Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay
Abstract
1. Introduction
2. Results
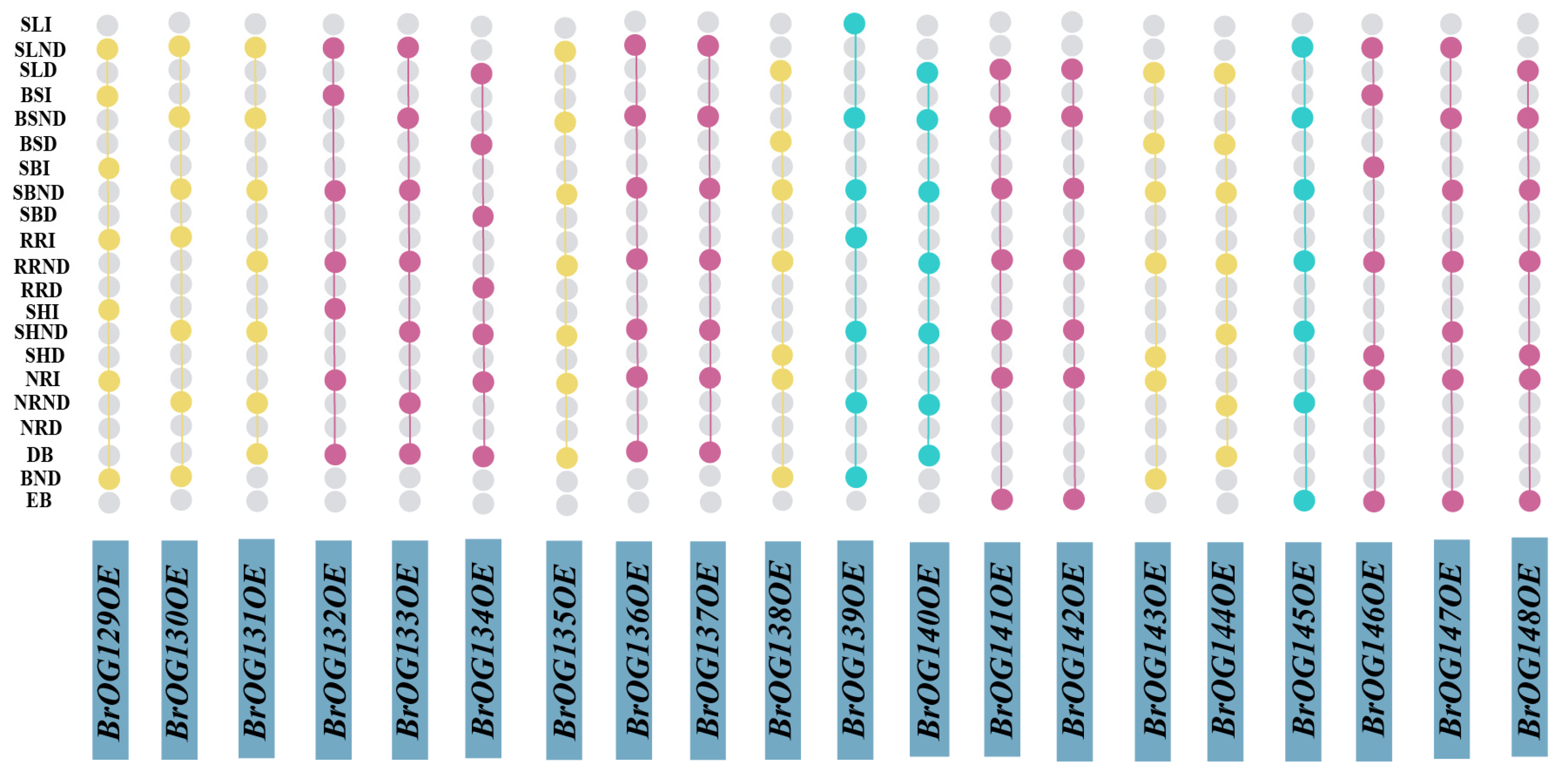
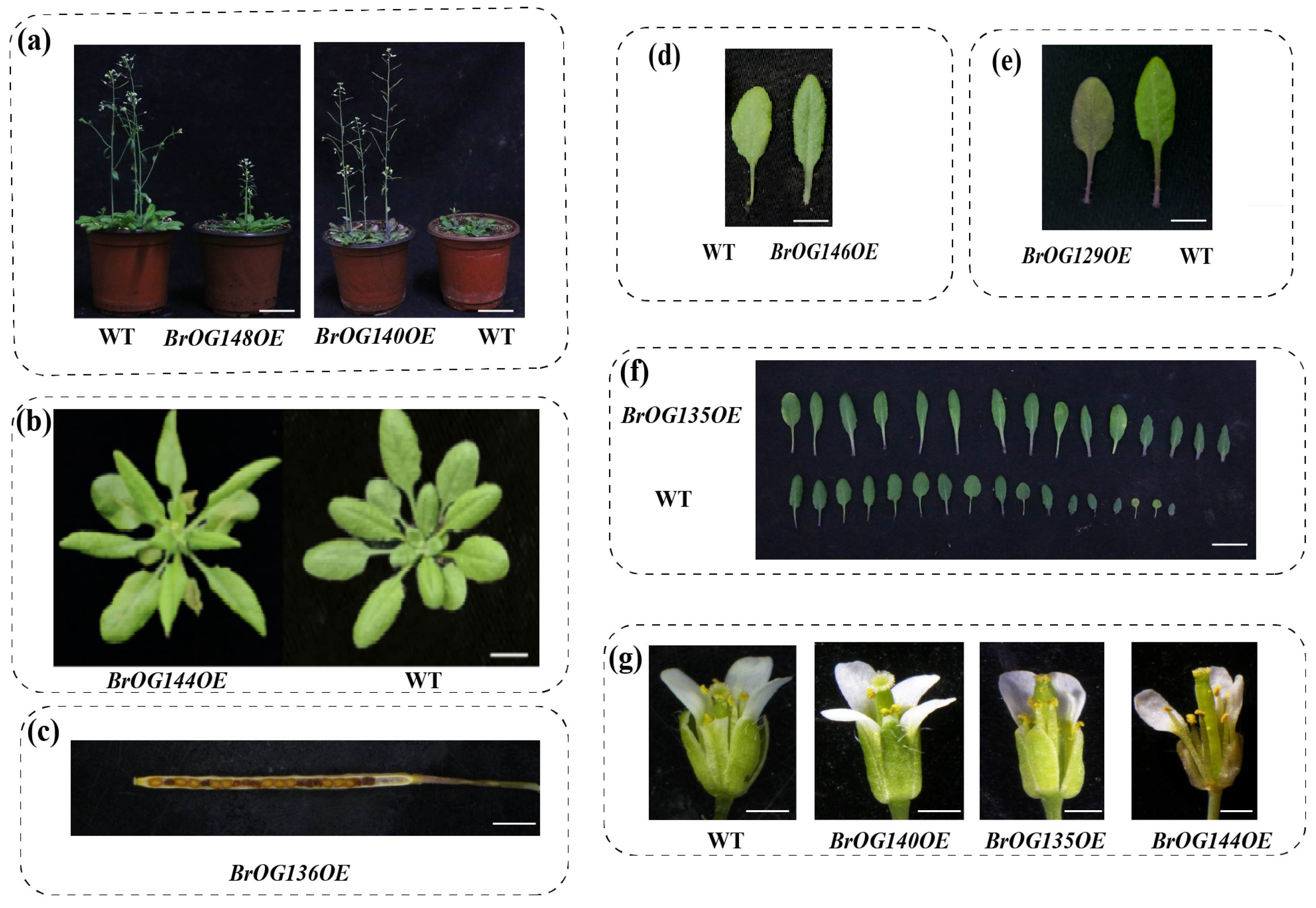
2.1. Construction and Phenotypic Investigation of a BrOGs Overexpression Library in Arabidopsis
2.2. Phenotypic Identification of BR4OE Lines
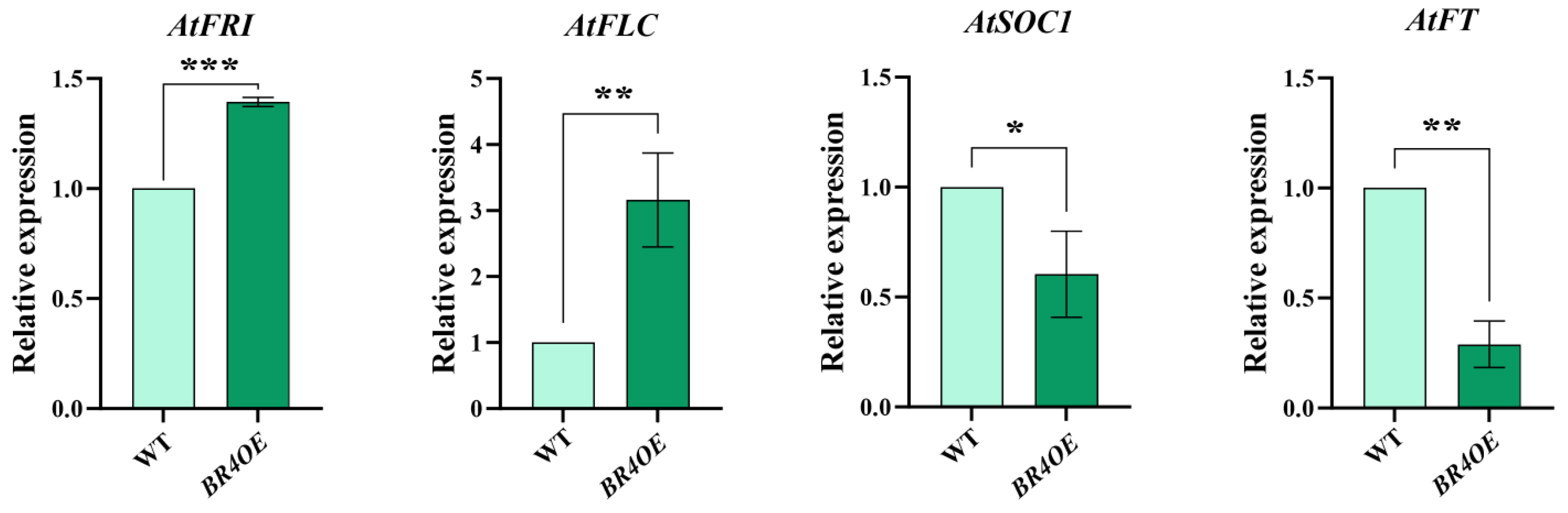
2.3. Expression Analysis of Key Flowering Genes
2.4. BR4 Gene Sequence Analysis
2.5. BR4 Promoter-Driven GUS Expression and Its Regulatory Patterns During the Flowering Stage
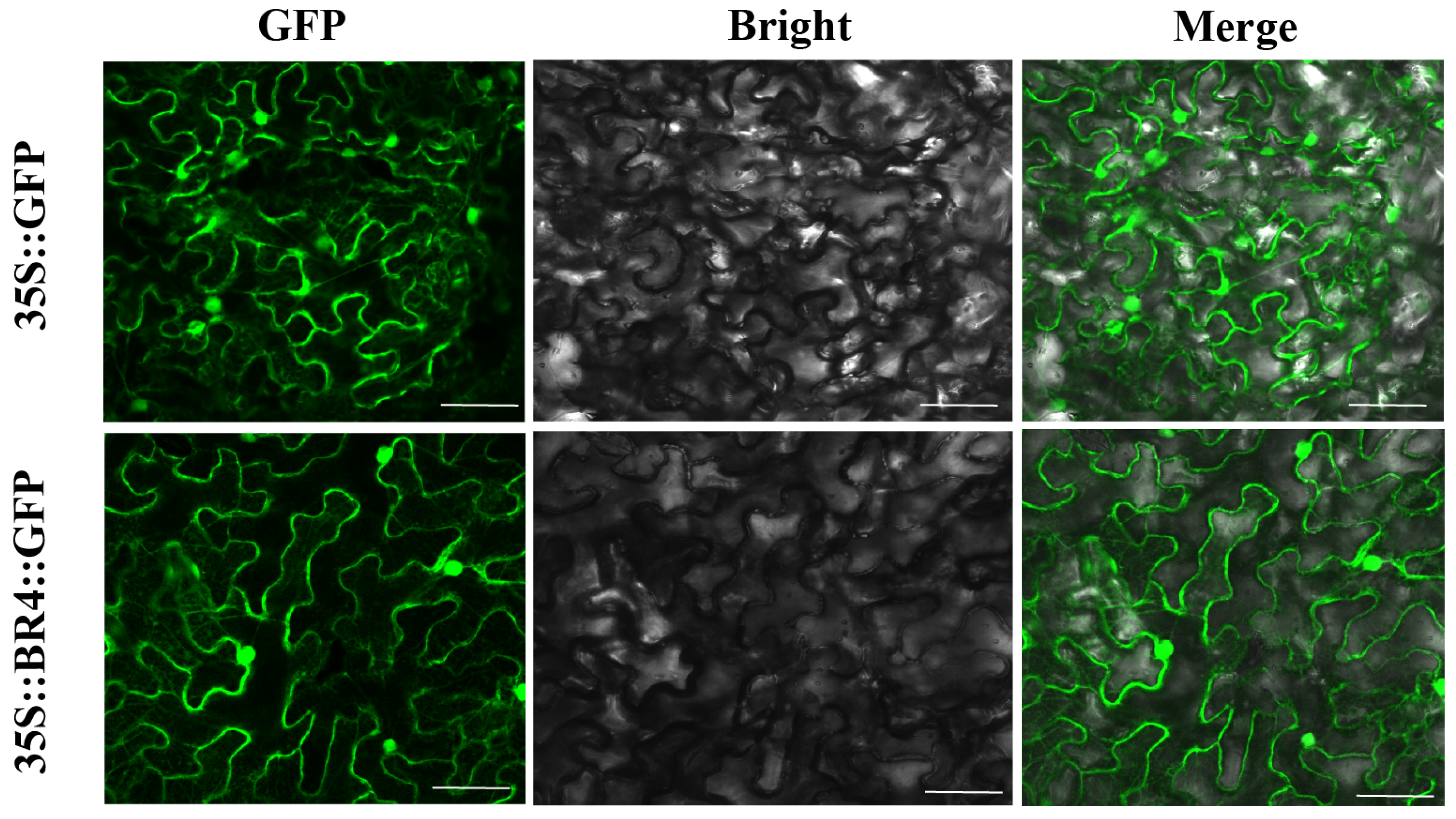
2.6. Subcellular Localization of the BR4 Protein
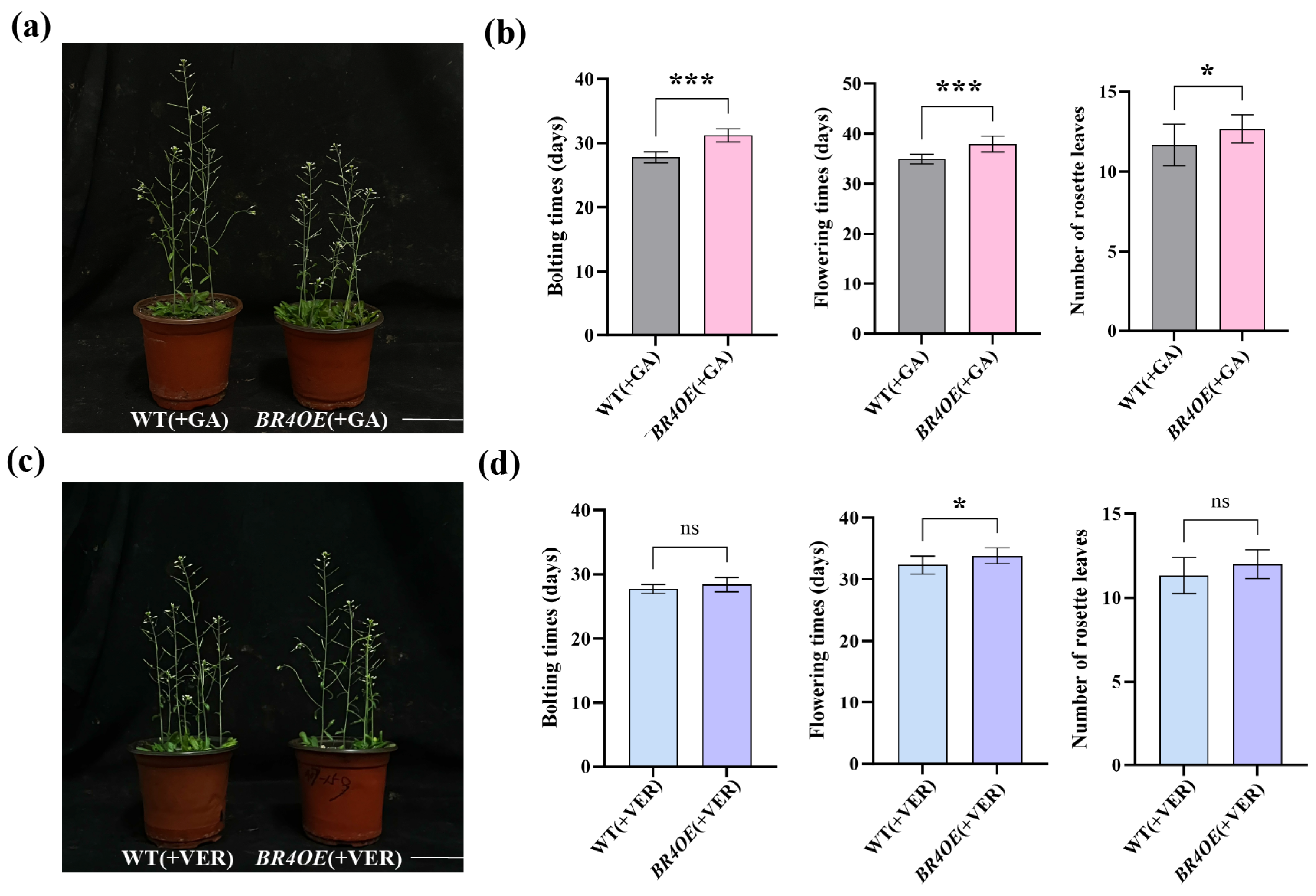
2.7. BR4 Regulates Flowering Time in Response to GA and Vernalization Treatments
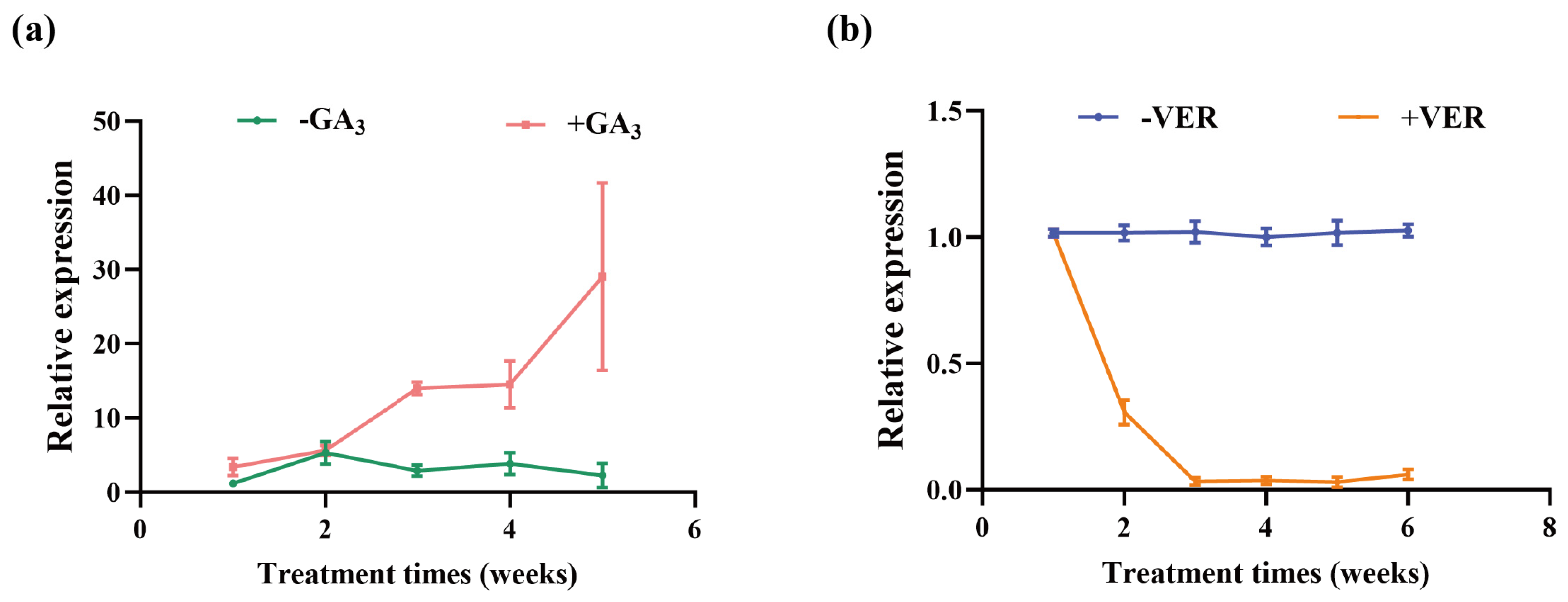
2.8. BR4 Expression Level Under Exogenous Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Cultivation Conditions
4.2. BrOGs Overexpression in Arabidopsis
4.3. Phenotypic Investigation of BrOGsOE-Overexpressing Plants
4.4. Analysis of BR4 Sequence
4.5. GUS Fusion Expression of the BR4 Promoter
4.6. Subcellular Localization Assays of BR4 Protein
4.7. GA3 and Vernalization Treatments of Arabidopsis WT and BR4OE
4.8. GA3 and Vernalization Treatment of Chinese Cabbage
4.9. qRT-PCR Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP1 | APETALA 1 |
| AtFUL | FRUITFULL |
| AtQQS | Arabidopsis Qua-Quine Starch |
| BR1 | BOLTING RESISTANCE 1 |
| BR2 | BOLTING RESISTANCE 2 |
| BR3 | BOLTING RESISTANCE 3 |
| BR4 | BOLTING RESISTANCE 4 |
| BrAGL9a | AGAMOUS-LIKE 9a |
| BraRGL1 | RGA-LIKE1 |
| BraXTH3 | Xyloglucan endotransferase genes |
| BrOGs | Brassica rapa orphan genes |
| BrSDG8 | SET DOMAIN GROUP 8 |
| BrVIN3.1 | VERNALISATION INSENTIVE 3.1 |
| FD | FLOWER LOCUS D |
| FLC | FLOWERING LOCUS C |
| FRI | FRIGIDA |
| FT | FLOWER LOCUS T |
| GA | Gibberellin |
| LFY | LEAFY |
| MS | Murashige and Skoog |
| OE | Overexpression |
| OGs | Orphan genes |
| PpARDT | ABA-RESPONSIVE DROUGHT TOLERANCE |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| TaFROG | FUSARIUM RESISTANCE ORPHAN GENE |
| TaSnRK1 | SUCROSE NON-FERMENTING1-RELATED KINASE1 |
| Ver | Vernalization |
References
- Yang, X.; Jawdy, S.; Tschaplinski, T.J.; Tuskan, G.A. Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics 2009, 93, 473–480. [Google Scholar] [CrossRef]
- Rdelsperger, C.; Prabh, N.; Sommer, R.J. New gene origin and deep taxon phylogenomics: Opportunities and challenges. Trends Genet. 2019, 35, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Yanbin, Y.; Daniel, F. Identification and investigation of ORFans in the viral world. BMC Genom. 2008, 9, 24. [Google Scholar]
- Yanbin, Y.; Daniel, F. On the origin of microbial ORFans: Quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 2006, 6, 63. [Google Scholar]
- Arendsee, Z.W.; Li, L.; Wurtele, E.S. Coming of age: Orphan genes in plants. Trends Plant Sci. 2014, 19, 698–708. [Google Scholar] [CrossRef]
- Campbell, M.Q.; Zhu, W.; Jiang, N.; Lin, H.N.; Shu, O.Y.; Childs, K.L.; Haas, B.J.; Hamilton, J.P.; Buell, C.R. Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol. 2007, 145, 1311–1322. [Google Scholar] [CrossRef]
- Cecilia, L.; Martin, K.; Armaity, D.; Ning, F.; Geeta, H.; Mathias, U.; Sanjay, N.; Svante, P.; Fredrik, P. Analysis of candidate genes for lineage-specific expression changes in humans and primates. J. Proteome Res. 2014, 13, 3596–3606. [Google Scholar]
- Sun, W.; Zhao, X.W.; Zhang, Z. Identification and evolution of the orphan genes in the domestic silkworm, bombyx mori. FEBS Lett. 2015, 589, 2731–2738. [Google Scholar] [CrossRef]
- Donoghue, M.T.; Keshavaiah, C.N.; Swamidatta, S.H.; Spillane, C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 2011, 11, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Eisenberg, D. Finding families for genomic ORFans. Plant Sci. 1999, 15, 759–762. [Google Scholar] [CrossRef]
- Xu, Y.T.; Wu, G.Z.; Hao, B.H.; Chen, L.L.; Deng, X.X.; Xu, Q. Identification, characterization and expression analysis of lineage-specific genes within sweet orange (Citrus sinensis). BMC Genom. 2015, 16, 995. [Google Scholar] [CrossRef]
- Gao, Q.J.; Jin, X.; Xia, E.H.; Wu, X.W.; Gu, L.C.; Yan, H.W.; Xia, Y.C.; Li, S.W. Identification of orphan genes in unbalanced datasets based on ensemble learning. Front. Genet. 2020, 11, 820. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Syrkin, W.E. The QQS orphan gene of Arabidopsis modulates carbon and nitrogen allocation in soybean. Plant Biotechnol. 2015, 13, 177–187. [Google Scholar]
- Jiang, M.L.; Zhan, Z.X.; Li, H.Y.; Dong, X.S.; Chen, F.; Piao, Z.Y. Brassica rapa orphan genes largely affect soluble sugar metabolism. Hortic. Res. 2020, 7, 181. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Jiang, M.L.; Sun, S.R.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Chinese cabbage orphan gene BR3 confers bolting resistance to Arabidopsis through the gibberellin pathway. Front. Plant Sci. 2024, 15, 1518962. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Hong, J.; Zhao, Y.; Li, X.; Feng, X.; Wang, H.; Zhang, L.; Lin, M.; Cai, Y.; Han, Y. De novo gene integration into regulatory networks via interaction with conserved genes in peach. Hortic. Res. 2024, 11, uhae252. [Google Scholar] [CrossRef]
- Jiang, M.L.; Li, X.N.; Dong, X.S.; Zu, Y.; Zhan, Z.X.; Piao, Z.Y.; Lang, H. Research advances and prospects of orphan genes in plants. Front. Plant Sci. 2022, 13, 947129. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.M.; Pu, X.J.; Zhou, S.Z.; Li, P.; Luo, T.; Chen, Z.X.; Chen, S.L.; Liu, L. Orphan gene PpARDT positively involved in drought tolerance potentially by enhancing ABA response in Physcomitrium (Physcomitrella) patens. Plant Sci. 2022, 319, 111222. [Google Scholar] [CrossRef]
- Perochon, A.; Jianguang, J.; Kahla, A.; Arunachalam, C.; Scofield, S.R.; Bowden, S.; Wallington, E.; Doohan, F.M. TaFROG encodes a pooideae orphan protein that interacts with SnRK1 and enhances resistance to the mycotoxigenic fungus fusarium graminearum. Plant Physiol. 2015, 169, 2895–2906. [Google Scholar] [CrossRef]
- Jiang, M.L.; Zhang, Y.T.; Yang, X.L.; Li, X.N.; Lang, H. Brassica rapa orphan gene BR1 delays flowering time in Arabidopsis. Front. Plant Sci. 2023, 14, 1135684. [Google Scholar] [CrossRef]
- Fernando, A.; George, C. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar]
- Madeira, A.C.; Lars, H. FLC or not FLC: The other side of vernalization. J. Exp. Bot. 2008, 59, 1127–1135. [Google Scholar]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. Bioessays 2004, 26, 363–373. [Google Scholar] [CrossRef]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16 (Suppl. S1), S18–S31. [Google Scholar] [CrossRef]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.I.; Wang, J.G.; Poon, S.Y.; Su, C.L.; Wang, S.S.; Chiou, T.J. Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 2005, 137, 1037–1048. [Google Scholar] [CrossRef]
- Takada, S.; Akter, A.; Itabashi, E.; Nishida, N.; Shea, D.J.; Miyaji, N.; Mehraj, H.; Osabe, K.; Shimizu, M.; Takasaki-Yasuda, T.; et al. The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 2019, 9, 13843. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Fu, W.; Huang, S.N.; Gao, Y.; Zhang, M.D.; Qu, G.Y.; Wang, N.; Liu, Z.Y.; Feng, H. Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 2020, 133, 2937–2948. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.Y.; Su, T.B.; Li, P.R.; Wang, W.H.; Zhao, X.Y.; Yu, Y.J.; Zhang, D.S.; Yu, S.C.; Zhang, F.L. A histone H4 gene prevents drought-induced bolting in Chinese cabbage by attenuating the expression of flowering genes. J. Exp. Bot. 2020, 72, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Quijada, P.; Sung, S.B.; Lukens, L.; Amasino, R.; Osborn, T.C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 2002, 162, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Wei, K.Y.; Gao, B.Z.; Liu, J.H.; Liang, J.L.; Cheng, F.; Wang, X.W.; Wu, J. BrFLC5: A weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 2018, 131, 2107–2116. [Google Scholar] [CrossRef]
- Zu, Y.; Jiang, M.L.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Orphan gene BR2 positively regulates bolting resistance through the vernalization pathway in Chinese cabbage. Hortic. Res. 2024, 11, uhae216. [Google Scholar] [CrossRef]
- Jiang, M.L.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Construction and evaluation of Brassica rapa orphan genes overexpression library. Front. Plant Sci. 2025, 16, 1532449. [Google Scholar] [CrossRef]
- Jiang, M.L.; Dong, X.S.; Lang, H.; Pang, W.X.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Mining of Brassica-Specific Genes (BSGs) and their induction in different developmental stages and under Plasmodiophora brassicae stress in Brassica rapa. Int. J. Mol. Sci. 2018, 19, 2064. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Bäurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef]
- Seki, M.; Kamei, A.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 2003, 14, 194–199. [Google Scholar] [CrossRef]
- Li, X.; Huang, L.; Lu, J.H.; Cheng, Y.H.; You, Q.B.; Wang, L.J.; Song, X.J.; Zhou, X.A.; Jiao, Y.Q. Large-scale investigation of soybean gene functions by overexpressing a full-length soybean cDNA library in Arabidopsis. Front. Plant Sci. 2018, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Huang, S.N.; Liu, Z.Y. Fine mapping of Brebm6, a gene conferring the early-bolting phenotype in Chinese cabbage (Brassica rapa ssp. pekinensis). Veg. Res. 2021, 1, 7. [Google Scholar]
- Kim, S.Y.; Michaels, S.D. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 2006, 133, 4699–4707. [Google Scholar] [CrossRef] [PubMed]
- Song, S.W.; Lei, Y.L.; Huang, X.M.; Su, W.; Chen, R.Y.; Hao, Y.W. Crosstalk of cold and gibberellin effects on bolting and flowering in flowering Chinese cabbage. J. Integr. Agric. 2019, 18, 992–1000. [Google Scholar] [CrossRef]
- Wang, Y.D.; Song, S.W.; Hao, Y.W.; Chen, C.M.; Ou, X.; He, B.; Zhang, J.W.; Jiang, Z.H.; Li, C.M.; Zhang, S.W.; et al. Role of BraRGL1 in regulation of Brassica rapa bolting and flowering. Hortic. Res. 2023, 10, uhad119. [Google Scholar] [CrossRef]
- Wang, Y.D.; Huang, X.; Huang, X.M.; Su, W.; Hao, Y.W.; Liu, H.C.; Chen, R.Y.; Song, S.W. BcSOC1 promotes bolting and stem elongation in flowering Chinese cabbage. Int. J. Mol. Sci. 2022, 23, 3459. [Google Scholar] [CrossRef]
- Ding, L.; Kim, S.Y.; Michaels, S.D. FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol. 2013, 163, 243–252. [Google Scholar] [CrossRef][Green Version]
- Zhan, Z.X.; Liu, H.S.; Yang, Y.; Liu, S.; Li, X.N.; Piao, Z.Y. Identification and characterization of putative effectors from Plasmodiophora brassicae that suppress or induce cell death in Nicotiana benthamiana. Front. Plant Sci. 2022, 13, 881992. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Jiang, M.L.; Ma, J.J.; Chen, J.J.; Kong, L.Y.; Zhan, Z.X.; Li, X.N.; Piao, Z.Y. Metabolomics and transcriptomics reveal the function of trigonelline and its synthesis gene BrNANMT in clubroot susceptibility of Brassica rapa. In Plant Cell Environment; Wiley: Oxford, UK, 2025. [Google Scholar] [CrossRef]
- Zhang, M.D.; Zhu, M.; Lang, H.; Wang, W.M.; Li, X.N.; Jiang, M.L. Genome-wide identification, characterization, and expression analysis of orphan genes within coriander. Plants 2025, 14, 778. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, R.; Zhang, R.; Li, X.; Jiang, M. Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay. Plants 2025, 14, 1947. https://doi.org/10.3390/plants14131947
Liao R, Zhang R, Li X, Jiang M. Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay. Plants. 2025; 14(13):1947. https://doi.org/10.3390/plants14131947
Chicago/Turabian StyleLiao, Ruiqi, Ruiqi Zhang, Xiaonan Li, and Mingliang Jiang. 2025. "Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay" Plants 14, no. 13: 1947. https://doi.org/10.3390/plants14131947
APA StyleLiao, R., Zhang, R., Li, X., & Jiang, M. (2025). Construction of an Overexpression Library for Chinese Cabbage Orphan Genes in Arabidopsis and Functional Analysis of BOLTING RESISTANCE 4-Mediated Flowering Delay. Plants, 14(13), 1947. https://doi.org/10.3390/plants14131947





