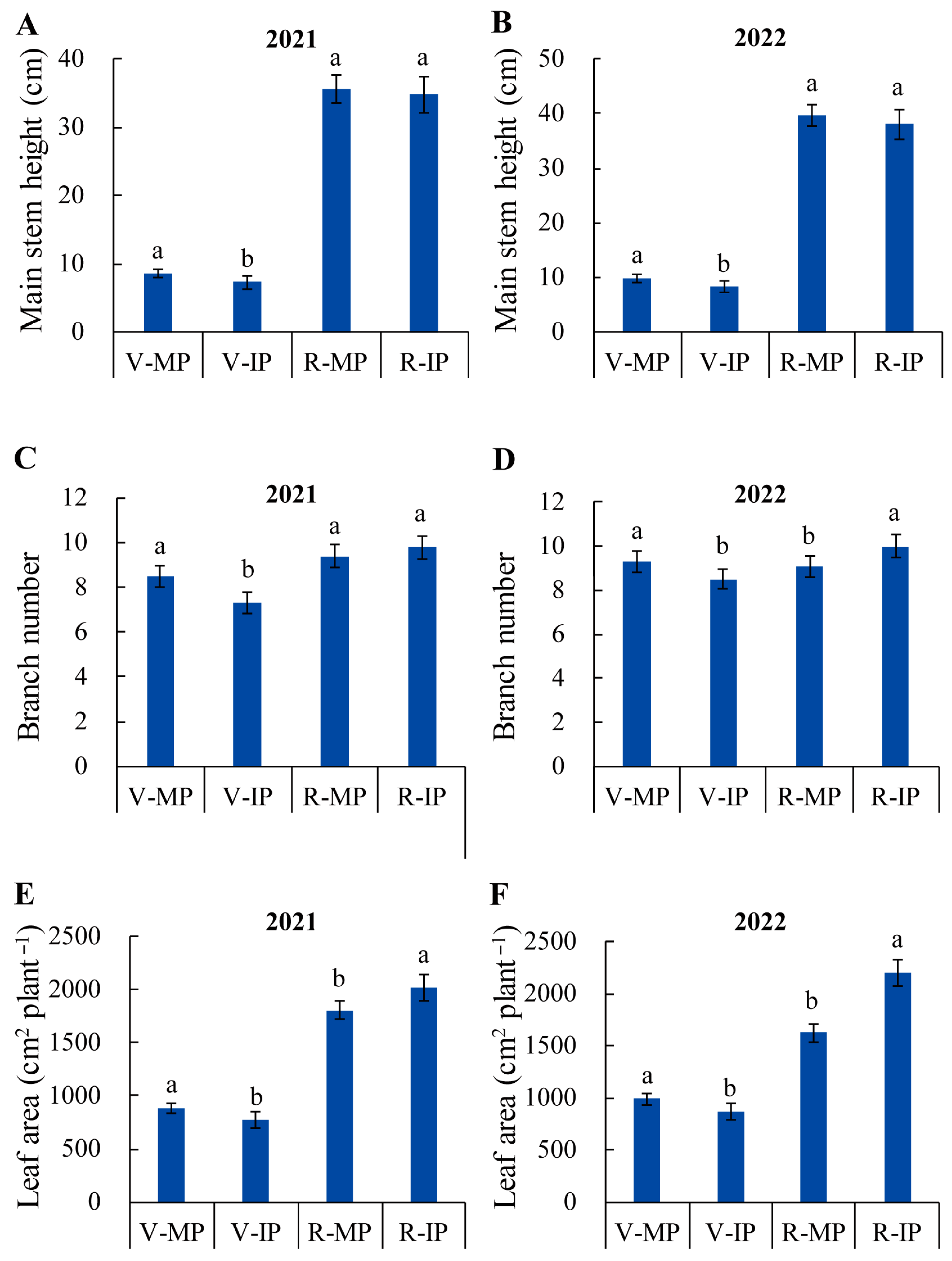





| Year | Treatment | Pod Yield (kg/hm2) | Pod Number/plant | Pod Weight (g/plant) | Biomass (g/plant) | Harvest Index | 100 Kernel Weight (g) |
|---|---|---|---|---|---|---|---|
| 2021 | MP | 6304 ± 20 b | 18.8 ± 1.0 b | 29.6 ± 1.6 b | 58.3 ± 3.0 b | 0.508 ± 0.02 a | 86.5 ± 3.5 b |
| IP | 6814 ± 21 a | 20.2 ± 1.2 a | 33.2 ± 1.7 a | 64.7 ± 4.2 a | 0.513 ± 0.03 a | 92.4 ± 4.0 a | |
| 2022 | MP | 7124 ± 29 b | 22.5 ± 1.2 b | 36.0 ± 1.8 b | 69.0 ± 3.8 b | 0.521 ± 0.01 a | 87.7 ± 4.2 b |
| IP | 7730 ± 29 a | 24.6 ± 1.3 a | 40.48 ± 2.1 a | 77.2 ± 4.3 a | 0.524 ± 0.02 a | 95.5 ± 3.6 a |
| Year | Treatment | WC % | EC (us/cm) | pH | Na+ (mg/kg) | K+ (mg/kg) | NO3− (mg/kg) | NH4+ (mg/kg) | PO43− (mg/kg) | TOC (g/kg) |
|---|---|---|---|---|---|---|---|---|---|---|
| 2021 | V_MP | 31.4 a | 682.4 a | 7.93 a | 282.5 a | 283.7 b | 20.5 a | 11.9 a | 82.3 a | 8.64 b |
| V_IP | 27.7 b | 450.2 b | 7.98 a | 206.1 b | 364.3 a | 18.7 b | 12.1 a | 70.7 b | 9.43 a | |
| R_MP | 15.1 a | 386.6 a | 8.06 a | 132.8 a | 179.6 b | 13.7 b | 7.3 b | 35.8 b | 6.70 b | |
| R_IP | 14.9 a | 304.2 b | 8.04 a | 118.2 b | 289.2 a | 15.1 a | 8.7 a | 40.3 a | 8.45 a | |
| 2022 | V_MP | 22.3 a | 576.7 a | 7.91 a | 244.1 a | 274.7 b | 22.4 a | 12.6 b | 89.6 a | 8.36 b |
| V_IP | 19.3 b | 354.2 b | 7.96 a | 183.1 b | 341.9 a | 19.6 b | 14.1 a | 66.1 b | 9.58 a | |
| R_MP | 12.3 a | 297.2 a | 8.13 a | 102.4 a | 157.7 b | 12.8 b | 8.2 b | 38.0 b | 6.13 b | |
| R_IP | 13.7 a | 230.9 b | 8.09 a | 88.1 b | 282.2 a | 13.6 a | 9.4 a | 43.7 a | 8.62 a |
| WC | EC | pH | Na+ | K+ | NO3− | NH4+ | PO43− | TOC | PY | PDW | BI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | 1 | |||||||||||
| EC | 0.859 * | 1 | ||||||||||
| pH | −0.754 | −0.828 * | 1 | |||||||||
| Na+ | 0.78 | 0.948 ** | −0.772 | 1 | ||||||||
| K+ | −0.775 | −0.686 | 0.537 | −0.507 | 1 | |||||||
| NO3 | 0.834 * | 0.501 | −0.329 | 0.432 | −0.539 | 1 | ||||||
| NH4+ | −0.508 | −0.471 | 0.586 | −0.264 | 0.278 | −0.434 | 1 | |||||
| PO43− | 0.919 ** | 0.951 ** | −0.892 * | 0.832 * | −0.792 | 0.578 | −0.609 | 1 | ||||
| TOC | −0.757 | −0.575 | 0.634 | −0.399 | 0.932 ** | −0.515 | 0.337 | −0.761 | 1 | |||
| PY | −0.828 * | −0.978 ** | 0.899 * | −0.942 ** | 0.681 | −0.401 | 0.413 | −0.947 ** | 0.624 | 1 | ||
| PDW | −0.719 | −0.922 ** | 0.684 | −0.917 * | 0.712 | −0.311 | 0.119 | −0.823 * | 0.555 | 0.927 ** | 1 | |
| BI | −0.906 * | −0.885 * | 0.592 | −0.856 * | 0.798 | −0.706 | 0.21 | −0.841 * | 0.654 | 0.838 * | 0.885 * | 1 |
| WC | EC | pH | Na+ | K+ | NO3− | NH4+ | PO43− | TOC | PY | PDW | BI | |
| WC | 1 | |||||||||||
| EC | −0.205 | 1 | ||||||||||
| pH | −0.712 | 0.104 | 1 | |||||||||
| Na+ | −0.568 | 0.735 | 0.452 | 1 | ||||||||
| K+ | 0.684 | −0.681 | −0.735 | −0.638 | 1 | |||||||
| NO3− | 0.913 * | −0.331 | −0.663 | −0.756 | 0.650 | 1 | ||||||
| NH4+ | 0.709 | −0.414 | −0.866 * | −0.441 | 0.945 ** | 0.601 | 1 | |||||
| PO43− | 0.503 | −0.615 | −0.002 | −0.347 | 0.572 | 0.476 | 0.404 | 1 | ||||
| TOC | 0.779 | −0.691 | −0.641 | −0.673 | 0.965 ** | 0.755 | 0.878 * | 0.735 | 1 | |||
| PY | 0.752 | −0.422 | −0.771 | −0.430 | 0.920 ** | 0.698 | 0.945 ** | 0.594 | 0.918 ** | 1 | ||
| PDW | 0.891 * | −0.45 | −0.637 | −0.536 | 0.852 * | 0.837 * | 0.823 * | 0.759 | 0.940 ** | 0.927 ** | 1 | |
| BI | 0.672 | −0.808 | −0.483 | −0.731 | 0.910 * | 0.726 | 0.754 | 0.803 | 0.969 ** | 0.838 * | 0.885 * | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, X.; Fu, R.; Li, J.; Gu, Y.; Yi, K.; Li, M.; Chen, C.; Zhang, H.; Li, J.; Ma, L.; et al. Quinoa–Peanut Relay Intercropping Promotes Peanut Productivity Through the Temporal Optimization of Soil Physicochemical Properties and Microbial Community Composition in Saline Soil. Plants 2025, 14, 2102. https://doi.org/10.3390/plants14142102
Liang X, Fu R, Li J, Gu Y, Yi K, Li M, Chen C, Zhang H, Li J, Ma L, et al. Quinoa–Peanut Relay Intercropping Promotes Peanut Productivity Through the Temporal Optimization of Soil Physicochemical Properties and Microbial Community Composition in Saline Soil. Plants. 2025; 14(14):2102. https://doi.org/10.3390/plants14142102
Chicago/Turabian StyleLiang, Xiaoyan, Rao Fu, Jiajia Li, Yinyu Gu, Kuihua Yi, Meng Li, Chuanjie Chen, Haiyang Zhang, Junlin Li, Lan Ma, and et al. 2025. "Quinoa–Peanut Relay Intercropping Promotes Peanut Productivity Through the Temporal Optimization of Soil Physicochemical Properties and Microbial Community Composition in Saline Soil" Plants 14, no. 14: 2102. https://doi.org/10.3390/plants14142102
APA StyleLiang, X., Fu, R., Li, J., Gu, Y., Yi, K., Li, M., Chen, C., Zhang, H., Li, J., Ma, L., Song, Y., Wang, X., Zhang, J., Wan, S., & Zhang, H. (2025). Quinoa–Peanut Relay Intercropping Promotes Peanut Productivity Through the Temporal Optimization of Soil Physicochemical Properties and Microbial Community Composition in Saline Soil. Plants, 14(14), 2102. https://doi.org/10.3390/plants14142102






