Genome-Wide Analysis of the FNSII Gene Family and the Role of CitFNSII-1 in Flavonoid Synthesis in Citrus
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Tree of FNSII Genes in Citrus
2.2. Chromosomal Localization, Synteny Analysis, and Characterizations of the FNSII Genes in Citrus
2.3. Conserved Motif and Gene Structure Analysis of the FNSII Genes in Citrus
2.4. Expression of CitFNSII-1 in Citrus
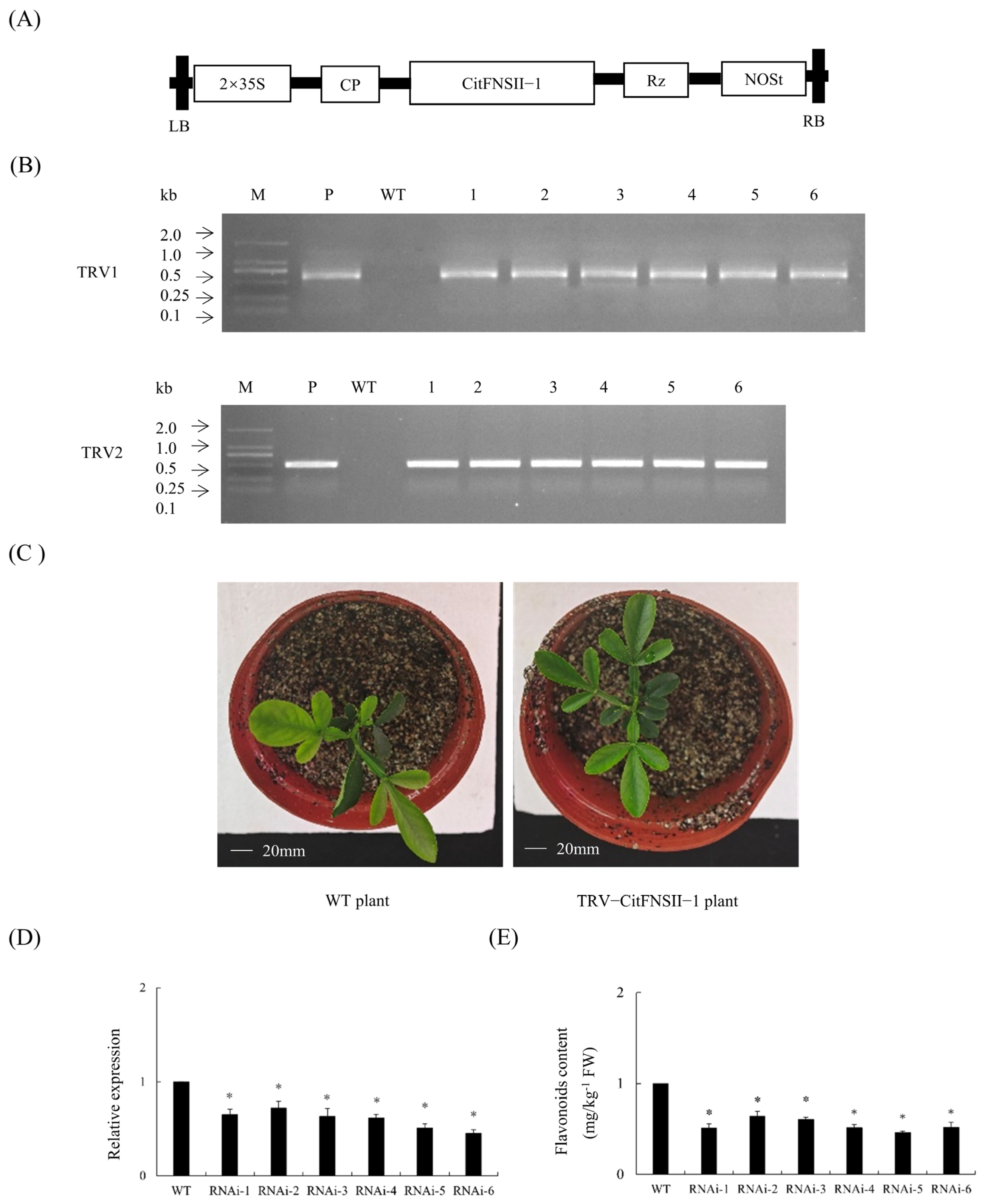
2.5. Generation of TRV-CitFNSII-1 Plants
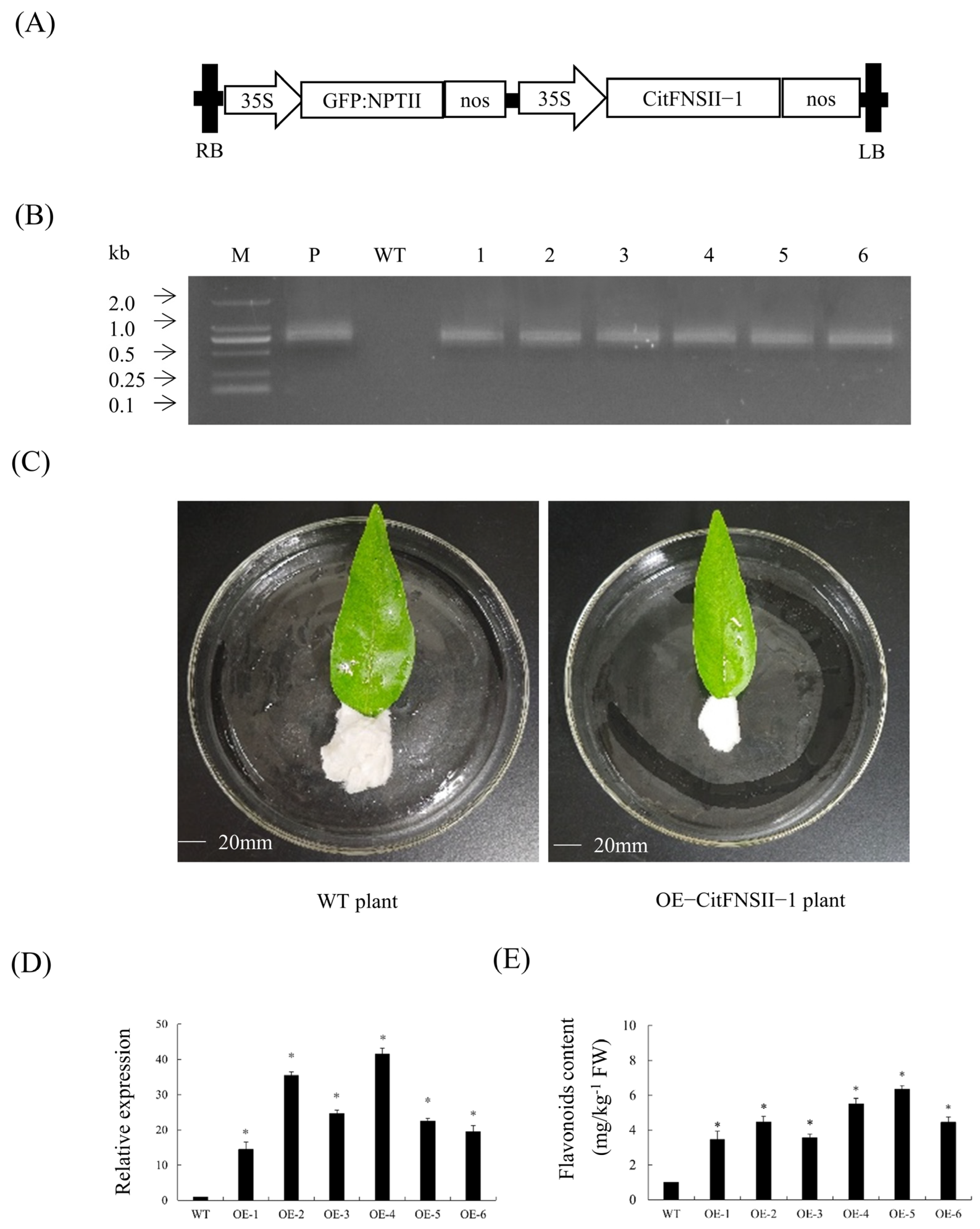
2.6. Transient Overexpression Analysis of CitFNSII-1 in Citrus Leaves
2.7. Overexpression of CitFNSII-1 in Transgenic Hairy Roots
2.8. Identification and Editing Efficiency Analysis of CitFNSII-1 in CRISPR/Cas9-Edited Citrus Hairy Roots
2.9. Characteristics of Changes in SA, MeSA, JA, and MeJA Content in CitFNSII-1 Transgenic Hairy Roots
2.10. CitFNSII-1 Interacts with CHI-1
2.11. Expression of CHI-1 in Citrus
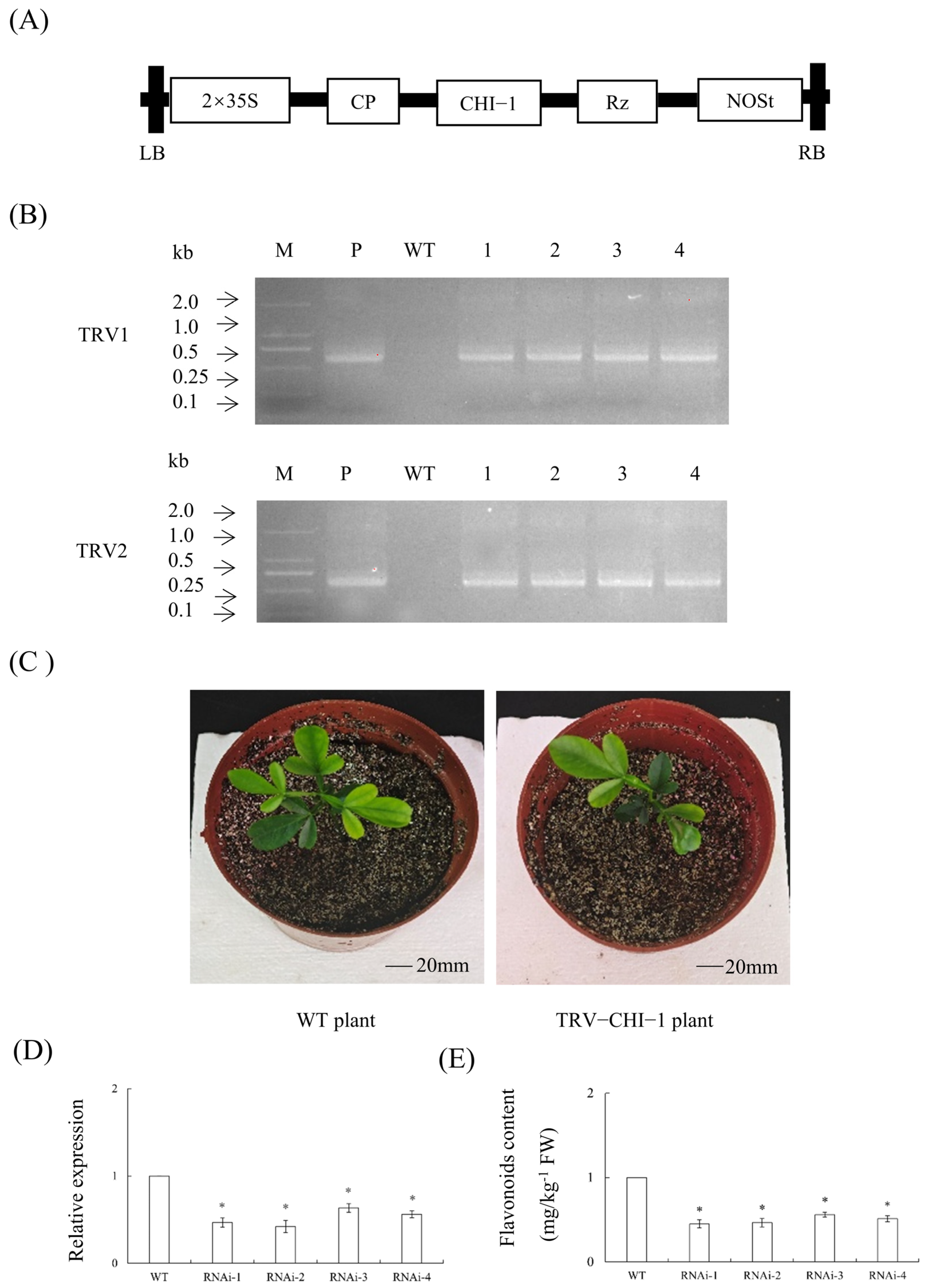
2.12. Generation of TRV-CHI-1 Plants
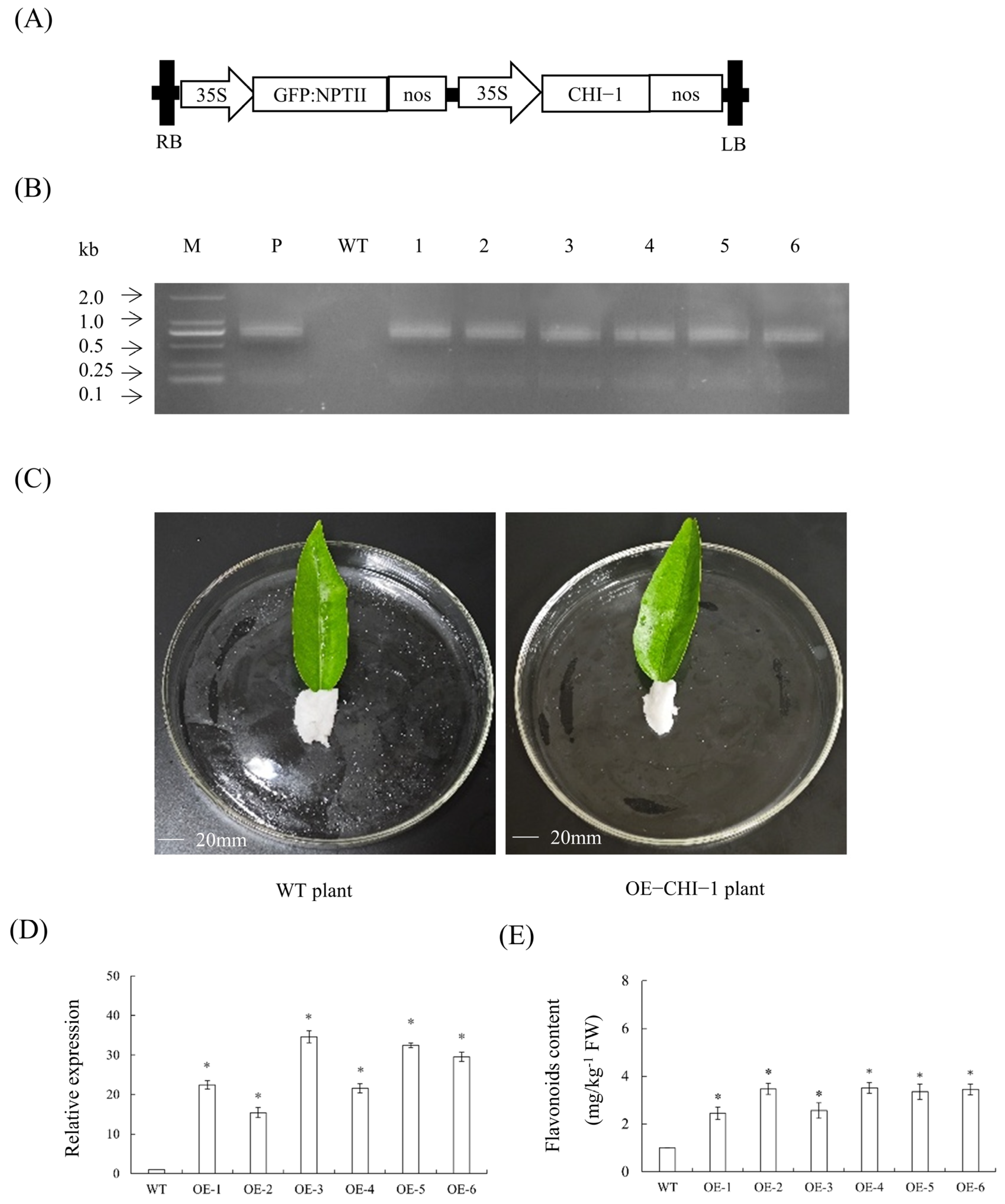
2.13. Transient Overexpression Analysis of CHI-1 in Citrus Leaves
2.14. Overexpression of CHI-1 in Transgenic Hairy Roots
2.15. Identification and Editing Efficiency Analysis of CHI-1 in CRISPR/Cas9-Edited Citrus Hairy Roots
2.16. Characteristics of Changes in SA, MeSA, JA, and MeJA Content in CHI-1 Transgenic Hairy Roots
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Microbial Strains, and Growth Conditions
4.2. Identification of the FNSII Genes in Citrus
4.3. Phylogenetic Tree, Multiple Sequence Alignment, and Characterizations Analysis of the FNSII Proteins
4.4. Chromosomal Localization, Gene Structure, Conserved Motif, and Synteny Analysis of the FNSII Genes in Citrus
4.5. Cloning and Sequence Analysis of Genes and Promoter
4.6. RT-qPCR Analysis
4.7. Exogenous MeSA and MeJA Treatment of Citrus Leaves and Fruit
4.8. Vectors Construction
4.9. Citrus Transformation
4.10. Determination of Hormone Content
4.11. Flavonoids Extraction and Measurement
4.12. Identification of Positive Transgenic Plants and Detection of Editing Efficiency
4.13. Homology Modeling and Molecular Docking
4.14. Yeast Two-Hybrid (Y2H) Assay
4.15. Bimolecular Fluorescence Complementation Assay
4.16. Luciferase Complementation Assay
4.17. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, F.; Wang, M.; Liu, X.; Xu, Y.; Zhu, S.; Shen, W.; Zhao, X. Identification of Putative Genes Involved in Limonoids Biosynthesis in Citrus by Comparative Transcriptomic Analysis. Front. Plant Sci. 2017, 8, 782. [Google Scholar] [CrossRef] [PubMed]
- Durand-Hulak, M.; Dugrand, A.; Duval, T.; Bidel, L.P.; Jay-Allemand, C.; Froelicher, Y.; Bourgaud, F.; Fanciullino, A.L. Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC-MS approach. Ann. Bot. 2015, 115, 861–877. [Google Scholar] [CrossRef]
- Torregrosa, C.; Cluzet, S.; Fournier, J.; Huguet, T.; Gamas, P.; Prospéri, J.M.; Esquerré-Tugayé, M.T.; Dumas, B.; Jacquet, C. Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol. Plant Microbe Interact. 2004, 17, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J.; Hu, H.; Almqvist, J.; Gao, X.; Wang, L. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef] [PubMed]
- Bondonno, N.P.; Dalgaard, F.; Kyrø, C.; Murray, K.; Bondonno, C.P.; Lewis, J.R.; Croft, K.D.; Gislason, G.; Scalbert, A.; Cassidy, A.; et al. Flavonoid intake is associated with lower mortality in the Danish Diet Cancer and Health Cohort. Nat. Commun. 2019, 10, 3651. [Google Scholar] [CrossRef]
- Peng, Z.; Zhang, H.; Li, W.; Yuan, Z.; Xie, Z.; Zhang, H.; Cheng, Y.; Chen, J.; Xu, J. Comparative profiling and natural variation of polymethoxylated flavones in various citrus germplasms. Food Chem. 2021, 354, 129499. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, Y.; Sun, T.; Zhang, C.; Liu, X.; Li, Y. Genome-Wide Classification and Evolutionary Analysis Reveal Diverged Patterns of Chalcone Isomerase in Plants. Biomolecules. 2022, 12, 961. [Google Scholar] [CrossRef]
- Parmenter, B.H.; Thompson, A.S.; Bondonno, N.P.; Jennings, A.; Murray, K.; Perez-Cornago, A.; Hodgson, J.M.; Tresserra-Rimbau, A.; Kühn, T.; Cassidy, A. High diversity of dietary flavonoid intake is associated with a lower risk of all-cause mortality and major chronic diseases. Nat. Food. [CrossRef]
- Du, Y.; Chu, H.; Wang, M.; Chu, I.K.; Lo, C. Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene (SbFNSII) in sorghum. J. Exp. Bot. 2010, 61, 983–994. [Google Scholar] [CrossRef]
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant Cell Environ. 2019, 42, 495–508. [Google Scholar] [CrossRef]
- Zheng, J.; Zhao, C.; Liao, Z.; Liu, X.; Gong, Q.; Zhou, C.; Liu, Y.; Wang, Y.; Cao, J.; Liu, L.; et al. Functional characterization of two flavone synthase II members in citrus. Hortic Res. 2023, 10, uhad113. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, C.; Li, H.; Lu, S. Identification and Characterization of Flavonoid Biosynthetic Enzyme Genes in Salvia miltiorrhiza (Lamiaceae). Molecules. 2018, 23, 1467. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Wang, J.; Pei, T.; Gao, R.; Xiang, C.; Chen, J.; Zhang, C.; Xiao, Y.; Li, Q.; Wu, Z.; et al. Functional evolution and diversification of CYP82D subfamily members have shaped flavonoid diversification in the genus Scutellaria. Plant Commun. 2025, 6, 101134. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, J.; Cui, M.Y.; Liu, J.; Fang, Y.; Yan, M.; Qiu, W.; Shang, H.; Xu, Z.; Yidiresi, R.; et al. The Reference Genome Sequence of Scutellaria baicalensis Provides Insights into the Evolution of Wogonin Biosynthesis. Mol. Plant. 2019, 12, 935–950. [Google Scholar] [CrossRef]
- Su, D.; Wu, M.; Wang, H.; Shu, P.; Song, H.; Deng, H.; Yu, S.; Garcia-Caparros, P.; Bouzayen, M.; Zhang, Y.; et al. Bi-functional transcription factor SlbHLH95 regulates fruits flavonoid metabolism and grey mould resistance in tomato. Plant Biotechnol. J. 2025, 23, 2083–2094. [Google Scholar] [CrossRef]
- Wu, Z.; Singh, S.K.; Lyu, R.; Pattanaik, S.; Wang, Y.; Li, Y.; Yuan, L.; Liu, Y. Metabolic engineering to enhance the accumulation of bioactive flavonoids licochalcone A and echinatin in Glycyrrhiza inflata (Licorice) hairy roots. Front. Plant Sci. 2022, 13, 932594. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cao, L.; Jiao, B.; Yang, H.; Ma, C.; Liang, Y. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion (Allium cepa L.). Hortic. Res. 2022, 9, uhac128. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, Y.; Wang, G.; Hill, L.; Weng, J.K.; Chen, X.Y.; Xue, H.; Martin, C. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2016, 2, e1501780. [Google Scholar] [CrossRef]
- Zou, X.; Zhao, K.; Liu, Y.; Du, M.; Zheng, L.; Wang, S.; Xu, L.; Peng, A.; He, Y.; Long, Q.; et al. Overexpression of Salicylic Acid Carboxyl Methyltransferase (CsSAMT1) Enhances Tolerance to Huanglongbing Disease in Wanjincheng Orange (Citrus sinensis (L.) Osbeck). Int. J. Mol. Sci. 2021, 22, 2803. [Google Scholar] [CrossRef]
- Wang, W.; Li, T.; Chen, Q.; Yao, S.; Deng, L.; Zeng, K. CsWRKY25 Improves Resistance of Citrus Fruit to Penicillium digitatum via Modulating Reactive Oxygen Species Production. Front. Plant Sci. 2022, 12, 818198. [Google Scholar] [CrossRef]
- Mizuno, H.; Yazawa, T.; Kasuga, S.; Sawada, Y.; Kanamori, H.; Ogo, Y.; Hirai, M.Y.; Matsumoto, T.; Kawahigashi, H. Expression of Flavone Synthase II and Flavonoid 3’-Hydroxylase Is Associated with Color Variation in Tan-Colored Injured Leaves of Sorghum. Front. Plant Sci. 2016, 7, 1718. [Google Scholar] [CrossRef]
- Tian, S.; Yang, Y.; Wu, T.; Luo, C.; Li, X.; Zhao, X.; Xi, W.; Liu, X.; Zeng, M. Functional Characterization of a Flavone Synthase That Participates in a Kumquat Flavone Metabolon. Front. Plant Sci. 2022, 13, 826780. [Google Scholar] [CrossRef]
- Luo, C.; Liu, L.; Zhao, J.; Xu, Y.; Liu, H.; Chen, D.; Cheng, X.; Gao, J.; Hong, B.; Huang, C.; et al. CmHY5 functions in apigenin biosynthesis by regulating flavone synthase II expression in chrysanthemum flowers. Planta 2022, 257, 7. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, Z.; Li, J.; Qi, Y.; Zhang, X.; Shen, C. The key role of cytochrome P450s in the biosynthesis of plant derived natural products. Plant Physiol Biochem. 2025, 222, 109695. [Google Scholar] [CrossRef]
- Lam, P.Y.; Zhu, F.Y.; Chan, W.L.; Liu, H.; Lo, C. Cytochrome P450 93G1 Is a Flavone Synthase II That Channels Flavanones to the Biosynthesis of Tricin O-Linked Conjugates in Rice. Plant Physiol. 2014, 165, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Fujino, N.; Tenma, N.; Waki, T.; Ito, K.; Komatsuzaki, Y.; Sugiyama, K.; Yamazaki, T.; Yoshida, S.; Hatayama, M.; Yamashita, S.; et al. Physical interactions among flavonoid enzymes in snapdragon and torenia reveal the diversity in the flavonoid metabolon organization of different plant species. Plant J. 2018, 94, 372–392. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Escobar, C.A.; Priego-Capote, F.; Robles Olvera, V.J.; Luque de Castro, M.D. Targeted Analysis of the Concentration Changes of Phenolic Compounds in Persian Lime (Citrus latifolia) during Fruit Growth. J. Agric. Food Chem. 2018, 66, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Seoka, M.; Ma, G.; Zhang, L.; Yahata, M.; Yamawaki, K.; Kan, T.; Kato, M. Expression and functional analysis of the nobiletin biosynthesis-related gene CitOMT in citrus fruit. Sci. Rep. 2020, 10, 15288. [Google Scholar] [CrossRef]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Luo, P.; Shen, Y.; Jin, S.; Huang, S.; Cheng, X.; Wang, Z.; Li, P.; Zhao, J.; Bao, M.; Ning, G. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016, 245, 35–49. [Google Scholar] [CrossRef]
- Yang, W.; Xu, X.; Li, Y.; Wang, Y.; Li, M.; Wang, Y.; Ding, X.; Chu, Z. Rutin-Mediated Priming of Plant Resistance to Three Bacterial Pathogens Initiating the Early SA Signal Pathway. PLoS ONE. 2016, 11, e0146910. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.P.; Zhang, L.; Yan, P.; Ahammed, G.J.; Han, W.Y. Methyl Salicylate Enhances Flavonoid Biosynthesis in Tea Leaves by Stimulating the Phenylpropanoid Pathway. Molecules 2019, 24, 362. [Google Scholar] [CrossRef]
- Ramabulana, A.T.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Profiling of Altered Metabolomic States in Bidens pilosa Leaves in Response to Treatment by Methyl Jasmonate and Methyl Salicylate. Plants 2020, 9, 1275. [Google Scholar] [CrossRef]
- Gondor, O.K.; Janda, T.; Soós, V.; Pál, M.; Majláth, I.; Adak, M.K.; Balázs, E.; Szalai, G. Salicylic Acid Induction of Flavonoid Biosynthesis Pathways in Wheat Varies by Treatment. Front. Plant Sci. 2016, 7, 1447. [Google Scholar] [CrossRef]
- Cheng, B.; Xu, L.; Bilal, M.S.; Huang, Q.; Niu, D.; Ma, H.; Zhou, S.; Peng, A.; Wei, G.; Chen, F.; et al. Small RNAs contribute to citrus Huanglongbing tolerance by manipulating methyl salicylate signaling and exogenous methyl salicylate primes citrus groves from emerging infection. Plant J. 2023, 116, 1309–1324. [Google Scholar] [CrossRef]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inzé, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.J.; Chen, J.B.; Cao, J.P.; Li, X.; Sun, C.D. Citrus flavonoids and their antioxidant evaluation. Crit. Rev. Food Sci. Nutr. 2022, 62, 3833–3854. [Google Scholar] [CrossRef] [PubMed]
- Ngaki, M.N.; Louie, G.V.; Philippe, R.N.; Manning, G.; Pojer, F.; Bowman, M.E.; Li, L.; Larsen, E.; Wurtele, E.S.; Noel, J.P. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 2012, 485, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Mehdy, M.C.; Lamb, C.J. Chalcone isomerase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection. EMBO J. 1987, 6, 1527–1533. [Google Scholar] [CrossRef]
- Jiang, W.; Yin, Q.; Wu, R.; Zheng, G.; Liu, J.; Dixon, R.A.; Pang, Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 7165–7179. [Google Scholar] [CrossRef]
- Shimada, N.; Aoki, T.; Sato, S.; Nakamura, Y.; Tabata, S.; Ayabe, S. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; McRoberts, J.; Shi, F.; Moreno, J.E.; Jones, A.D.; Howe, G.A. The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 2014, 164, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, L.C.; Walker, J.F.; Ng, J.; Deanna, R.; Dunbar-Wallis, A.; Backes, A.; Pezzi, P.H.; Palchetti, M.V.; Robertson, H.M.; Monaghan, A.; et al. Transcription Factors Evolve Faster Than Their Structural Gene Targets in the Flavonoid Pigment Pathway. Mol. Biol. Evol. 2022, 39, msac044. [Google Scholar] [CrossRef] [PubMed]
- Longevity, O.M.A.C. Retracted: Two Myricetin-Derived Flavonols from Morella rubra Leaves as Potent α-Glucosidase Inhibitors and Structure-Activity Relationship Study by Computational Chemistry. Oxid. Med. Cell Longev. 2024, 2024, 9849172. [Google Scholar] [CrossRef]
- Li, J.; Yu, Q.; Liu, C.; Zhang, N.; Xu, W. Flavonoids as key players in cold tolerance: Molecular insights and applications in horticultural crops. Hortic. Res. 2025, 12, uhae366. [Google Scholar] [CrossRef]
- Tian, X.; Hu, M.; Yang, J.; Yin, Y.; Fang, W. Ultraviolet-B Radiation Stimulates Flavonoid Biosynthesis and Antioxidant Systems in Buckwheat Sprouts. Foods. 2024, 13, 3650. [Google Scholar] [CrossRef]
- Deng, H.; Wu, M.; Wu, Y.; Xiao, X.; Gao, Z.; Li, H.; Hu, N.; Gao, Y.; Grierson, D.; Liu, M. SlMYC2-SlMYB12 module orchestrates a hierarchical transcriptional cascade that regulates fruit flavonoid metabolism in tomato. Plant Biotechnol. J. 2025, 23, 477. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, J.; Johnson, M.; Mandal, R.; Cruz, M.; Martínez-Huélamo, M.; Andres-Lacueva, C.; Wishart, D.S. A Comprehensive LC-MS Metabolomics Assay for Quantitative Analysis of Serum and Plasma. Metabolites. 2024, 14, 622. [Google Scholar] [CrossRef]
- Mou, J.; Zhang, Z.; Qiu, H.; Lu, Y.; Zhu, X.; Fan, Z.; Zhang, Q.; Ye, J.; Fernie, A.R.; Cheng, Y.; et al. Multiomics-based dissection of citrus flavonoid metabolism using a Citrus reticulata × Poncirus trifoliata population. Hortic. Res. 2021, 8, 56. [Google Scholar] [CrossRef]
- Baldassari, S.; Klingler, E.; Teijeiro, L.G.; Doladilhe, M.; Raoux, C.; Roig-Puiggros, S.; Bizzotto, S.; Couturier, J.; Gilbert, A.; Sami, L.; et al. Single-cell genotyping and transcriptomic profiling of mosaic focal cortical dysplasia. Nat. Neurosci. 2025, 28, 964–972. [Google Scholar] [CrossRef]
- Munir, S.; Khan, M.R.; Song, J.; Munir, S.; Zhang, Y.; Ye, Z.; Wang, T. Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2016, 102, 167–179. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, M.; Liu, X.; Xia, Y.; Hu, R.; Xia, Q.; Jing, D.; Guo, Q. Genome-wide analysis of the WOX gene family and the role of EjWUSa in regulating flowering in loquat (Eriobotrya japonica). Front. Plant Sci. 2022, 13, 1024515. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, W.; Wang, P.; Wang, D.; Shi, M.; Xia, Y.; He, Q.; Dang, J.; Guo, Q.; Jing, D.; Liang, G. EjFRI, FRIGIDA (FRI) Ortholog from Eriobotrya japonica, Delays Flowering in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1087. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-wide investigation of AP2/ERF gene family in the desert legume Eremosparton songoricum: Identification, classification, evolution, and expression profiling under drought stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, Y.; Yang, D.; Liu, H.; Zhang, Y.; Wang, X.; Bai, F.; Cheng, S. Foliar methyl jasmonate (MeJA) application increased 2-acetyl-1-Pyrroline (2-AP) content and modulated antioxidant attributes and yield formation in fragrant rice. J. Plant Physiol. 2023, 282, 153946. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant. 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Tang, T.; Yu, X.; Yang, H.; Gao, Q.; Ji, H.; Wang, Y.; Yan, G.; Peng, Y.; Luo, H.; Liu, K.; et al. Development and Validation of an Effective CRISPR/Cas9 Vector for Efficiently Isolating Positive Transformants and Transgene-Free Mutants in a Wide Range of Plant Species. Front. Plant Sci. 2018, 9, 1533. [Google Scholar] [CrossRef]
- Wang, S.; Yang, C.; Tu, H.; Zhou, J.; Liu, X.; Cheng, Y.; Luo, J.; Deng, X.; Zhang, H.; Xu, J. Characterization and Metabolic Diversity of Flavonoids in Citrus Species. Sci. Rep. 2017, 7, 10549. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 2021, 19, 671–688. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, H.; Wei, W.; Han, Z.; Zuo, J.; He, Q. Expression characteristics of CsESA1 in citrus and analysis of its interacting protein. Plant Signal Behav. 2025, 20, 2439249. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, S.; Dong, L.; Qu, R.; Zheng, L.; He, Y.; Chen, S.; Zou, X. Overexpression of a “Candidatus Liberibacter Asiaticus” Effector Gene CaLasSDE115 Contributes to Early Colonization in Citrus sinensis. Front. Microbiol. 2022, 12, 797841. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, H.; Xian, W.; Liu, Z.; Yuan, Y.; Fan, J.; Xiang, S.; Yang, X.; Liu, Y.; Liu, S.; et al. Duplication and sub-functionalization of flavonoid biosynthesis genes plays important role in Leguminosae root nodule symbiosis evolution. J. Integr. Plant Biol. 2024, 66, 2191–2207. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Wu, K.; Ding, Y.; Tang, M.; Zhang, X.; Pan, Y.; Wu, L.; Su, C.; Hong, Z.; et al. The DnaJ1 heat shock protein interacts with the flavanone 3-hydroxylase-like protein F3HL to synergistically enhance drought tolerance by scavenging reactive oxygen species in tomato. Plant J. 2025, 121, e70097. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Chen, B.; Luo, L.; Zhong, Q.; Teo, C.H.; Huang, S. Genome-Wide Analysis of the FNSII Gene Family and the Role of CitFNSII-1 in Flavonoid Synthesis in Citrus. Plants 2025, 14, 1936. https://doi.org/10.3390/plants14131936
Liu X, Chen B, Luo L, Zhong Q, Teo CH, Huang S. Genome-Wide Analysis of the FNSII Gene Family and the Role of CitFNSII-1 in Flavonoid Synthesis in Citrus. Plants. 2025; 14(13):1936. https://doi.org/10.3390/plants14131936
Chicago/Turabian StyleLiu, Xinya, Beibei Chen, Ling Luo, Qi Zhong, Chee How Teo, and Shengjia Huang. 2025. "Genome-Wide Analysis of the FNSII Gene Family and the Role of CitFNSII-1 in Flavonoid Synthesis in Citrus" Plants 14, no. 13: 1936. https://doi.org/10.3390/plants14131936
APA StyleLiu, X., Chen, B., Luo, L., Zhong, Q., Teo, C. H., & Huang, S. (2025). Genome-Wide Analysis of the FNSII Gene Family and the Role of CitFNSII-1 in Flavonoid Synthesis in Citrus. Plants, 14(13), 1936. https://doi.org/10.3390/plants14131936




