Analysis of Genetic Diversity and Core Germplasm Construction of Castanea crenata Siebold and Zucc. Using Simple Sequence Repeat Markers and Morphological Traits
Abstract
1. Introduction
2. Results and Analysis
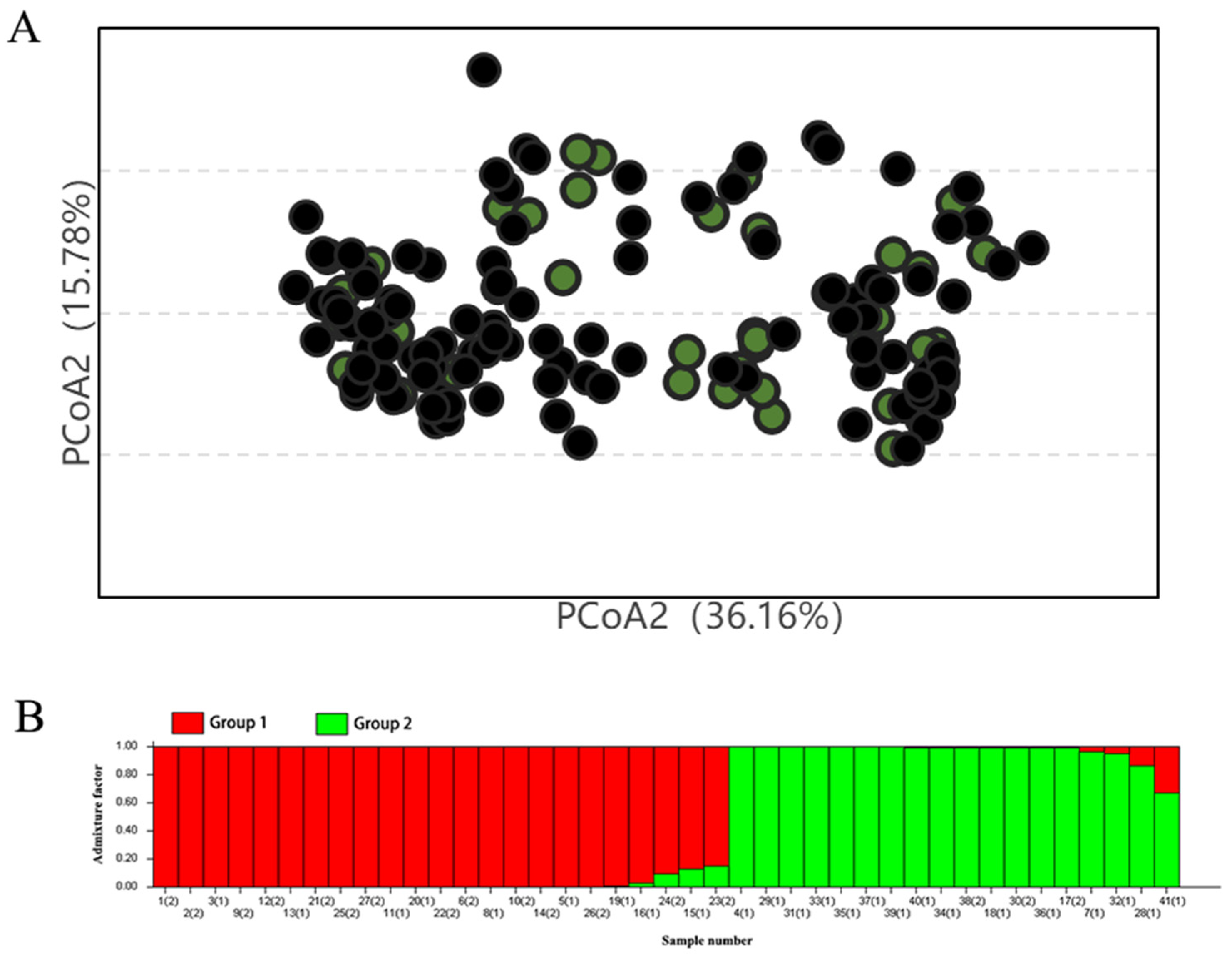
2.1. Cluster Analysis of the Original Chestnut Germplasm (Varieties and Lines)
2.2. Genetic Diversity of the Japanese Chestnut Germplasm Resources
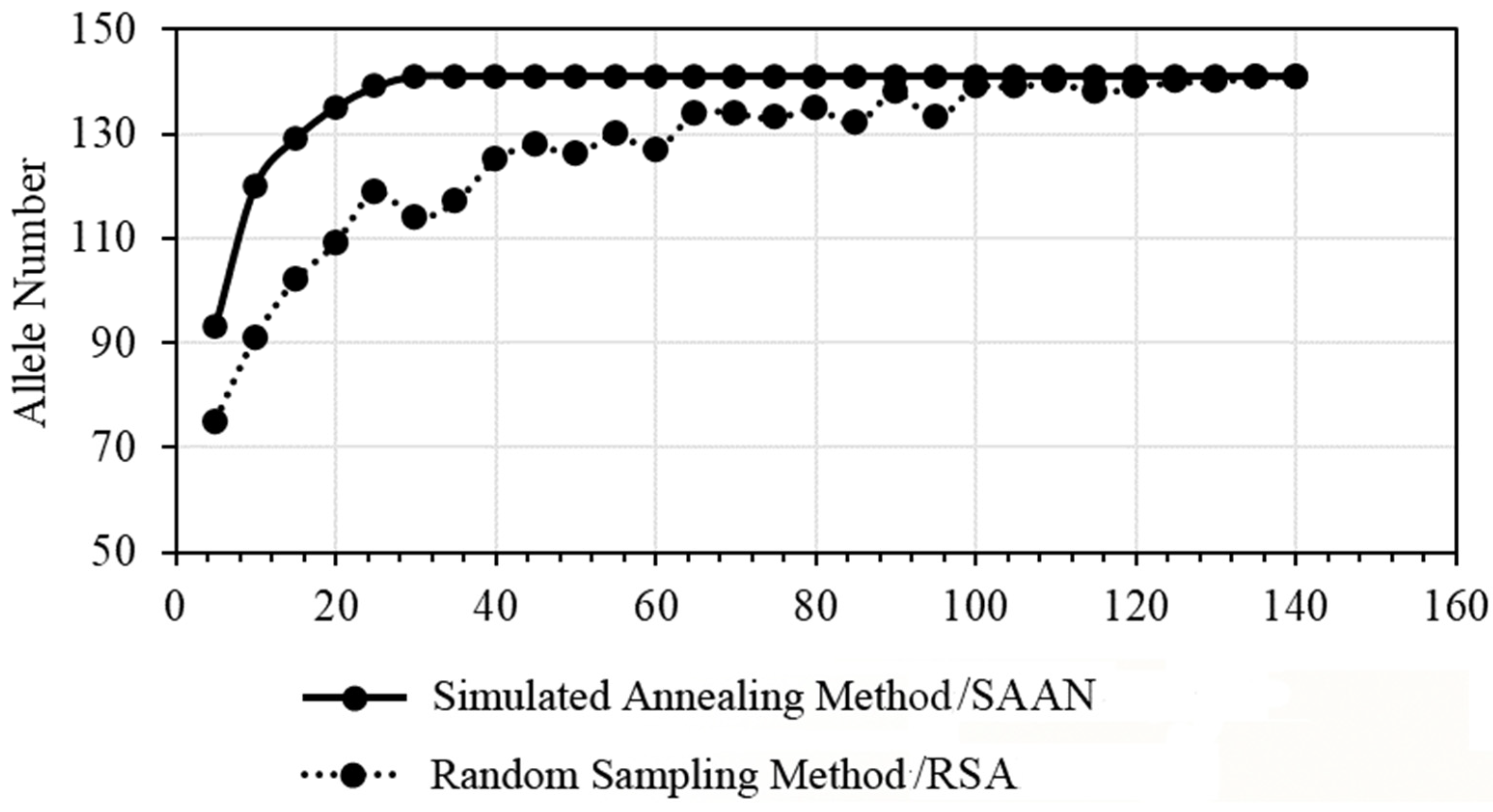
2.3. Selection of the Core Germplasm Resources
2.4. Analysis of the Genetic Diversity in Core Germplasm
2.5. Validation of the Core Germplasm Rationality
2.6. Genetic Diversity Parameters Comparison
3. Discussion
3.1. Analysis of the Genetic Diversity in Japanese Chestnut
3.2. Analysis of the Core Germplasm Construction for Japanese Chestnut
4. Materials and Methods
4.1. Plant Materials and Experimental Site Conditions
4.2. Experimental Methods
4.2.1. DNA Extraction
4.2.2. Fluorescent Capillary Electrophoresis SSR-PCR
4.2.3. Data Analysis
4.2.4. Construction of the Core Germplasm
- (1)
- Selection of the Core Germplasm Based on a Simulated Annealing Algorithm
- (2)
- Selection of the Core Germplasm Based on a Random Search Algorithm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| GBS | genotyping by sequencing |
| GD | genetic distance |
| MAF | major allele frequency |
| PCoA | principal coordinate analysis |
| PCR | polymerase chain reaction |
| PIC | polymorphism information content |
| SNP | single nucleotide polymorphism |
| SSR | simple sequence repeat |
References
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- UPOV. UPOV/INF/17/2. In Guidelines for DNA-Profiling: Molecular Marker Selection and Database Construction; UPOV: Geneva, Switzerland, 2021; pp. 3–4. [Google Scholar]
- Li, D.B.; Xu, N.; Qin, X.Q.; Li, H.L.; Hou, Y.J.; Qiu, H.Y.; Zhang, S.W.; Zhu, J.H.; Peng, H.X. Genetic diversity and construction of core collections of litchi (Litchi chinensis Sonn.) germplasm originated and introduced in Guangxi. J. South. Agric. 2020, 51, 15371544. [Google Scholar]
- Zou, L.F. Establishment of the Core Collection of Male Germplasm Resources of Actinidia eriantha and Analysis of Its Genetic Diversity. Master’s Thesis, Jiangxi Agricultural University, Nanchang, China, 2019. [Google Scholar]
- Nishio, S.; Hayashi, T.; Yamamoto, T.; Terakami, S.; Iwata, H.; Imai, A.; Takada, N.; Kato, H.; Saito, T. Bayesian genome-wide association study of nut traits in Japanese chestnut. Mol. Breed. 2018, 38, 99. [Google Scholar] [CrossRef]
- Nishio, S.; Ruan, S.; Sawamura, Y.; Terakami, S.; Inoue, E. Genetic evidence that Chinese chestnut cultivars in Japan are derived from two divergent genetic structures that originated in China. PLoS ONE 2020, 15, e0235354. [Google Scholar] [CrossRef]
- Ji, F.Y.; Wei, W.; Yang, L.; Wang, G.P.; Zhang, Q.; Xing, Y.; Zhang, S.; Liu, Z.; Cao, Q.; Qin, L. Construction of a SNP-based high-density genetic map using genotyping by sequencing (GBS) and QTL analysis of nut traits in Chinese chestnut (Castanea mollissima Blume). Front. Plant Sci. 2018, 9, 816. [Google Scholar] [CrossRef]
- Kang, M.J.; Kim, T.D.; Lee, S.A.; Lee, H.R.; Park, E.J. The complete chloroplast genome of Castanea crenata Sieb. & Zucc. Mitochondrial DNA Part B 2019, 4, 3864–3865. [Google Scholar]
- Li, J.; Gao, G.; Li, B.; Li, B.; Lu, Q. Genetic analysis of Prunus salicina L. by random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR). Genet. Res. 2022, 2022, 2409324. [Google Scholar] [CrossRef]
- Liu, S.; Nie, H.; Li, Y.R.; Liu, H.T.; Zhang, Q.; Wang, X.F.; Tian, S.L.; Cao, Q.Q.; Qin, L.; Xing, Y. Construction of a core germplasm population of chestnut varieties/lines (lines) based on SSR fluorescent markers. J. Fruit. Tree 2023, 40, 230–241. [Google Scholar]
- Gur-Arie, R.; Cohen, C.J.; Eitan, Y.; Shelef, L.; Hallerman, E.M.; Kashi, Y. Simple sequence repeats in Escherichia coli: Abundance, distribution, composition, and polymorphism. Genome Res. 2000, 10, 62–71. [Google Scholar]
- Nie, X.H.; Wang, Z.H.; Liu, N.W.; Song, L.; Yan, B.Q.; Xing, Y.; Zhang, Q.; Fang, K.-F.; Zhao, Y.-L.; Chen, X.; et al. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume) accessions and selecting a core collection using SSR markers. J. Integr. Agric. 2021, 20, 1277–1286. [Google Scholar] [CrossRef]
- Yang, W.; Yang, B.; Lu, L.; Zhang, X.; Sun, J.; Wang, L.; Zheng, Z.; Liang, D.; Wang, K.; Yan, X.; et al. Utilizing SSR-based core collection development to improve conservation and utilization of Corylus L. genetic resources. PLoS ONE 2024, 19, e0312116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Liu, L.; Liang, W.J.; Zhang, Y.M. Chinese Fruit Trees—Chestnut and Hazelnut; China Forestry Press: Beijing, China, 2005; pp. 22–27. [Google Scholar]
- Gu, J.C.; Nie, X.H.; Cao, S.Y.; Zhang, Y.; Zhang, W.W.; Du, B.S.; Zheng, R.J.; Fang, K.F.; Qin, L.; Xing, Y. Study on the interspecific recognition of leaf morphology in Chestnut. J. Electron. Microsc. 2021, 40, 432–440. [Google Scholar]
- Zheng, R.J.; Xing, Y.; Wang, D.Y.; Shao, Y.; Xu, K.X. Selection and breeding of a new high-yielding, large-fruited and cold-resistant Japanese chestnut variety ‘Huangfeng’. Liaoning For. Sci. Technol. 2024, 3, 22–25+40. [Google Scholar]
- Nie, X.H.; Liu, S.; Wang, B.Y.; Li, Y.; Lian, M.Q.; Qin, L.; Zheng, R.J.; Xing, Y. Genetic structure analysis and fingerprinting of main planted Japanese chestnut varieties/lines (lines) in China based on SSR markers. J. Nucl. Agric. 2022, 36, 2104–2114. [Google Scholar]
- Inoue, E.; Ning, L.; Hara, H.; Ruan, S.; Anzai, H. Development of simple sequence repeat markers in chinese chestnut and their characterization in diverse chestnut cultivars. J. Am. Soc. Hortic. Sci. 2009, 134, 610–617. [Google Scholar] [CrossRef]
- Jiang, X.B.; Tang, D.; Gong, B.C. Genetic diversity and association analysis of Chinese chestnut (Castanea mollissima Blume) cultivars based on SSR markers. Braz. J. Bot. 2017, 40, 235–246. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Barreneche, T.; Mattioni, C.; Villani, F.; Díaz-Hernández, M.B.; Martín, L.M.; Martín, Á. Database of European chestnut cultivars and definition of a core collection using simple sequence repeats. Tree Genet. Genomes 2017, 13, 114. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Aslam, M.M.; Ditta, A.; Iqbal, R.; Mustafa, A.E.-Z.M.A.; Elshikh, M.S.; Uzair, M.; Aghayeva, S.; Qasim, M.; Ercisli, S.; et al. Evaluation of genetic diversity and population structure of the Chinese chestnut (Castanea mollissima) by using NR-SSR markers. Genet. Resour. Crop Evol. 2025, 72, 2445–2457. [Google Scholar] [CrossRef]
- Zheng, R.J. Selection and breeding of Japanese chestnut species ‘Daidanbo’ and key cultivation technology. Liaoning For. Sci. Technol. 2020, 3, 71–72. [Google Scholar]
- Zheng, R.J.; Zheng, J.L.; You, W.Z.; Chen, X.Z. Research on the development status of economic forestry industry in liaoning province (I)-overview of the development of chestnut industry in liaoning province, existing problems and suggestions. Liaoning For. Sci. Technol. 2018, 5, 40–43+47. [Google Scholar]
- Chen, X.Z. Investigation and thinking of Liaoning chestnut industry. Prot. For. Sci. Technol. 2017, 2, 81–84. [Google Scholar]
- Muller, M.; Nelson, C.D.; Gai, L.O. Analysis of environment-marker associations in American chestnut. Forests 2018, 9, 695. [Google Scholar] [CrossRef]
- Terakami, S.; Nishio, S.; Kato, H.; Takada, N.; Saito, T.; Yamamoto, T. Genetic mapping of the dominant gene controlling weeping habit in Japanese chestnut (Castanea crenata Sieb.et Zucc.). Tree Genet. Genomes 2021, 17, 16. [Google Scholar] [CrossRef]
- Egan, L.M.; Conaty, W.C.; Stiller, W.N. Core collections: Is there any value for cotton breeding. Front. Plant Sci. 2022, 13, 895155. [Google Scholar] [CrossRef] [PubMed]
- Brown, A. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Zhao, W.G.; Cho, G.T.; Ma, K.H.; Chung, J.W.; Gwag, J.G.; Park, Y.J. Development of an allele-mining set in rice using a heuristic algorithm and SSR genotype data with least redundancy for the post-genomic era. Mol. Breed. 2010, 26, 639–651. [Google Scholar] [CrossRef]
- Bataillon, T.M.; David, J.L.; Schoen, D.J. Neutral genetic markers and conservation genetics: Simulated germplasm collections. Genetics 1996, 144, 409–417. [Google Scholar] [CrossRef]
- Li, G.S.; Zhang, L.J.; Bai, C.K. Chinese Cornus officinalis: Genetic resources, genetic diversity and core collection. Genet. Resour. Crop Evol. 2012, 59, 1659–1671. [Google Scholar] [CrossRef]
- Vaiman, D.; Mercier, D.; Moazami-Goudarzi, K.; Eggen, A.; Ciampolini, R.; Lépingle, A.; Velmala, R.; Kaukinen, J.; Varvio, S.L.; Martin, P.; et al. A set of 99 cattle microsatellites: Characterization, synteny mapping, and polymorphism. Mamm. Genome 1994, 5, 288–297. [Google Scholar] [CrossRef]
- Liu, K.J.; Muse, S.V. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population geneticsoftware for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multi-locus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]


| Locus | GD | MAF | Na | Ne | I | Ho | He | F | PIC |
|---|---|---|---|---|---|---|---|---|---|
| CmSI0509 | 0.800 | 0.254 | 6.000 | 4.998 | 1.684 | 0.687 | 0.800 | 0.142 | 0.770 |
| CmSI0561 | 0.429 | 0.729 | 5.000 | 1.751 | 0.820 | 0.380 | 0.429 | 0.114 | 0.386 |
| CmSI0614 | 0.741 | 0.445 | 10.000 | 3.860 | 1.676 | 0.593 | 0.741 | 0.199 | 0.714 |
| CmSI0658 | 0.768 | 0.410 | 11.000 | 4.314 | 1.813 | 0.629 | 0.768 | 0.181 | 0.744 |
| CmSI0735 | 0.656 | 0.479 | 4.000 | 2.905 | 1.199 | 0.681 | 0.656 | −0.038 | 0.598 |
| CmSI0742 | 0.719 | 0.423 | 7.000 | 3.557 | 1.426 | 0.458 | 0.719 | 0.363 | 0.676 |
| CmSI0853 | 0.805 | 0.320 | 9.000 | 5.134 | 1.829 | 0.755 | 0.805 | 0.062 | 0.781 |
| CmSI0871 | 0.862 | 0.234 | 12.000 | 7.270 | 2.162 | 0.660 | 0.862 | 0.235 | 0.848 |
| CmSI0883 | 0.736 | 0.441 | 10.000 | 3.787 | 1.658 | 0.627 | 0.736 | 0.147 | 0.706 |
| CmSI0922 | 0.880 | 0.201 | 11.000 | 8.313 | 2.236 | 0.838 | 0.880 | 0.047 | 0.868 |
| CmSI0930 | 0.794 | 0.302 | 9.000 | 4.844 | 1.773 | 0.691 | 0.794 | 0.130 | 0.766 |
| CmSI0938 | 0.751 | 0.341 | 8.000 | 4.021 | 1.506 | 0.644 | 0.751 | 0.142 | 0.709 |
| CmSI0396 | 0.561 | 0.496 | 4.000 | 2.276 | 0.937 | 0.627 | 0.561 | −0.118 | 0.465 |
| CmSI0800 | 0.800 | 0.371 | 10.000 | 4.995 | 1.873 | 0.741 | 0.800 | 0.074 | 0.779 |
| CmSI0881 | 0.793 | 0.336 | 11.000 | 4.824 | 1.876 | 0.445 | 0.793 | 0.438 | 0.767 |
| CmSI0702 | 0.813 | 0.277 | 7.000 | 5.353 | 1.787 | 0.810 | 0.813 | 0.004 | 0.788 |
| CmSI0617 | 0.665 | 0.516 | 7.000 | 2.986 | 1.370 | 0.611 | 0.665 | 0.081 | 0.626 |
| Mean | 0.740 | 0.387 | 8.294 | 4.423 | 1.625 | 0.640 | 0.740 | 0.130 | 0.705 |
| Source of Variance | Sum of Squares (df) | Sum of Squares (SS) | Mean Square (MS) | Estimated Variance | Variation (%) | p |
|---|---|---|---|---|---|---|
| Among Pops | 1 | 57.443 | 57.443 | 0.440 | 7% | <0.01 |
| Within Pops | 141 | 1775.381 | 4.975 | 6.305 | 93% | <0.01 |
| Total | 142 | 1832.824 | 6.745 | 100% |
| Germplasm | Collection Number | Total Allele Points | Na | Ne | MAF | Ho | He | PIC | GD |
|---|---|---|---|---|---|---|---|---|---|
| Original germplasm | 142 | 141 | 8.29 | 4.423 | 0.388 | 0.64 | 0.74 | 0.71 | 0.740 |
| Core germplasm | 41 | 141 | 7.29 | 4.236 | 0.405 | 0.63 | 0.73 | 0.69 | 0.727 |
| Retention rate% | 28.87% | 100.00% | 87.94% | 95.77% | 104.38% | 98.44% | 98.65% | 97.18% | 97.30% |
| t-value | 1.219 | 0.914 | 1.693 | 0.4923 | 0.335 | 2.154 | 0.916 | ||
| p-value | 0.332 | 0.750 | 0.1098 | 0.6292 | 0.739 | 0.0469 | 0.739 |
| Locus | GD | MAF | Na | Ne | I | Ho | He | F | PIC |
|---|---|---|---|---|---|---|---|---|---|
| CmSI0509 | 0.805 | 0.232 | 6.000 | 5.117 | 1.673 | 0.561 | 0.805 | 0.303 | 0.775 |
| CmSI0561 | 0.438 | 0.716 | 4.000 | 1.779 | 0.798 | 0.459 | 0.438 | −0.049 | 0.387 |
| CmSI0614 | 0.747 | 0.438 | 8.000 | 3.959 | 1.672 | 0.583 | 0.747 | 0.220 | 0.721 |
| CmSI0658 | 0.715 | 0.482 | 8.000 | 3.508 | 1.605 | 0.714 | 0.715 | 0.001 | 0.688 |
| CmSI0735 | 0.651 | 0.500 | 4.000 | 2.861 | 1.196 | 0.659 | 0.651 | −0.012 | 0.596 |
| CmSI0742 | 0.664 | 0.512 | 7.000 | 2.978 | 1.342 | 0.366 | 0.664 | 0.449 | 0.623 |
| CmSI0853 | 0.755 | 0.425 | 8.000 | 4.087 | 1.684 | 0.775 | 0.755 | −0.026 | 0.729 |
| CmSI0871 | 0.858 | 0.207 | 11.000 | 7.063 | 2.099 | 0.683 | 0.858 | 0.204 | 0.842 |
| CmSI0883 | 0.682 | 0.500 | 7.000 | 3.141 | 1.443 | 0.567 | 0.682 | 0.169 | 0.645 |
| CmSI0922 | 0.888 | 0.159 | 11.000 | 8.894 | 2.275 | 0.756 | 0.888 | 0.148 | 0.877 |
| CmSI0930 | 0.814 | 0.317 | 9.000 | 5.379 | 1.885 | 0.780 | 0.814 | 0.041 | 0.792 |
| CmSI0938 | 0.747 | 0.295 | 5.000 | 3.956 | 1.436 | 0.564 | 0.747 | 0.245 | 0.702 |
| CmSI0396 | 0.574 | 0.500 | 4.000 | 2.349 | 0.980 | 0.683 | 0.574 | −0.189 | 0.485 |
| CmSI0800 | 0.778 | 0.410 | 8.000 | 4.513 | 1.803 | 0.744 | 0.778 | 0.045 | 0.758 |
| CmSI0881 | 0.767 | 0.392 | 10.000 | 4.285 | 1.776 | 0.486 | 0.767 | 0.365 | 0.739 |
| CmSI0702 | 0.804 | 0.295 | 7.000 | 5.104 | 1.745 | 0.769 | 0.804 | 0.043 | 0.777 |
| CmSI0617 | 0.670 | 0.500 | 7.000 | 3.032 | 1.362 | 0.611 | 0.670 | 0.088 | 0.627 |
| Mean | 0.727 | 0.405 | 7.294 | 4.236 | 1.575 | 0.633 | 0.727 | 0.120 | 0.692 |
| Number | Cultivars (Lines) | Origin Region | Number | Cultivars (Lines) | Origin Region |
|---|---|---|---|---|---|
| 1 | Youmo | Japan | 22 | 1703 | Dalian, Liaoning |
| 2 | Fangyangyu | Japan | 23 | 1404 | Dalian, Liaoning |
| 3 | Liaoli No. 10 | Fengcheng, Liaoning | 24 | Shanda | Republic of Korea |
| 4 | 08-3 | Kuandin, Liaoning | 25 | Yuguang | Republic of Korea |
| 5 | Hemu Shisheng | Fengcheng, Liaoning | 26 | Dabao | Republic of Korea |
| 6 | Guojian | Japan | 27 | Guangyin | Republic of Korea |
| 7 | Kuanyou 9113 | Kuandin, Liaoning | 28 | Danze | Japan |
| 8 | Dongshi No. 2 | Donggang, Liaoning | 29 | Wuci | Japan |
| 9 | Chuyun | Japan | 30 | Nonglin No. 8 | Japan |
| 10 | Gaojiangan | Japan | 31 | 1207 | Dalian, Liaoning |
| 11 | Liaolizaofeng | Dalian, Liaoning | 32 | 1121 | Dalian, Liaoning |
| 12 | Liping | Japan | 33 | Dacheng No. 3 | N. Korea |
| 13 | Baoyingduguo | Fengcheng, Liaoning | 34 | 1201 | Dalian, Liaoning |
| 14 | Yinji | Japan | 35 | Hongqiduguo | Fengcheng, Liaoning |
| 15 | Liaodan No. 61 | Kuandin, Liaoning | 36 | 1113 | Dalian, Liaoning |
| 16 | 1601 | Dalian, Liaoning | 37 | 1120 | Dalian, Liaoning |
| 17 | Zhubo | Japan | 38 | 1111 | Dalian, Liaoning |
| 18 | 02-6 | Kuandin, Liaoning | 39 | 9716 | Kuandin, Liaoning |
| 19 | 0420 | Kuandin, Liaoning | 40 | 1066 | Dalian, Liaoning |
| 20 | Danli No. 3 | Japan | 41 | 04-3 | Kuandin, Liaoning |
| 21 | 1502 | Dalian, Liaoning |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Nie, X.; Liu, J.; Chu, S.; Liu, H.; Xu, K.; Shao, Y.; Wang, Z.; Zheng, R.; Xing, Y. Analysis of Genetic Diversity and Core Germplasm Construction of Castanea crenata Siebold and Zucc. Using Simple Sequence Repeat Markers and Morphological Traits. Plants 2025, 14, 1998. https://doi.org/10.3390/plants14131998
Cui Y, Nie X, Liu J, Chu S, Liu H, Xu K, Shao Y, Wang Z, Zheng R, Xing Y. Analysis of Genetic Diversity and Core Germplasm Construction of Castanea crenata Siebold and Zucc. Using Simple Sequence Repeat Markers and Morphological Traits. Plants. 2025; 14(13):1998. https://doi.org/10.3390/plants14131998
Chicago/Turabian StyleCui, Yanhong, Xinghua Nie, Juanjuan Liu, Shihui Chu, Hanqi Liu, Kaiyuan Xu, Yi Shao, Zhannan Wang, Ruijie Zheng, and Yu Xing. 2025. "Analysis of Genetic Diversity and Core Germplasm Construction of Castanea crenata Siebold and Zucc. Using Simple Sequence Repeat Markers and Morphological Traits" Plants 14, no. 13: 1998. https://doi.org/10.3390/plants14131998
APA StyleCui, Y., Nie, X., Liu, J., Chu, S., Liu, H., Xu, K., Shao, Y., Wang, Z., Zheng, R., & Xing, Y. (2025). Analysis of Genetic Diversity and Core Germplasm Construction of Castanea crenata Siebold and Zucc. Using Simple Sequence Repeat Markers and Morphological Traits. Plants, 14(13), 1998. https://doi.org/10.3390/plants14131998






