Plant Defense Proteins: Recent Discoveries and Applications
Abstract
1. Introduction
2. Plant Stressors and Defense Strategies
2.1. Pathogens
2.2. Environmental Stressors
3. Defense Protein Categories
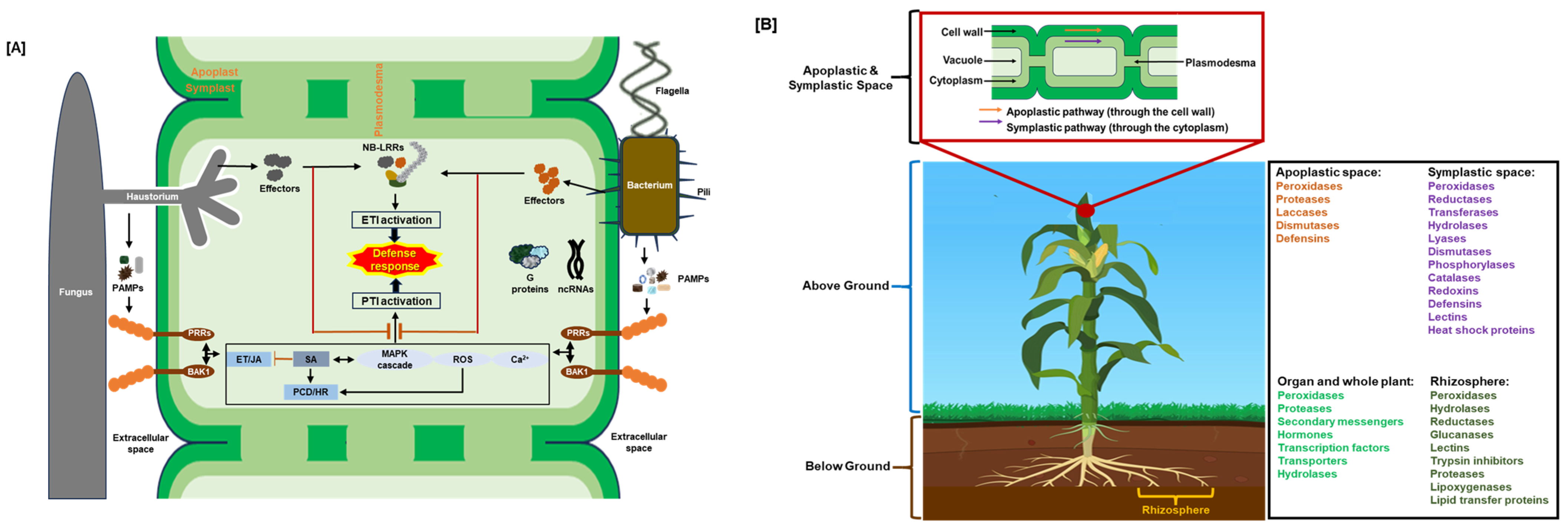
4. Defense Protein Localization
5. Examples of Known Critical Defense Proteins
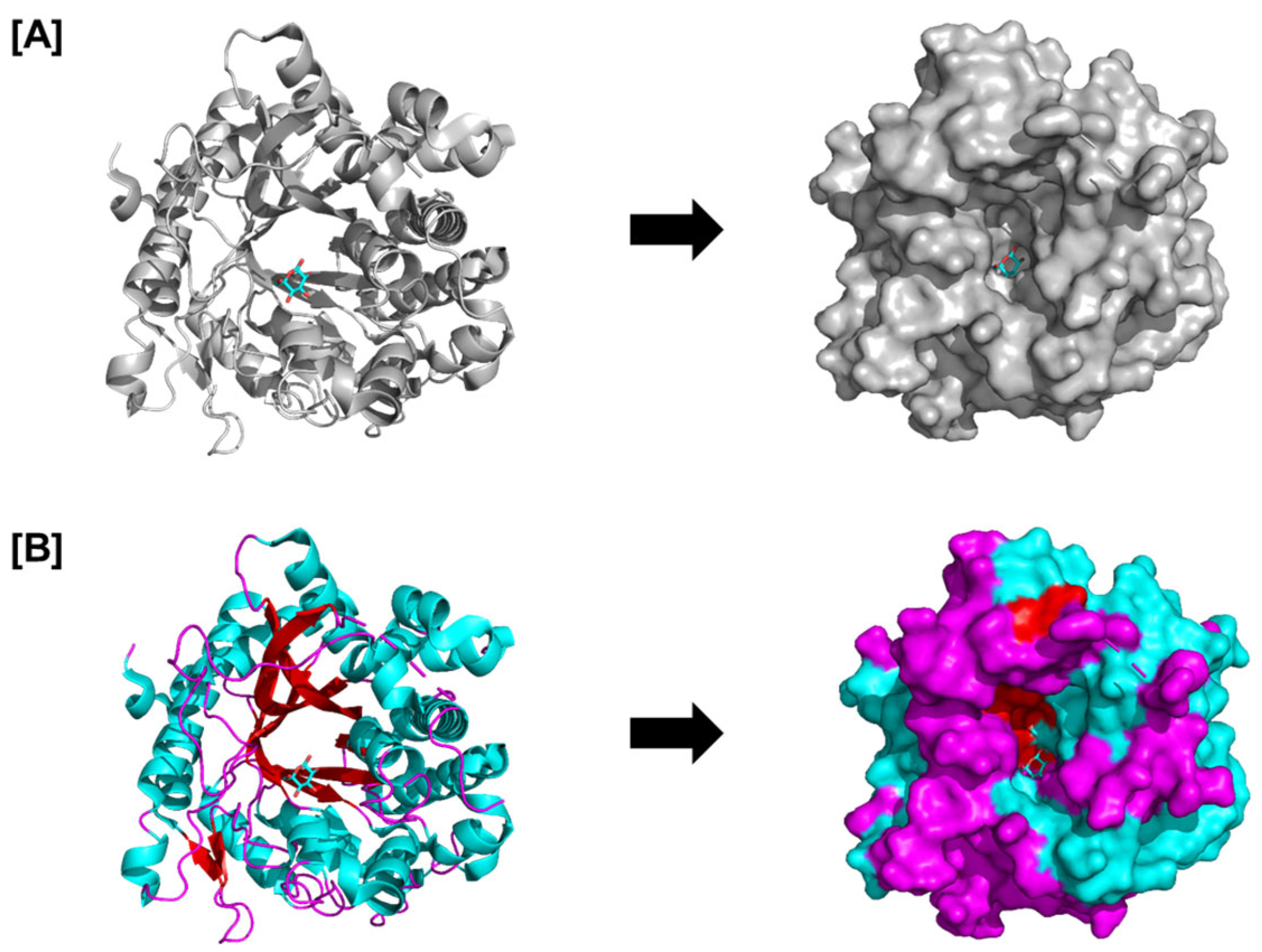
5.1. Glycosyl Hydrolases (GHs) in Archaeplastida
5.2. Jacalin-Related Lectins; Subcellular Localization
5.3. Plant Defense Initiated by Chloroplast Disruption
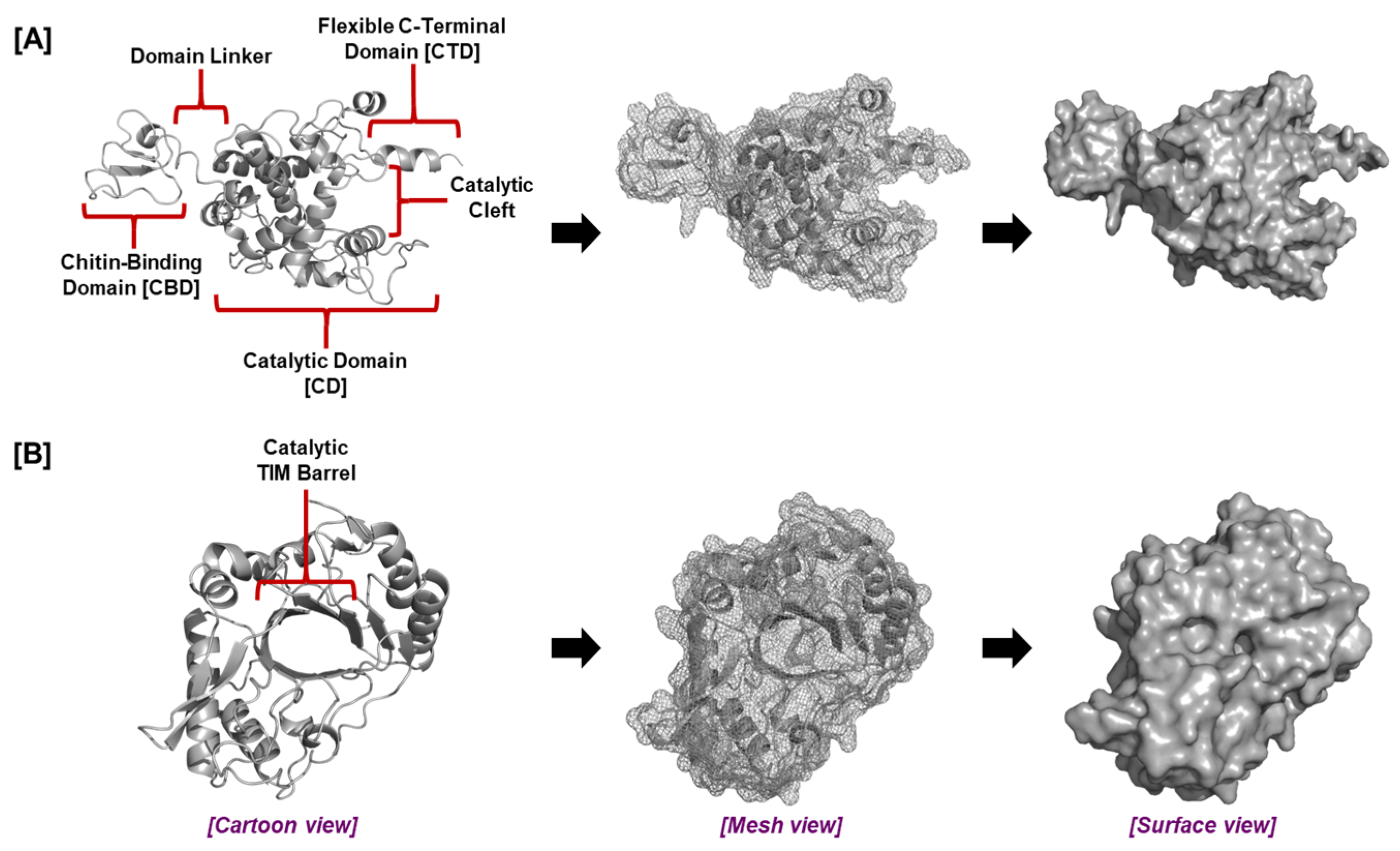
5.4. Rhizosphere-Associated Chitinases
6. Protein Functions in Defense
6.1. SWEET Proteins
6.2. Pathogen-Related Proteins (PR) Are Pivotal in Plant Defense
6.3. BAHD Acyltransferase in Herbivorous Pest Control
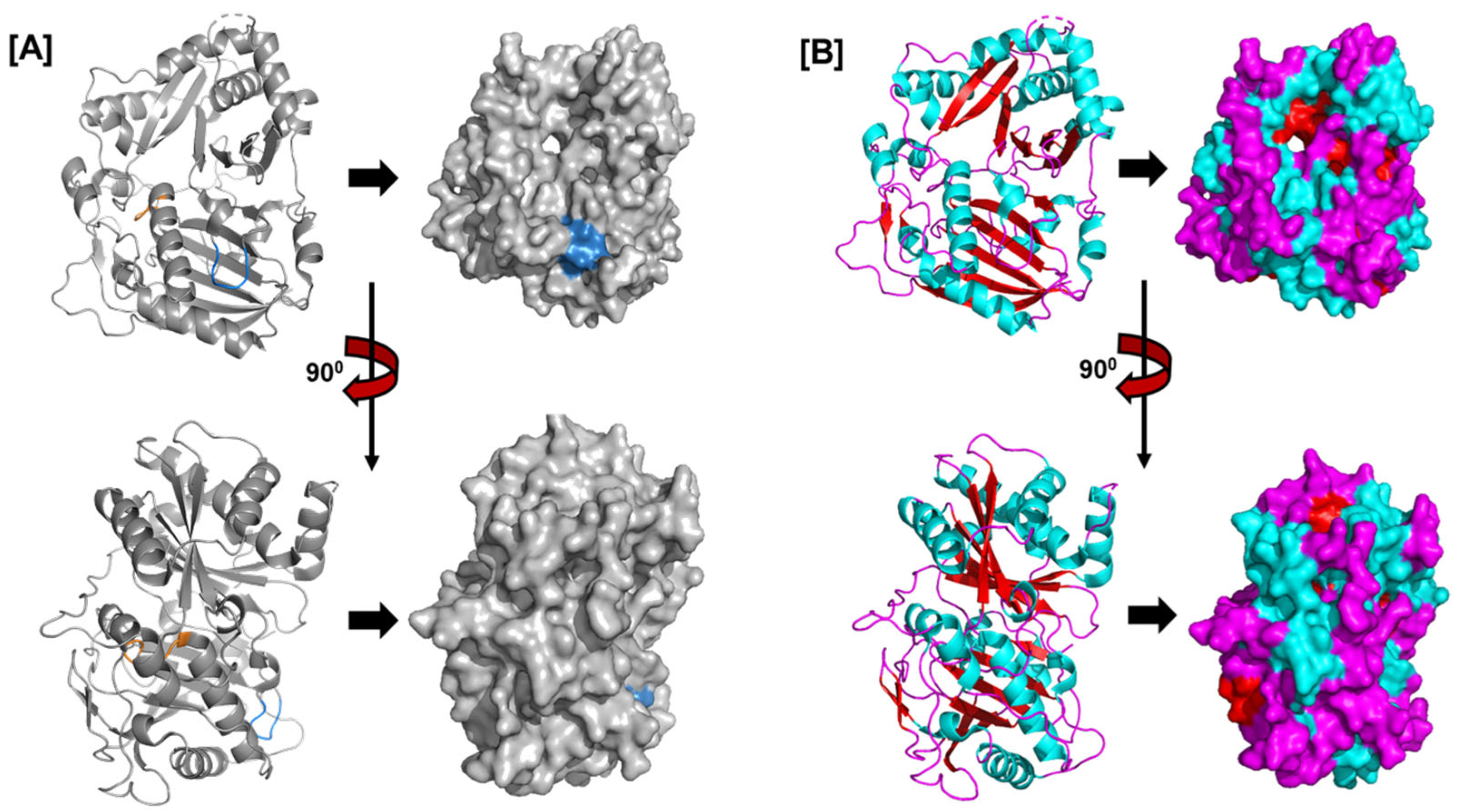
6.4. JALs in Protein Recognition and Defense
6.5. DNA Binding Proteins
6.6. Plant Defense Proteins in Viral Resistance
6.7. Class III Plant Peroxidases
6.8. Root Chitinases
6.9. G Proteins
7. Identification of Novel Plant Defense Proteins Through Omics Approaches
8. Applications of Plant Defense Proteins
8.1. Agricultural Applications
8.2. Applications in Biotechnology
8.3. Environmental Applications
9. Future Perspectives
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drummond, C.P.; Renner, T. Genomic insights into the evolution of plant chemical defense. Curr. Opin. Plant Biol. 2022, 68, 102254. [Google Scholar] [CrossRef]
- Negin, B.; Jander, G. Convergent and divergent evolution of plant chemical defenses. Curr. Opin. Plant Biol. 2023, 73, 102368. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.N.; Li, Y.T.; Wu, Y.Z.; Li, T.; Geng, R.; Cao, J.; Zhang, W.; Tan, X.L. Plant Disease Resistance-Related Signaling Pathways: Recent Progress and Future Prospects. Int. J. Mol. Sci. 2022, 23, 16200. [Google Scholar] [CrossRef]
- Buziashvili, A.; Yemets, A. Lactoferrin and its role in biotechnological strategies for plant defense against pathogens. Transgenic Res. 2023, 32, 1–16. [Google Scholar] [CrossRef]
- Pritchard, L.; Birch, P.R. The zigzag model of plant-microbe interactions: Is it time to move on? Mol. Plant Pathol. 2014, 15, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, J. The plant immune system—zig-zag model. Postepy Biochem. 2022, 68, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, J.; Zhou, J.M. From molecule to cell: The expanding frontiers of plant immunity. J. Genet. Genom. 2024, 51, 680–690. [Google Scholar] [CrossRef]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Du, B.; Haensch, R.; Alfarraj, S.; Rennenberg, H. Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. Camb. Philos. Soc. 2024, 99, 1524–1536. [Google Scholar] [CrossRef]
- Georgieva, M.; Vassileva, V. Stress Management in Plants: Examining Provisional and Unique Dose-Dependent Responses. Int. J. Mol. Sci. 2023, 24, 5105. [Google Scholar] [CrossRef]
- Brosset, A.; Blande, J.D. Volatile-mediated plant-plant interactions: Volatile organic compounds as modulators of receiver plant defence, growth, and reproduction. J. Exp. Bot. 2022, 73, 511–528. [Google Scholar] [CrossRef]
- Vlot, A.C.; Rosenkranz, M. Volatile compounds-the language of all kingdoms? J. Exp. Bot. 2022, 73, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, J.; Shi, J.; Khashi, U.R.M.; Liu, H.; Wei, Z.; Wu, F.; Dini-Andreote, F. Volatile-mediated interspecific plant interaction promotes root colonization by beneficial bacteria via induced shifts in root exudation. Microbiome 2024, 12, 207. [Google Scholar] [CrossRef]
- Remick, B.C.; Gaidt, M.M.; Vance, R.E. Effector-Triggered Immunity. Annu. Rev. Immunol. 2023, 41, 453–481. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef]
- Zhao, S.; Li, M.; Ren, X.; Wang, C.; Sun, X.; Sun, M.; Yu, X.; Wang, X. Enhancement of broad-spectrum disease resistance in wheat through key genes involved in systemic acquired resistance. Front. Plant Sci. 2024, 15, 1355178. [Google Scholar] [CrossRef] [PubMed]
- Lonjon, F.; Lai, Y.; Askari, N.; Aiyar, N.; Bundalovic-Torma, C.; Laflamme, B.; Wang, P.W.; Desveaux, D.; Guttman, D.S. The effector-triggered immunity landscape of tomato against Pseudomonas syringae. Nat. Commun. 2024, 15, 5102. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Shen, S.; Meng, L.; Sun, X.; Jin, Y.; Liu, Y.; Liu, D.; Ma, L.; Wang, H. Evasion of wheat resistance gene Lr15 recognition by the leaf rust fungus is attributed to the coincidence of natural mutations and deletion in AvrLr15 gene. Mol. Plant Pathol. 2024, 25, e13490. [Google Scholar] [CrossRef]
- Coomber, A.; Saville, A.; Ristaino, J.B. Evolution of Phytophthora infestans on its potato host since the Irish potato famine. Nat. Commun. 2024, 15, 6488. [Google Scholar] [CrossRef]
- Vo, K.T.X.; Yi, Q.; Jeon, J.S. Engineering effector-triggered immunity in rice: Obstacles and perspectives. Plant Cell Environ. 2023, 46, 1143–1156. [Google Scholar] [CrossRef]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, M.; Vishwakarma, K.; Kumar, V.; Arif, N.; Das, S.; Johnson, R.; Janeeshma, E.; Puthur, J.T.; Aliniaeifard, S.; Chauhan, D.K.; et al. Metal/Metalloid-Based Nanomaterials for Plant Abiotic Stress Tolerance: An Overview of the Mechanisms. Plants 2022, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.A.; Sabeem, M.; Kutty, M.S.; Rahman, S.; Alneyadi, M.K.; Alkaabi, A.B.; Almeqbali, E.S.; Brini, F.; Vijayan, R.; Masmoudi, K. Enzyme stabilization and thermotolerance function of the intrinsically disordered LEA2 proteins from date palm. Sci. Rep. 2023, 13, 11878. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Several lines of antioxidant defense against oxidative stress: Antioxidant enzymes, nanomaterials with multiple enzyme-mimicking activities, and low-molecular-weight antioxidants. Arch. Toxicol. 2024, 98, 1323–1367. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, Y.; Zhang, G.; Yang, Z.; Xu, W.; Chen, Q. The Applications and Mechanisms of Superoxide Dismutase in Medicine, Food, and Cosmetics. Antioxidants 2023, 12, 1675. [Google Scholar] [CrossRef]
- Movahedi, A.; Dzinyela, R.; Aghaei-Dargiri, S.; Alhassan, A.R.; Yang, L.; Xu, C. Advanced Study of Drought-Responsive Protein Pathways in Plants. Agronomy 2023, 13, 849. [Google Scholar] [CrossRef]
- Inam, S.; Muhammad, A.; Irum, S.; Rehman, N.; Riaz, A.; Uzair, M.; Khan, M.R. Genome editing for improvement of biotic and abiotic stress tolerance in cereals. Funct. Plant Biol. 2024, 51, FP24092. [Google Scholar] [CrossRef]
- Kumari, K.; Gusain, S.; Joshi, R. Engineering cold resilience: Implementing gene editing tools for plant cold stress tolerance. Planta 2024, 261, 2. [Google Scholar] [CrossRef]
- Srivastava, V.; De Guzman, C.; Fernandes, S.B. Beat the heat: Breeding, genomics, and gene editing for high nighttime temperature tolerance in rice. Curr. Opin. Plant Biol. 2024, 82, 102659. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Kamran, M.; Burdiak, P.; Karpiński, S. Crosstalk Between Abiotic and Biotic Stresses Responses and the Role of Chloroplast Retrograde Signaling in the Cross-Tolerance Phenomena in Plants. Cells 2025, 14, 176. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.; Franco, O.L. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants 2023, 12, 2226. [Google Scholar] [CrossRef] [PubMed]
- Mani, T.; Joshi, J.B.; Priyadharshini, R.; Sharmila, J.S.; Uthandi, S. Flagellin, a plant-defense-activating protein identified from Xanthomonas axonopodis pv. Dieffenbachiae invokes defense response in tobacco. BMC Microbiol. 2023, 23, 284. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X.; Zhang, J.; Huang, L.; Shi, Z.; Tian, Z.; Sha, A.; Lu, G. Thaumatin-like protein family genes VfTLP4-3 and VfTLP5 are critical for faba bean’s response to drought stress at the seedling stage. Plant Physiol. Biochem. 2024, 206, 108243. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Michaud, J.M.; Madani, A.; Fraser, J.S. A language model beats alphafold2 on orphans. Nat. Biotechnol. 2022, 40, 1576–1577. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Shobade, S.O.; Zabotina, O.A.; Nilsen-Hamilton, M. Plant root associated chitinases: Structures and functions. Front. Plant Sci. 2024, 15, 1344142. [Google Scholar] [CrossRef]
- Anwaar, S.; Jabeen, N.; Ahmad, K.S.; Shafique, S.; Irum, S.; Ismail, H.; Khan, S.U.; Tahir, A.; Mehmood, N.; Gleason, M.L. Cloning of maize chitinase 1 gene and its expression in genetically transformed rice to confer resistance against rice blast caused by Pyricularia oryzae. PLoS ONE 2024, 19, e0291939. [Google Scholar] [CrossRef]
- Ezzine, A.; Ben Hadj Mohamed, S.; Bezzine, S.; Aoudi, Y.; Hajlaoui, M.R.; Baciou, L.; Smaali, I. Improved Expression of a Thermostable GH18 Bacterial Chitinase in Two Different Escherichia coli Strains and Its Potential Use in Plant Protection and Biocontrol of Phytopathogenic Fungi. Mol. Biotechnol. 2024, 66, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Riyazuddin, R.; Csiszár, J. Plant Glutathione Peroxidases: Non-Heme Peroxidases with Large Functional Flexibility as a Core Component of ROS-Processing Mechanisms and Signalling. Antioxidants 2022, 11, 1624. [Google Scholar] [CrossRef] [PubMed]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Freitas, C.D.T.; Costa, J.H.; Germano, T.A.; de, O.R.R.; Ramos, M.V.; Bezerra, L.P. Class III plant peroxidases: From classification to physiological functions. Int. J. Biol. Macromol. 2024, 263, 130306. [Google Scholar] [CrossRef]
- Misas-Villamil, J.C.; van der Hoorn, R.A.; Doehlemann, G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016, 212, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Moloi, S.J.; Ngara, R. The roles of plant proteases and protease inhibitors in drought response: A review. Front. Plant Sci. 2023, 14, 1165845. [Google Scholar] [CrossRef]
- Tampakaki, A.P.; Bastaki, M.; Mansfield, J.W.; Panopoulos, N.J. Molecular determinants required for the avirulence function of AvrPphB in bean and other plants. Mol. Plant Microbe Interact. 2002, 15, 292–300. [Google Scholar] [CrossRef][Green Version]
- Warren, R.F.; Henk, A.; Mowery, P.; Holub, E.; Innes, R.W. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 1998, 10, 1439–1452. [Google Scholar] [CrossRef][Green Version]
- Shao, F.; Golstein, C.; Ade, J.; Stoutemyer, M.; Dixon, J.E.; Innes, R.W. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 2003, 301, 1230–1233. [Google Scholar] [CrossRef]
- Shao, F.; Merritt, P.M.; Bao, Z.; Innes, R.W.; Dixon, J.E. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 2002, 109, 575–588. [Google Scholar] [CrossRef]
- Lin, Y.H.; Xu, M.Y.; Hsu, C.C.; Damei, F.A.; Lee, H.C.; Tsai, W.L.; Hoang, C.V.; Chiang, Y.R.; Ma, L.S. Ustilago maydis PR-1-like protein has evolved two distinct domains for dual virulence activities. Nat. Commun. 2023, 14, 5755. [Google Scholar] [CrossRef] [PubMed]
- Pečenková, T.; Pleskot, R.; Žárský, V. Subcellular Localization of Arabidopsis Pathogenesis-Related 1 (PR1) Protein. Int. J. Mol. Sci. 2017, 18, 825. [Google Scholar] [CrossRef] [PubMed]
- Pečenková, T.; Pejchar, P.; Moravec, T.; Drs, M.; Haluška, S.; Šantrůček, J.; Potocká, A.; Žárský, V.; Potocký, M. Immunity functions of Arabidopsis pathogenesis-related 1 are coupled but not confined to its C-terminus processing and trafficking. Mol. Plant Pathol. 2022, 23, 664–678. [Google Scholar] [CrossRef]
- Bian, J.; Chen, R.; Gu, S.; Wang, W.; Yang, X. Quantitative proteomics analysis identified new interacting proteins of JAL30 in Arabidopsis. J. Proteom. 2024, 297, 105127. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef]
- Woo, J.Y.; Kim, Y.J.; Paek, K.H. CaLecRK-S.5, a pepper L-type lectin receptor kinase gene, accelerates Phytophthora elicitin-mediated defense response. Biochem. Biophys. Res. Commun. 2020, 524, 951–956. [Google Scholar] [CrossRef]
- Bay, G.; Lee, C.; Chen, C.; Mahal, N.K.; Castellano, M.J.; Hofmockel, K.S.; Halverson, L.J. Agricultural Management Affects the Active Rhizosphere Bacterial Community Composition and Nitrification. mSystems 2021, 6, e0065121. [Google Scholar] [CrossRef]
- Amador, V.C.; Santos-Silva, C.A.D.; Vilela, L.M.B.; Oliveira-Lima, M.; de Santana Rêgo, M.; Roldan-Filho, R.S.; Oliveira-Silva, R.L.; Lemos, A.B.; de Oliveira, W.D.; Ferreira-Neto, J.R.C.; et al. Lipid Transfer Proteins (LTPs)-Structure, Diversity and Roles beyond Antimicrobial Activity. Antibiotics 2021, 10, 1281. [Google Scholar] [CrossRef]
- Han, Z.; Xiong, D.; Schneiter, R.; Tian, C. The function of plant PR1 and other members of the CAP protein superfamily in plant-pathogen interactions. Mol. Plant Pathol. 2023, 24, 651–668. [Google Scholar] [CrossRef]
- Chiu, T.; Poucet, T.; Li, Y. The potential of plant proteins as antifungal agents for agricultural applications. Synth. Syst. Biotechnol. 2022, 7, 1075–1083. [Google Scholar] [CrossRef]
- Karimian, P.; Trusov, Y.; Botella, J.R. Conserved Role of Heterotrimeric G Proteins in Plant Defense and Cell Death Progression. Genes 2024, 15, 115. [Google Scholar] [CrossRef]
- Mann, M.M.; Vigil, T.N.; Felton, S.M.; Fahy, W.E.; Kinkeade, M.A.; Kartseva, V.K.; Rowson, M.-J.C.; Frost, A.J.; Berger, B.W. Proteins in Synthetic Biology with Agricultural and Environmental Applications. SynBio 2023, 1, 77–88. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, Y.; Han, S.; Wang, J.; Liu, Y.; Yin, J. Structure, evolution, and roles of SWEET proteins in growth and stress responses in plants. Int. J. Biol. Macromol. 2024, 263, 130441. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, X.; Li, M.; Lun, Z.; Yan, X.; Yin, C.; Yuan, G.; Wang, X.; Liu, N.; Liu, D.; et al. Plant immunity suppression by an exo-β-1,3-glucanase and an elongation factor 1α of the rice blast fungus. Nat. Commun. 2023, 14, 5491. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Park, C.J.; Kim, S.K.; Kim, K.J.; Paek, K.H. In vivo binding of hot pepper bZIP transcription factor CabZIP1 to the G-box region of pathogenesis-related protein 1 promoter. Biochem. Biophys. Res. Commun. 2006, 344, 55–62. [Google Scholar] [CrossRef]
- Brummell, D.A.; Catala, C.; Lashbrook, C.C.; Bennett, A.B. A membrane-anchored E-type endo-1,4-beta-glucanase is localized on Golgi and plasma membranes of higher plants. Proc. Natl. Acad. Sci. USA 1997, 94, 4794–4799. [Google Scholar] [CrossRef]
- Punja, Z.K.; Zhang, Y.Y. Plant chitinases and their roles in resistance to fungal diseases. J. Nematol. 1993, 25, 526–540. [Google Scholar]
- Ferreira, R.B.; Monteiro, S.; Freitas, R.; Santos, C.N.; Chen, Z.; Batista, L.M.; Duarte, J.; Borges, A.; Teixeira, A.R. The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 2007, 8, 677–700. [Google Scholar] [CrossRef]
- Garcia, T.B.; Soares, A.A.; Costa, J.H.; Costa, H.P.S.; Neto, J.X.S.; Rocha-Bezerra, L.C.B.; Silva, F.D.A.; Arantes, M.R.; Sousa, D.O.B.; Vasconcelos, I.M.; et al. Gene expression and spatiotemporal localization of antifungal chitin-binding proteins during Moringa oleifera seed development and germination. Planta 2019, 249, 1503–1519. [Google Scholar] [CrossRef]
- Honorato, L.; Bonilla, J.J.A.; Piffer, A.C.; Nimrichter, L. Fungal Extracellular Vesicles as a Potential Strategy for Vaccine Development. Curr. Top. Microbiol. Immunol. 2021, 432, 121–138. [Google Scholar] [CrossRef]
- Jovanović, S.; Kukavica, B.; Vidović, M.; Morina, F.; Menckhoff, L. Class III Peroxidases: Functions, Localization and Redox Regulation of Isoenzymes; Springer International Publishing: Cham, Switzerland, 2018; pp. 269–300. [Google Scholar]
- Petre, B.; Major, I.; Rouhier, N.; Duplessis, S. Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol. 2011, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Z.; Yin, W.; Xu, K.; Wang, S.; Shang, Q.; Sa, W.; Liang, J.; Wang, L. Genome-wide analysis of the Thaumatin-like gene family in Qingke (Hordeum vulgare L. var. nudum) uncovers candidates in-volved in plant defense against biotic and abiotic stresses. Front. Plant Sci. 2022, 13, 912296. [Google Scholar] [CrossRef]
- Weerawanich, K.; Webster, G.; Ma, J.K.; Phoolcharoen, W.; Sirikantaramas, S. Gene expression analysis, subcellular localization, and in planta antimicrobial activity of rice (Oryza sativa L.) defensin 7 and 8. Plant Physiol. Biochem. 2018, 124, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhang, Y.; Xu, Y.; Bai, M.; Luo, W.; Wang, B.; Niu, Y.; Zhao, Y.; Li, S.; Weng, Y.; et al. Cyclophilin OsCYP20-2 with a novel variant integrates defense and cell elongation for chilling response in rice. New Phytol. 2020, 225, 2453–2467. [Google Scholar] [CrossRef]
- Singh, H.; Kaur, K.; Singh, M.; Kaur, G.; Singh, P. Plant Cyclophilins: Multifaceted Proteins With Versatile Roles. Front. Plant Sci. 2020, 11, 585212. [Google Scholar] [CrossRef]
- De Zaeytijd, J.; Van Damme, E.J. Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins 2017, 9, 123. [Google Scholar] [CrossRef]
- Dougherty, K.; Hudak, K.A. Phylogeny and domain architecture of plant ribosome inactivating proteins. Phytochemistry 2022, 202, 113337. [Google Scholar] [CrossRef]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef]
- Fang, C.; Wu, S.; Li, Z.; Pan, S.; Wu, Y.; An, X.; Long, Y.; Wei, X.; Wan, X. A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize. Int. J. Mol. Sci. 2023, 24, 1660. [Google Scholar] [CrossRef]
- Liang, R.; You, L.; Dong, F.; Zhao, X.; Zhao, J. Identification of Hydroxyproline-Containing Proteins and Hydroxylation of Proline Residues in Rice. Front. Plant Sci. 2020, 11, 1207. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chen, X.M.; Zhang, Y.; Cho, Y.H.; Wang, A.R.; Yeung, E.C.; Zeng, X.; Guo, S.X.; Lee, Y.I. Immunolocalization and Changes of Hydroxyproline-Rich Glycoproteins During Symbiotic Germination of Dendrobium officinale. Front. Plant Sci. 2018, 9, 552. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Sajib, S.D.; Jui, Z.S.; Arabia, S.; Islam, T.; Ghosh, A. Genome-wide identification of glutathione S-transferase gene family in pepper, its classification, and expression profiling under different anatomical and environmental conditions. Sci. Rep. 2019, 9, 9101. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Sylvestre-Gonon, E.; Law, S.R.; Schwartz, M.; Robe, K.; Keech, O.; Didierjean, C.; Dubos, C.; Rouhier, N.; Hecker, A. Functional, Structural and Biochemical Features of Plant Serinyl-Glutathione Transferases. Front. Plant Sci. 2019, 10, 608. [Google Scholar] [CrossRef] [PubMed]
- Alam, O.; Khan, L.U.; Khan, A.; Salmen, S.H.; Ansari, M.J.; Mehwish, F.; Ahmad, M.; Zaman, Q.U.; Wang, H.F. Functional characterisation of Dof gene family and expression analysis under abiotic stresses and melatonin-mediated tolerance in pitaya (Selenicereus undatus). Funct. Plant Biol. 2024, 51, FP23269. [Google Scholar] [CrossRef]
- Oliveira, E.G.; Filho, C.; Rodrigues, R.A.L. An overview of viral chitinases: General properties and biotechnological potential. Exp. Biol. Med. 2023, 248, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; Yang, C.; Mi, X.; Tang, M.; Liang, S.; Chen, Z. Genome-wide identification of tea plant (Camellia sinensis) BAHD acyltransferases reveals their role in response to herbivorous pests. BMC Plant Biol. 2024, 24, 229. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Z.; Zhuang, W.; Wang, T.; Xie, Y. Family characteristics, phylogenetic reconstruction, and potential applications of the plant BAHD acyltransferase family. Front. Plant Sci. 2023, 14, 1218914. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Li, M.; Yin, X.; Song, L. Overexpression of the NbZFP1 encoding a C3HC4-type zinc finger protein enhances antiviral activity of Nicotiana benthamiana. Gene 2024, 908, 148290. [Google Scholar] [CrossRef]
- Gautam, T.; Dutta, M.; Jaiswal, V.; Zinta, G.; Gahlaut, V.; Kumar, S. Emerging Roles of SWEET Sugar Transporters in Plant Development and Abiotic Stress Responses. Cells 2022, 11, 1303. [Google Scholar] [CrossRef]
- Kfoury, B.; Rodrigues, W.F.C.; Kim, S.J.; Brandizzi, F.; Del-Bem, L.E. Multiple horizontal gene transfer events have shaped plant glycosyl hydrolase diversity and function. New Phytol. 2024, 242, 809–824. [Google Scholar] [CrossRef]
- Esch, L.; Schaffrath, U. An Update on Jacalin-Like Lectins and Their Role in Plant Defense. Int. J. Mol. Sci. 2017, 18, 1592. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yin, X.; Wang, F.; Hu, S.; Liu, W.; Chen, L.; Dai, X.; Liang, M. OsJRL40, a Jacalin-Related Lectin Gene, Promotes Salt Stress Tolerance in Rice. Int. J. Mol. Sci. 2023, 24, 7441. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.J.; Liu, X.; Guo, L.; Luo, Y.; Zhang, B.; Cui, X.; Yang, K.; Cai, J.; Liu, F.; Ma, N.; et al. Discovery of plant chemical defence mediated by a two-component system involving β-glucosidase in Panax species. Nat. Commun. 2024, 15, 602. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Muthreich, N.; Liao, C.; Franz-Wachtel, M.; Schütz, W.; Zhang, F.; Hochholdinger, F.; Li, C. The Mucilage Proteome of Maize (Zea mays L.) Primary Roots. J. Proteome Res. 2010, 9, 2968–2976. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.C.; Stacey, G. Chitin signaling and plant disease resistance. Plant Signal Behav. 2008, 3, 831–833. [Google Scholar] [CrossRef]
- da Silva, D.M.G.; Pedrosa, F.R.; Ângela Taipa, M.; Costa, R.; Keller-Costa, T. Widespread occurrence of chitinase-encoding genes suggests the Endozoicomonadaceae family as a key player in chitin processing in the marine benthos. ISME Commun. 2023, 3, 109. [Google Scholar] [CrossRef]
- Huang, C.J.; Guo, S.H.; Chung, S.C.; Lin, Y.J.; Chen, C.Y. Analysis of the involvement of chitin-binding domain of ChiCW in antifungal activity, and engineering a novel chimeric chitinase with high enzyme and antifungal activities. J. Microbiol. Biotechnol. 2009, 19, 1169–1175. [Google Scholar]
- Onaga, S.; Taira, T. A new type of plant chitinase containing LysM domains from a fern (Pteris ryukyuensis): Roles of LysM domains in chitin binding and antifungal activity. Glycobiology 2008, 18, 414–423. [Google Scholar] [CrossRef]
- Yokoyama, S.; Iida, Y.; Kawasaki, Y.; Minami, Y.; Watanabe, K.; Yagi, F. The chitin-binding capability of Cy-AMP1 from cycad is essential to antifungal activity. J. Pept. Sci. 2009, 15, 492–497. [Google Scholar] [CrossRef]
- Bellasio, C.; Stuart-Williams, H.; Farquhar, G.D.; Flexas, J. C(4) maize and sorghum are more sensitive to rapid dehydration than C(3) wheat and sunflower. New Phytol. 2023, 240, 2239–2252. [Google Scholar] [CrossRef] [PubMed]
- Bellasio, C.; Farquhar, G.D. A leaf-level biochemical model simulating the introduction of C(2) and C(4) photosynthesis in C(3) rice: Gains, losses and metabolite fluxes. New Phytol. 2019, 223, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Osinde, C.; Sobhy, I.S.; Wari, D.; Dinh, S.T.; Hojo, Y.; Osibe, D.A.; Shinya, T.; Tugume, A.K.; Nsubuga, A.M.; Galis, I. Comparative analysis of sorghum (C4) and rice (C3) plant headspace volatiles induced by artificial herbivory. Plant Signal. Behav. 2023, 18, 2243064. [Google Scholar] [CrossRef]
- Yan, Y.; Ryu, Y.; Dechant, B.; Li, B.; Kim, J. Dark respiration explains nocturnal stomatal conductance in rice regardless of drought and nutrient stress. Plant Cell Environ. 2023, 46, 3748–3759. [Google Scholar] [CrossRef] [PubMed]
- Doan, L.C.; Dahanayake, J.N.; Mitchell-Koch, K.R.; Singh, A.K.; Vinh, N.Q. Probing Adaptation of Hydration and Protein Dynamics to Temperature. ACS Omega 2022, 7, 22020–22031. [Google Scholar] [CrossRef]
- Vallina Estrada, E.; Zhang, N.; Wennerström, H.; Danielsson, J.; Oliveberg, M. Diffusive intracellular interactions: On the role of protein net charge and functional adaptation. Curr. Opin. Struct. Biol. 2023, 81, 102625. [Google Scholar] [CrossRef]
- Di Savino, A.; Foerster, J.M.; Ullmann, G.M.; Ubbink, M. The Charge Distribution on a Protein Surface Determines Whether Productive or Futile Encounter Complexes Are Formed. Biochemistry 2021, 60, 747–755. [Google Scholar] [CrossRef]
- Anjali, A.; Fatima, U.; Manu, M.S.; Ramasamy, S.; Senthil-Kumar, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef]
- Gwon, S.; Park, J.; Huque, A.M.; Cheung, L.S. The Arabidopsis SWEET1 and SWEET2 uniporters recognize similar substrates while differing in subcellular localization. J. Biol. Chem. 2023, 299, 105389. [Google Scholar] [CrossRef]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant SWEETs: From sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Anuradha, C.; Mol, P.P.; Chandrasekar, A.; Backiyarani, S.; Thangavelu, R.; Selvarajan, R. Unveiling the dynamic expression of PR-1 during Musa spp. infection by Fusarium oxysporum fsp. Cubense: A cloning and characterization study. Mol. Biol. Rep. 2024, 51, 362. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef] [PubMed]
- Garzo, E.; Moreno, A.; Plaza, M.; Fereres, A. Feeding Behavior and Virus-Transmission Ability of Insect Vectors Exposed to Systemic Insecticides. Plants 2020, 9, 895. [Google Scholar] [CrossRef]
- Liang, P.; Zeng, Y.; Ning, J.; Wu, X.; Wang, W.; Ren, J.; Wu, Q.; Yang, X.; Wang, S.; Guo, Z.; et al. A plant virus manipulates both its host plant and the insect that facilitates its transmission. Sci. Adv. 2025, 11, eadr4563. [Google Scholar] [CrossRef]
- Barathi, S.; Sabapathi, N.; Kandasamy, S.; Lee, J. Present status of insecticide impacts and eco-friendly approaches for remediation-a review. Environ. Res. 2024, 240, 117432. [Google Scholar] [CrossRef]
- Shekhar, C.; Khosya, R.; Thakur, K.; Mahajan, D.; Kumar, R.; Kumar, S.; Sharma, A.K. A systematic review of pesticide exposure, associated risks, and long-term human health impacts. Toxicol. Rep. 2024, 13, 101840. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.F.; Ahmad, F.A.; Alsayegh, A.A.; Zeyaullah, M.; AlShahrani, A.M.; Muzammil, K.; Saati, A.A.; Wahab, S.; Elbendary, E.Y.; Kambal, N.; et al. Pesticides impacts on human health and the environment with their mechanisms of action and possible countermeasures. Heliyon 2024, 10, e29128. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. Embo J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; et al. Structural basis of NPR1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef]
- de Oliveira, F.K.; Santos, L.O.; Buffon, J.G. Mechanism of action, sources, and application of peroxidases. Food Res. Int. 2021, 143, 110266. [Google Scholar] [CrossRef]
- Ware, A.; Hess, S.; Gligor, D.; Numer, S.; Gregory, J.; Farmer, C.; Raner, G.M.; Medina, H.E. Identification of Plant Peroxidases Catalyzing the Degradation of Fluorinated Aromatics Using a Peroxidase Library Approach. Eng. Life Sci. 2024, 24, e202400054. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, X.; Liu, X.; Wu, X.; He, Z.; Gao, Z.; Wang, Z. Overexpression of NB-LRR Gene AtRPM1(D505V) Improved Drought and Salt Resistance and Decreased Cold Tolerance in Transgenic Rice. Agronomy 2024, 14, 1050. [Google Scholar] [CrossRef]
- Zhai, X.; Yan, X.; Zenda, T.; Wang, N.; Dong, A.; Yang, Q.; Zhong, Y.; Xing, Y.; Duan, H. Overexpression of the peroxidase gene ZmPRX1 increases maize seedling drought tolerance by promoting root development and lignification. Crop J. 2024, 12, 753–765. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, M.; Teng, Y.; Jia, S.; Yu, D.; Wei, T.; Chen, C.; Song, W. Overexpression of the Glutathione Peroxidase 5 (RcGPX5) Gene From Rhodiola crenulata Increases Drought Tolerance in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 1950. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Sun, Y.; Zhao, R.; Shan, Z.; Gai, J.; Li, Y. Overexpression of Peroxidase Gene GsPRX9 Confers Salt Tolerance in Soybean. Int. J. Mol. Sci. 2019, 20, 3745. [Google Scholar] [CrossRef]
- Jayaraman, K.; Sevanthi, A.M.; Raman, K.V.; Jiwani, G.; Solanke, A.U.; Mandal, P.K.; Mohapatra, T. Overexpression of a DUF740 family gene (LOC_Os04g59420) imparts enhanced climate resilience through multiple stress tolerance in rice. Front. Plant Sci. 2022, 13, 947312. [Google Scholar] [CrossRef]
- Hollis, T.; Honda, Y.; Fukamizo, T.; Marcotte, E.; Day, P.J.; Robertus, J.D. Kinetic analysis of barley chitinase. Arch. Biochem. Biophys. 1997, 344, 335–342. [Google Scholar] [CrossRef]
- Honda, Y.; Kitaoka, M.; Tokuyasu, K.; Sasaki, C.; Fukamizo, T.; Hayashi, K. Kinetic studies on the hydrolysis of N-acetylated and N-deacetylated derivatives of 4-methylumbelliferyl chitobioside by the family 18 chitinases ChiA and ChiB from Serratia marcescens. J. Biochem. 2003, 133, 253–258. [Google Scholar] [CrossRef]
- Kuo, C.J.; Liao, Y.C.; Yang, J.H.; Huang, L.C.; Chang, C.T.; Sung, H.Y. Cloning and characterization of an antifungal class III chitinase from suspension-cultured bamboo (Bambusa oldhamii) cells. J. Agric. Food Chem. 2008, 56, 11507–11514. [Google Scholar] [CrossRef]
- Patel, A.K.; Singh, V.K.; Yadav, R.P.; Moir, A.J.; Jagannadham, M.V. ICChI, a glycosylated chitinase from the latex of Ipomoea carnea. Phytochemistry 2009, 70, 1210–1216. [Google Scholar] [CrossRef]
- Sukprasirt, P.; Wititsuwannakul, R. A chitinolytic endochitinase and β-N-acetylglucosaminidase-based system from Hevea latex in generating N-acetylglucosamine from chitin. Phytochemistry 2014, 104, 5–11. [Google Scholar] [CrossRef]
- van Munster, J.M.; Sanders, P.; ten Kate, G.A.; Dijkhuizen, L.; van der Maarel, M.J. Kinetic characterization of Aspergillus niger chitinase CfcI using a HPAEC-PAD method for native chitin oligosaccharides. Carbohydr. Res. 2015, 407, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.; Sharma, N.; Kaur, M.; Gupta, R. Chitinases: Key players in plant defense mechanisms against fungal pathogens. Physiol. Mol. Plant Pathol. 2025, 138, 102664. [Google Scholar] [CrossRef]
- Ofoe, R. Signal transduction by plant heterotrimeric G-protein. Plant Biol. 2021, 23, 3–10. [Google Scholar] [CrossRef]
- Jain, A.; Sarsaiya, S.; Singh, R.; Gong, Q.; Wu, Q.; Shi, J. Omics approaches in understanding the benefits of plant-microbe interactions. Front. Microbiol. 2024, 15, 1391059. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Metabolomics-guided utilization of beneficial microbes for climate-resilient crops. Curr. Opin. Chem. Biol. 2024, 79, 102427. [Google Scholar] [CrossRef] [PubMed]
- Weidenhamer, J.D.; Cipollini, D.; Morris, K.; Gurusinghe, S.; Weston, L.A. Ecological realism and rigor in the study of plant-plant allelopathic interactions. Plant Soil 2023, 489, 1–39. [Google Scholar] [CrossRef]
- Alshareef, S.A. Metabolic analysis of the CAZy class glycosyltransferases in rhizospheric soil fungiome of the plant species Moringa oleifera. Saudi J. Biol. Sci. 2024, 31, 103956. [Google Scholar] [CrossRef]
- Gupta, P.; Elser, J.; Hooks, E.; D’Eustachio, P.; Jaiswal, P.; Naithani, S. Plant Reactome Knowledgebase: Empowering plant pathway exploration and OMICS data analysis. Nucleic Acids Res. 2024, 52, D1538–D1547. [Google Scholar] [CrossRef]
- Murmu, S.; Sinha, D.; Chaurasia, H.; Sharma, S.; Das, R.; Jha, G.K.; Archak, S. A review of artificial intelligence-assisted omics techniques in plant defense: Current trends and future directions. Front. Plant Sci. 2024, 15, 1292054. [Google Scholar] [CrossRef]
- Shen, D.; Wang, Y.; Chen, X.; Srivastava, V.; Toffolatti, S.L. Editorial: Advances in multi-omics study of filamentous plant pathogens. Front. Microbiol. 2022, 13, 998501. [Google Scholar] [CrossRef]
- Juurakko, C.L.; diCenzo, G.C.; Walker, V.K. Cold acclimation and prospects for cold-resilient crops. Plant Stress 2021, 2, 100028. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, R.; Wang, Y.; Zhang, L.; Yao, S. A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ. 2020, 43, 992–1007. [Google Scholar] [CrossRef] [PubMed]
- Wani, U.M.; Majeed, S.T.; Raja, V.; Wani, Z.A.; Jan, N.; Andrabi, K.I.; John, R. Ectopic expression of a novel cold-resistance protein 1 from Brassica oleracea promotes tolerance to chilling stress in transgenic tomato. Sci. Rep. 2021, 11, 16574. [Google Scholar] [CrossRef]
- Wang, Y.J.; Jiang, W.X.; Zhang, Y.S.; Cao, H.Y.; Zhang, Y.; Chen, X.L.; Li, C.Y.; Wang, P.; Zhang, Y.Z.; Song, X.Y.; et al. Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18. Front. Microbiol. 2019, 10, 2457. [Google Scholar] [CrossRef]
- Marini, G.; Poland, B.; Leininger, C.; Lukoyanova, N.; Spielbauer, D.; Barry, J.K.; Altier, D.; Lum, A.; Scolaro, E.; Ortega, C.P.; et al. Structural journey of an insecticidal protein against western corn rootworm. Nat. Commun. 2023, 14, 4171. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.Z.; Lum, A.; Schepers, E.; Liu, L.; Weston, R.T.; McGinness, B.S.; Heckert, M.J.; Xie, W.; Kassa, A.; Bruck, D.; et al. Novel insecticidal proteins from ferns resemble insecticidal proteins from Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2023, 120, e2306177120. [Google Scholar] [CrossRef]
- Brookes, G.; Barfoot, P. GM crop technology use 1996-2018: Farm income and production impacts. GM Crops Food 2020, 11, 242–261. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.; Ganapathi, T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014, 12, 541–553. [Google Scholar] [CrossRef]
- Datta, K.; Tu, J.; Oliva, N.; Ona, I.; Velazhahan, R.; Mew, T.W.; Muthukrishnan, S.; Datta, S.K. Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci. 2001, 160, 405–414. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.A.; Stukenbrock, E.H. Rapid emergence of pathogens in agro-ecosystems: Global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20160026. [Google Scholar] [CrossRef]
- Ponciano, G.; Yoshikawa, M.; Lee, J.L.; Ronald, P.C.; Whalen, M.C. Pathogenesis-related gene expression in rice is correlated with developmentally controlled Xa21-mediated resistance against Xanthomonas oryzae pv. oryzae. Physiol. Mol. Plant Pathol. 2006, 69, 131–139. [Google Scholar] [CrossRef]
- Hwang, S.H.; Lee, I.A.; Yie, S.W.; Hwang, D.J. Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 2008, 227, 1141–1150. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and Their Roles in Plant Development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.; San Segundo, B. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef]
- Kumar, J.; Ramlal, A.; Mallick, D.; Mishra, V. An Overview of Some Biopesticides and Their Importance in Plant Protection for Commercial Acceptance. Plants 2021, 10, 1185. [Google Scholar] [CrossRef]
- Wend, K.; Zorrilla, L.; Freimoser, F.M.; Gallet, A. Microbial pesticides—Challenges and future perspectives for testing and safety assessment with respect to human health. Environ. Health 2024, 23, 49. [Google Scholar] [CrossRef]
- Chavana, J.; Joshi, N.K. Toxicity and Risk of Biopesticides to Insect Pollinators in Urban and Agricultural Landscapes. Agrochemicals 2024, 3, 70–93. [Google Scholar] [CrossRef]
| S/N | Class | Localization | General Characteristics | Reports |
|---|---|---|---|---|
| 1 | PR-1 proteins | Intracellular and intercellular spaces, organ, and whole plant | Homology to the superfamily of cysteine-rich proteins (15–17 kDa) | [34,39,40,41,51,52,53,59,64,65] |
| 2 | β-glucanases | Rhizosphere, Golgi, and plasma membranes | 1,3-β-endoglucanase activity. Hydrolyze fungal cell wall structural 1,3-β-glucans | [8,64,66] |
| 3 | Chitinases | Apoplastic and symplastic space, organ and whole plant, rhizosphere | Glycoside hydrolase family (26–43 kDa) chitinases. Cleave cell wall chitin polymers in situ | [2,9,39,67] |
| 4 | Chitin-binding proteins | Apoplast, rhizosphere | Bind chitin (3.1–20 kDa) | [68,69,70] |
| 5 | Peroxidases | Cell wall, apoplastic and symplastic space, organ and whole plant, rhizosphere | Heme-containing monomeric glycoproteins that utilize either H2O2 or O2 to oxidize a wide variety of molecules | [2,9,42,44,71] |
| 6 | Thaumatin-like proteins | Apoplastic and symplastic space | Share significant amino acid homology to thaumatin. (~22 kDa) Some cause fungal cell permeability changes; others bind to 1,3-β-glucan and exhibit 1,3-β-glucanase activity | [72,73] |
| 7 | Defensins/thionins | Apoplastic and symplastic space | Low-molecular-mass, cysteine-rich proteins. Fungal inhibition probably occurs through an ion efflux mechanism | [2,9,74] |
| 8 | Cyclophilin-like proteins | Golgi, apoplastic, and symplastic space | Have peptidyl–prolyl cis–trans isomerase activity; are collectively known as immunophilins and include the FK-506-binding proteins, parvulins, and mungins | [75,76] |
| 9 | Ribosome-inactivating proteins (RIPs) | Apoplastic and symplastic space, organ, and whole plant | RNA N-glycosidases, depurinate rRNA. Inactivate fungal ribosomes in vitro and, presumably, in situ | [77,78] |
| 10 | Lipid transfer proteins (LTPs) | Rhizosphere, extracellular spaces | Molecular masses of ~8.7 kDa. Belong to the family of antimicrobial peptides (AMPs) | [2,9,58,79,80] |
| 11 | Protease inhibitors | Rhizosphere | Protein inhibitors of serine and cysteine proteases | [2,9,46] |
| 12 | Hydroxyproline-rich glycoproteins (HRGPs) | Cell wall and plasma membrane | Heavily glycosylated, includes extensins | [81,82] |
| 13 | Glutathione transferases (GSTs) | Cytosol, plasma membrane, endoplasmic reticulum, or apoplast under certain stress conditions | Play critical roles in plant defense, redox homeostasis, detoxification, and stress adaptation | [83,84,85] |
| 14 | Other proteins | Apoplastic and symplastic space, organ and whole plant, rhizosphere | Lectins, acyltransferases, SWEETs, DNA binding proteins, viridin, snakin-1, zinc finger proteins, G-proteins, flagellins | [4,33,54,58,61,63,86,87,88,89,90,91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shobade, S.O.; Nilsen-Hamilton, M.; Zabotina, O.A. Plant Defense Proteins: Recent Discoveries and Applications. Plants 2025, 14, 2069. https://doi.org/10.3390/plants14132069
Shobade SO, Nilsen-Hamilton M, Zabotina OA. Plant Defense Proteins: Recent Discoveries and Applications. Plants. 2025; 14(13):2069. https://doi.org/10.3390/plants14132069
Chicago/Turabian StyleShobade, Samuel O., Marit Nilsen-Hamilton, and Olga A. Zabotina. 2025. "Plant Defense Proteins: Recent Discoveries and Applications" Plants 14, no. 13: 2069. https://doi.org/10.3390/plants14132069
APA StyleShobade, S. O., Nilsen-Hamilton, M., & Zabotina, O. A. (2025). Plant Defense Proteins: Recent Discoveries and Applications. Plants, 14(13), 2069. https://doi.org/10.3390/plants14132069







