Resource-Efficient Cotton Network: A Lightweight Deep Learning Framework for Cotton Disease and Pest Classification
Abstract
1. Introduction
- We collected and constructed a dataset comprising several common cotton diseases and pests by integrating the most publicly available open-source datasets with field-collected images. The resulting dataset, named the Common Cotton Diseases and Pests Huge Dataset 11 (CCDPHD-11), includes 11 categories (10 types of diseased or pest-infested cotton leaves and 1 category of healthy leaves) and contains a total of 18,953 images.
- While many datasets used in cotton disease and pest research remain unpublished, the few that are open-sourced are often limited by small sample sizes and poor diversity, which hampers research progress. In contrast, CCDPHD-11 offers a relatively large sample size and high diversity compared to existing datasets. Moreover, our dataset will be made publicly available to promote further research in automated cotton disease and pest diagnosis.
- We propose the Resource-efficient Cotton Network (RF-Cott-Net), a lightweight deep learning model based on the MobileViTv2 architecture. By integrating quantization-aware training (QAT) and an early exit mechanism, RF-Cott-Net achieves a well-balanced trade-off between model compactness and performance. Experimental results demonstrate the model’s strong effectiveness in cotton disease and pest diagnostic tasks.
- On the proposed CCDPHD-11 dataset, our method achieves an accuracy of 98.4% across 11 classes, including healthy cotton leaves. The model size has a size of just 4.8 MB, with a 4.9 M parameter count, 310 M FLOPs, and inference latency of only 3.8 ms. These results demonstrate that RF-Cott-Net offers a practical solution for efficient, rapid, and low-cost on-device cotton disease and pest diagnosis.
2. Methods and Materials
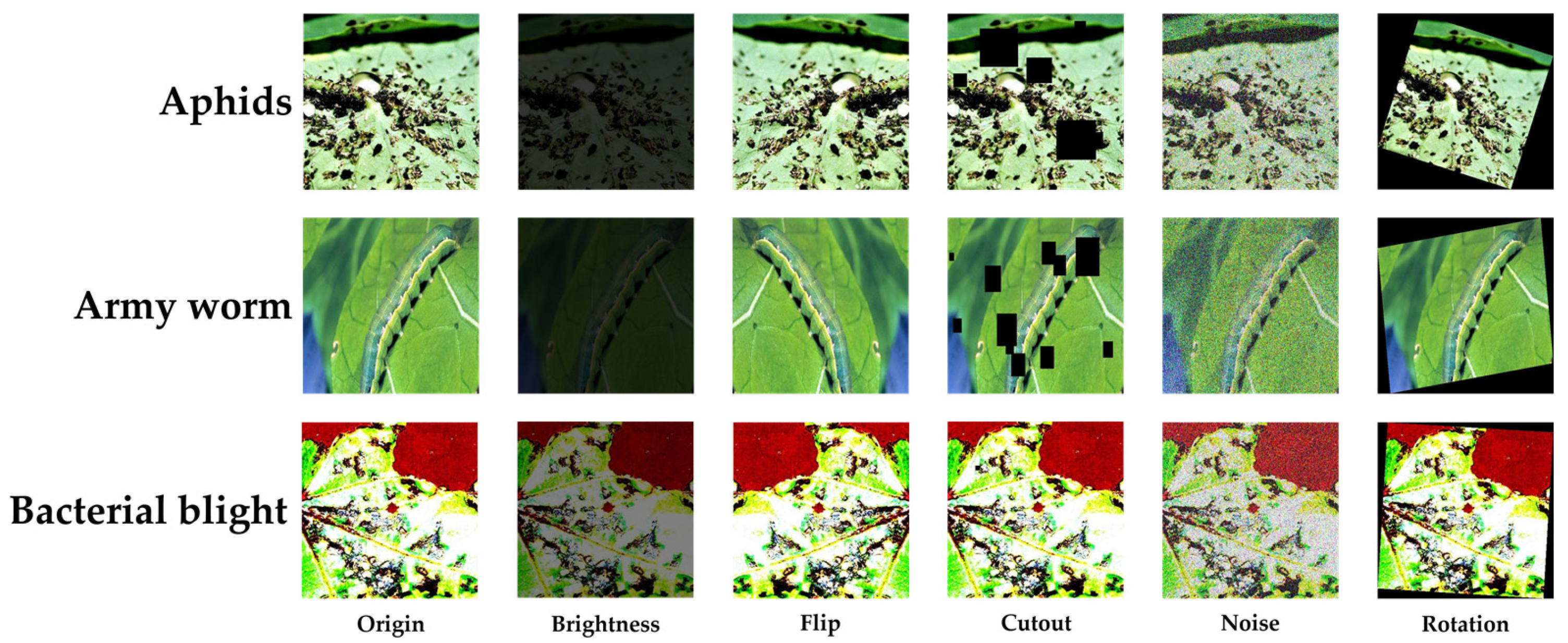
2.1. Dataset
2.2. MobileViTv2
2.3. Early Exit Mechanism
2.4. Quantization-Aware Training
2.5. Proposed Model (RF-Cott-Net)
2.5.1. Optimization in the Training Process
2.5.2. Optimization in Inference Process
| Algorithm 1 RF-Cott-Net Inference (Input: X) |
| for i = 1 to n do |
| zi = fi (X; θ) |
| if entropy (zi) < S then |
| return zi |
| end if |
| end for |
| return zn |
2.5.3. Structural Compression and Deployment Adaptability Enhancement
3. Experiments and Evaluation Metrics
3.1. Model Training Device and Parameters’ Setup
3.2. Model Evaluation Experiment
3.2.1. Baseline Models for Comparative Evaluation
3.2.2. Evaluation Metrics
3.3. Ablation Experiment
4. Results
4.1. Results of Model Evaluation Experiment
4.2. Results of Alation Experiment
5. Discussion
5.1. Advantages
5.2. Disadvantages
5.3. Future Perspectives
- Data expansion through synthetic generation: Future studies may leverage generative adversarial networks and related techniques to synthesize early-stage or rare disease and pest images [54], thereby enriching the dataset and improving the model’s ability to recognize mild infections.
- Cross-domain adaptation: Given the substantial environmental variability across different cotton-growing regions, future work could explore domain adaptation and transfer learning strategies to improve the model’s robustness and adaptability under diverse geographical and environmental conditions.
- Platform migration and edge deployment: RF-Cott-Net may be further validated through deployment in UAVs, handheld devices, mobile applications, or compact UGVs. These platforms impose stringent constraints on inference speed and energy consumption, and the lightweight design proposed in this study lays the groundwork for practical deployment in such scenarios.
- Moving towards a full pipeline for cotton disease and pest control: Future research could expand RF-Cott-Net into a comprehensive management system that integrates early warning, risk assessment, and decision support for cotton disease and pest control, thereby enhancing its utility and level of intelligence.
- Multimodal data fusion for comprehensive diagnosis: Future research could incorporate multimodal data sources—such as visible light, hyperspectral, thermal, and meteorological data—to enhance the comprehensiveness and precision of cotton disease and pest detection. By fusing heterogeneous data modalities, the system could better capture subtle physiological and environmental indicators, thereby improving early-stage diagnosis, disease differentiation, and stress factor attribution [55].
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, G.; Huang, J.-Q.; Chen, X.-Y.; Zhu, Y.-X. Recent Advances and Future Perspectives in Cotton Research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef] [PubMed]
- Scarpin, G.J.; Bhattarai, A.; Hand, L.C.; Snider, J.L.; Roberts, P.M.; Bastos, L.M. Cotton Lint Yield and Quality Variability in Georgia, USA: Understanding Genotypic and Environmental Interactions. Field Crops Res. 2025, 325, 109822. [Google Scholar] [CrossRef]
- Khan, M.A.; Wahid, A.; Ahmad, M.; Tahir, M.T.; Ahmed, M.; Ahmad, S.; Hasanuzzaman, M. World Cotton Production and Consumption: An Overview. In Cotton Production and Uses: Agronomy, Crop Protection, and Postharvest Technologies; Ahmad, S., Hasanuzzaman, M., Eds.; Springer: Singapore, 2020; pp. 1–7. ISBN 978-981-15-1472-2. [Google Scholar]
- Jimenez Madrid, A.M.; Munoz, G.; Wilkerson, T.; Chee, P.W.; Kemerait, R. Identification of Ramulariopsis Pseudoglycines Causing Areolate Mildew of Cotton in Georgia and First Detection of QoI Resistant Isolates in the United States. Plant Dis. 2025. [Google Scholar] [CrossRef]
- Edula, S.R.; Bag, S.; Milner, H.; Kumar, M.; Suassuna, N.D.; Chee, P.W.; Kemerait, R.C.; Hand, L.C.; Snider, J.L.; Srinivasan, R.; et al. Cotton Leafroll Dwarf Disease: An Enigmatic Viral Disease in Cotton. Mol. Plant Pathol. 2023, 24, 513–526. [Google Scholar] [CrossRef]
- Wubben, M.J.; Khanal, S.; Gaudin, A.G.; Callahan, F.E.; McCarty, J.C.; Jenkins, J.N.; Nichols, R.L.; Chee, P.W. Transcriptome Profiling and RNA-Seq SNP Analysis of Reniform Nematode (Rotylenchulus reniformis) Resistant Cotton (Gossypium hirsutum) Identifies Activated Defense Pathways and Candidate Resistance Genes. Front. Plant Sci. 2025, 16, 1532943. [Google Scholar] [CrossRef] [PubMed]
- Khanal, S.; Kumar, P.; da Silva, M.B.; Singh, R.; Suassuna, N.; Jones, D.C.; Davis, R.F.; Chee, P.W. Time-Course RNA-Seq Analysis of Upland Cotton (Gossypium hirsutum L.) Responses to Southern Root-Knot Nematode (Meloidogyne incognita) during Compatible and Incompatible Interactions. BMC Genom. 2025, 26, 183. [Google Scholar] [CrossRef]
- Ellsworth, P.C.; Fournier, A. Theory versus Practice: Are Insecticide Mixtures in Arizona Cotton Used for Resistance Management? Pest Manag. Sci. 2025, 81, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.-M.; Tu, Y.-H.; Li, T.; Ni, Y.; Wang, R.-F.; Wang, H. Deep Learning for Sustainable Agriculture: A Systematic Review on Applications in Lettuce Cultivation. Sustainability 2025, 17, 3190. [Google Scholar] [CrossRef]
- Qiu, P.; Zheng, B.; Yuan, H.; Yang, Z.; Lindsey, K.; Wang, Y.; Ming, Y.; Zhang, L.; Hu, Q.; Shaban, M.; et al. The Elicitor VP2 from Verticillium Dahliae Triggers Defence Response in Cotton. Plant Biotechnol. J. 2024, 22, 497–511. [Google Scholar] [CrossRef]
- Rani, M.; Murali-Baskaran, R.K. Synthetic Elicitors-Induced Defense in Crops against Herbivory: A Review. Plant Sci. 2025, 352, 112387. [Google Scholar] [CrossRef]
- Liu, E.M.; Huang, J. Risk Preferences and Pesticide Use by Cotton Farmers in China. J. Dev. Econ. 2013, 103, 202–215. [Google Scholar] [CrossRef]
- Zhou, W.; Li, M.; Achal, V. A Comprehensive Review on Environmental and Human Health Impacts of Chemical Pesticide Usage. Emerg. Contam. 2024, 11, 100410. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Chee, P.W.; Paterson, A.H.; Meng, C.; Zhang, J.; Ma, P.; Robertson, J.S.; Adhikari, J. High Resolution 3D Terrestrial LiDAR for Cotton Plant Main Stalk and Node Detection. Comput. Electron. Agric. 2021, 187, 106276. [Google Scholar] [CrossRef]
- Manavalan, R. Towards an Intelligent Approaches for Cotton Diseases Detection: A Review. Comput. Electron. Agric. 2022, 200, 107255. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, R.; Wang, M.; Lai, T.; Zhang, M. Self-Supervised Transformer-Based Pre-Training Method with General Plant Infection Dataset. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV); Springer: Singapore, 2024; pp. 189–202. [Google Scholar]
- Rothe, P.R.; Kshirsagar, R.V. Automated Extraction of Digital Images Features of Three Kinds of Cotton Leaf Diseases. In Proceedings of the 2014 International Conference on Electronics, Communication and Computational Engineering (ICECCE), Hosur, India, 17–18 November 2014; pp. 67–71. [Google Scholar]
- Jenifa, A.; Ramalakshmi, R.; Ramachandran, V. Classification of Cotton Leaf Disease Using Multi-Support Vector Machine. In Proceedings of the 2019 IEEE International Conference on Intelligent Techniques in Control, Optimization and Signal Processing (INCOS), Tamilnadu, India, 11–13 April 2019; pp. 1–4. [Google Scholar]
- Kurale, N.G.; Vaidya, M.V. Classification of Leaf Disease Using Texture Feature and Neural Network Classifier. In Proceedings of the 2018 International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 11–12 July 2018; pp. 1–6. [Google Scholar]
- Ebrahimi, M.; Khoshtaghaza, M.H.; Minaei, S.; Jamshidi, B. Vision-Based Pest Detection Based on SVM Classification Method. Comput. Electron. Agric. 2017, 137, 52–58. [Google Scholar] [CrossRef]
- Fu, H.; Zhao, H.; Song, R.; Yang, Y.; Li, Z.; Zhang, S. Cotton Aphid Infestation Monitoring Using Sentinel-2 MSI Imagery Coupled with Derivative of Ratio Spectroscopy and Random Forest Algorithm. Front. Plant Sci. 2022, 13, 1029529. [Google Scholar] [CrossRef]
- Wang, R.-F.; Su, W.-H. The Application of Deep Learning in the Whole Potato Production Chain: A Comprehensive Review. Agriculture 2024, 14, 1225. [Google Scholar] [CrossRef]
- Yang, Z.-Y.; Xia, W.-K.; Chu, H.-Q.; Su, W.-H.; Wang, R.-F.; Wang, H. A Comprehensive Review of Deep Learning Applications in Cotton Industry: From Field Monitoring to Smart Processing. Plants 2025, 14, 1481. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, R.-F. The Heterogeneous Network Community Detection Model Based on Self-Attention. Symmetry 2025, 17, 432. [Google Scholar] [CrossRef]
- Cui, K.; Zhu, R.; Wang, M.; Tang, W.; Larsen, G.D.; Pauca, V.P.; Alqahtani, S.; Yang, F.; Segurado, D.; Lutz, D.; et al. Detection and Geographic Localization of Natural Objects in the Wild: A Case Study on Palms. arXiv 2025, arXiv:2502.13023. [Google Scholar]
- Pan, C.-H.; Qu, Y.; Yao, Y.; Wang, M.-J.-S. HybridGNN: A Self-Supervised Graph Neural Network for Efficient Maximum Matching in Bipartite Graphs. Symmetry 2024, 16, 1631. [Google Scholar] [CrossRef]
- Zhao, C.-T.; Wang, R.-F.; Tu, Y.-H.; Pang, X.-X.; Su, W.-H. Automatic Lettuce Weed Detection and Classification Based on Optimized Convolutional Neural Networks for Robotic Weed Control. Agronomy 2024, 14, 2838. [Google Scholar] [CrossRef]
- Hu, P.; Cai, C.; Yi, H.; Zhao, J.; Feng, Y.; Wang, Q. Aiding Airway Obstruction Diagnosis with Computational Fluid Dynamics and Convolutional Neural Network: A New Perspective and Numerical Case Study. J. Fluids Eng. 2022, 144, 081206. [Google Scholar] [CrossRef]
- Cui, K.; Tang, W.; Zhu, R.; Wang, M.; Larsen, G.D.; Pauca, V.P.; Alqahtani, S.; Yang, F.; Segurado, D.; Fine, P.; et al. Real-Time Localization and Bimodal Point Pattern Analysis of Palms Using Uav Imagery. arXiv 2024, arXiv:2410.11124. [Google Scholar]
- Wu, A.-Q.; Li, K.-L.; Song, Z.-Y.; Lou, X.; Hu, P.; Yang, W.; Wang, R.-F. Deep Learning for Sustainable Aquaculture: Opportunities and Challenges. Sustainability 2025, 17, 5084. [Google Scholar] [CrossRef]
- Qiu, K.; Zhang, Y.; Ren, Z.; Li, M.; Wang, Q.; Feng, Y.; Chen, F. SpemNet: A Cotton Disease and Pest Identification Method Based on Efficient Multi-Scale Attention and Stacking Patch Embedding. Insects 2024, 15, 667. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, J.; Zhang, F.; Zhao, S.; Liu, W.; He, R.; Dong, H.; Hong, Q.; Tan, C.; Li, P. Residual Swin Transformer for Classifying the Types of Cotton Pests in Complex Background. Front. Plant Sci. 2024, 15, 1445418. [Google Scholar] [CrossRef]
- Faisal, H.M.; Aqib, M.; Rehman, S.U.; Mahmood, K.; Obregon, S.A.; Iglesias, R.C.; Ashraf, I. Detection of Cotton Crops Diseases Using Customized Deep Learning Model. Sci. Rep. 2025, 15, 10766. [Google Scholar] [CrossRef]
- Feng, H.; Chen, X.; Duan, Z. LCDDN-YOLO: Lightweight Cotton Disease Detection in Natural Environment, Based on Improved YOLOv8. Agriculture 2025, 15, 421. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, B.; Hu, Y.; Li, C.; Li, Y. Accurate Cotton Diseases and Pests Detection in Complex Background Based on an Improved YOLOX Model. Comput. Electron. Agric. 2022, 203, 107484. [Google Scholar] [CrossRef]
- Kakade, K.J.; More, V.A.; Shinde, M.; Suryawanshi, K.; Shinde, G.U. Design of Precision Agriculture System Using Automating Pink Bollworm Detection in Cotton Crops: AI Based Digital Approach for Sustainable Pest Management. In Proceedings of the 2025 1st International Conference on AIML-Applications for Engineering & Technology (ICAET), Pune, India, 16–17 January 2025; pp. 1–6. [Google Scholar]
- Mehta, S.; Rastegari, M. Mobilevit: Light-Weight, General-Purpose, and Mobile-Friendly Vision Transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar]
- Mehta, S.; Rastegari, M. Separable Self-Attention for Mobile Vision Transformers. arXiv 2022, arXiv:2206.02680. [Google Scholar]
- Wadekar, S.N.; Chaurasia, A. Mobilevitv3: Mobile-Friendly Vision Transformer with Simple and Effective Fusion of Local, Global and Input Features. arXiv 2022, arXiv:2209.15159. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.-C.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Xin, J.; Tang, R.; Lee, J.; Yu, Y.; Lin, J. DeeBERT: Dynamic Early Exiting for Accelerating BERT Inference. arXiv 2020, arXiv:2004.12993. [Google Scholar]
- Korol, G.; Beck, A.C.S. IoT–Edge Splitting with Pruned Early-Exit CNNs for Adaptive Inference. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2025. [Google Scholar] [CrossRef]
- Gilbert, M.S.; Pacheco, R.G.; Couto, R.S.; Fladenmuller, A.; Dias de Amorim, M.; de Campos, M.L.R.; Campista, M.E.M. Early-Exit Criteria for Edge Semantic Segmentation. In Proceedings of the 2025 IEEE International Conference on Machine Learning for Communications and Networking, Barcelona, Spain, 26–29 May 2025. [Google Scholar]
- Jacob, B.; Kligys, S.; Chen, B.; Zhu, M.; Tang, M.; Howard, A.; Adam, H.; Kalenichenko, D. Quantization and Training of Neural Networks for Efficient Integer-Arithmetic-Only Inference. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2704–2713. [Google Scholar]
- Xiao, G.; Lin, J.; Seznec, M.; Wu, H.; Demouth, J.; Han, S. SmoothQuant: Accurate and Efficient Post-Training Quantization for Large Language Models. In Proceedings of the 40th International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; pp. 38087–38099. [Google Scholar]
- Shi, H.; Cheng, X.; Mao, W.; Wang, Z. P2-ViT: Power-of-Two Post-Training Quantization and Acceleration for Fully Quantized Vision Transformer. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2024, 32, 1704–1717. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jegou, H. Training Data-Efficient Image Transformers & Distillation through Attention. In Proceedings of the 38th International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Pan, J.; Bulat, A.; Tan, F.; Zhu, X.; Dudziak, L.; Li, H.; Tzimiropoulos, G.; Martinez, B. EdgeViTs: Competing Light-Weight CNNs on Mobile Devices with Vision Transformers. In Proceedings of the Computer Vision—ECCV 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2022; pp. 294–311. [Google Scholar]
- Wang, R.-F.; Tu, Y.-H.; Chen, Z.-Q.; Zhao, C.-T.; Su, W.-H. A Lettpoint-Yolov11l Based Intelligent Robot for Precision Intra-Row Weeds Control in Lettuce. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=5162748 (accessed on 1 May 2025).
- Bishshash, P.; Nirob, A.S.; Shikder, H.; Sarower, A.H.; Bhuiyan, T.; Noori, S.R.H. A Comprehensive Cotton Leaf Disease Dataset for Enhanced Detection and Classification. Data Brief 2024, 57, 110913. [Google Scholar] [CrossRef]
- Paul Joshua, K.; Alex, S.A.; Mageswari, M.; Jothilakshmi, R. Enhanced Conditional Self-Attention Generative Adversarial Network for Detecting Cotton Plant Disease in IoT-Enabled Crop Management. Wirel. Netw. 2025, 31, 299–313. [Google Scholar] [CrossRef]
- Yang, Z.-X.; Li, Y.; Wang, R.-F.; Hu, P.; Su, W.-H. Deep Learning in Multimodal Fusion for Sustainable Plant Care: A Comprehensive Review. Sustainability 2025, 17, 5255. [Google Scholar] [CrossRef]






| Class | Training Set | Test Set | Total |
|---|---|---|---|
| Healthy | 5155 | 1289 | 6444 |
| Aphids | 1265 | 317 | 1582 |
| Amy worm | 1088 | 272 | 1360 |
| Bacterial blight | 1981 | 496 | 2477 |
| Cotton curl virus | 2024 | 506 | 2530 |
| Fusarium wilt | 704 | 177 | 881 |
| Herbicide growth damage | 224 | 56 | 280 |
| Leaf redding | 462 | 116 | 578 |
| Leaf variegation | 92 | 24 | 116 |
| Powdery mildew | 1080 | 270 | 1350 |
| Target spot | 1084 | 271 | 1355 |
| Total | 15,159 | 3794 | 18,953 |
| Model | Type | Storage Efficiency | Params | FLOPs | Latency (ms) | Accuracy (%) | F1 (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|---|---|---|---|---|
| EfficientNet-B0 | CNN | 15.6 MB | 3.8 M | 495 M | 10.6 | 97.4 | 97.4 | 97.5 | 97.3 |
| MobileNetV3 | CNN | 16.3 MB | 4.2 M | 280 M | 18.5 | 98.1 | 98.1 | 98.2 | 98.0 |
| ResNet-18 | CNN | 42.7 MB | 11.2 M | 2.4 G | 3.4 | 97.9 | 97.9 | 98.0 | 97.8 |
| MobileViT-S | Hybrid | 19 MB | 4.9 M | 1.9 G | 14.5 | 98.1 | 98.1 | 98.2 | 98.0 |
| MobileViTv2-0.75 | Hybrid | 9.6 MB | 2.5 M | 1.0 G | 9.8 | 96.6 | 96.6 | 96.8 | 96.4 |
| MobileViTv2-1.0 | Hybrid | 16.9 MB | 4.3 M | 1.9 G | 10.3 | 98.7 | 98.7 | 98.8 | 98.6 |
| DeiT-Tiny | Transformer | 21.2 MB | 5.5 M | 1.1 G | 12.3 | 92.1 | 92.1 | 92.3 | 92.0 |
| EdgeViT-XXS | Transformer | 14.8 MB | 4.1 M | 0.6 G | 9.5 | 94.2 | 94.2 | 94.3 | 94.1 |
| RF-Cott-Net | Hybrid | 4.8 MB | 4.9 M | 310 M | 3.8 | 98.4 | 98.4 | 98.5 | 98.3 |
| Model | Params | FLOPs | Latency (ms) | Accuracy (%) | F1 (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|---|---|---|
| MobileViTv2 + early exit | 4.4 M | 1.5 G | 8.7 | 98.5 | 98.5 | 98.6 | 98.4 |
| MobileViTv2 + QAT | 4.3 M | 470 M | 5.2 | 98.5 | 98.5 | 98.6 | 98.4 |
| RF-Cott-Net | 4.9 M | 310 M | 3.8 | 98.4 | 98.4 | 98.5 | 98.3 |
| Dataset | Class | Ava | Samp | Ave | Year | Reference |
|---|---|---|---|---|---|---|
| NA | 4 | No | 240 | 60 | 2019 | [18] |
| CottonInsect | 6 | Yes | 3225 | 537.5 | 2021 | [31] |
| NA | 3 | No | 2705 | 901.6 | 2024 | [32] |
| NA | 8 | No | 6712 | 839 | 2025 | [34] |
| NA | 5 | No | 5760 | 1152 | 2022 | [35] |
| SAR-CLD-2024 | 7 | Yes | 7000 | 1000 | 2024 | [53] |
| CCDPHD-11 | 11 | Yes | 18,953 | 1723 | 2025 | \ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhang, H.-W.; Dai, Y.-Q.; Cui, K.; Wang, H.; Chee, P.W.; Wang, R.-F. Resource-Efficient Cotton Network: A Lightweight Deep Learning Framework for Cotton Disease and Pest Classification. Plants 2025, 14, 2082. https://doi.org/10.3390/plants14132082
Wang Z, Zhang H-W, Dai Y-Q, Cui K, Wang H, Chee PW, Wang R-F. Resource-Efficient Cotton Network: A Lightweight Deep Learning Framework for Cotton Disease and Pest Classification. Plants. 2025; 14(13):2082. https://doi.org/10.3390/plants14132082
Chicago/Turabian StyleWang, Zhengle, Heng-Wei Zhang, Ying-Qiang Dai, Kangning Cui, Haihua Wang, Peng W. Chee, and Rui-Feng Wang. 2025. "Resource-Efficient Cotton Network: A Lightweight Deep Learning Framework for Cotton Disease and Pest Classification" Plants 14, no. 13: 2082. https://doi.org/10.3390/plants14132082
APA StyleWang, Z., Zhang, H.-W., Dai, Y.-Q., Cui, K., Wang, H., Chee, P. W., & Wang, R.-F. (2025). Resource-Efficient Cotton Network: A Lightweight Deep Learning Framework for Cotton Disease and Pest Classification. Plants, 14(13), 2082. https://doi.org/10.3390/plants14132082







