GAG Protein of Arabidopsis thaliana LTR Retrotransposon Forms Retrosome-like Cytoplasmic Granules and Activates Stress Response Genes
Abstract
1. Introduction
2. Results
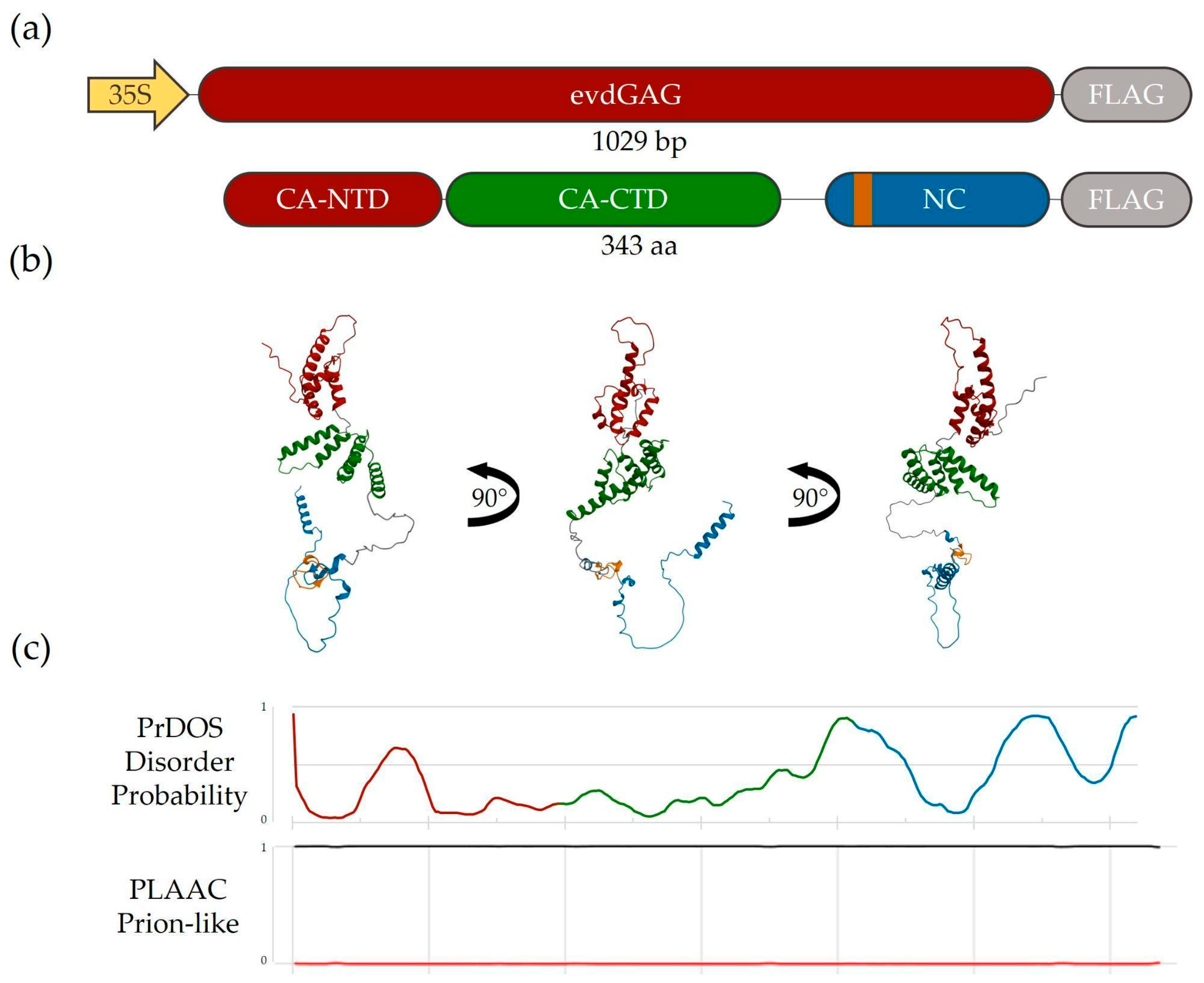
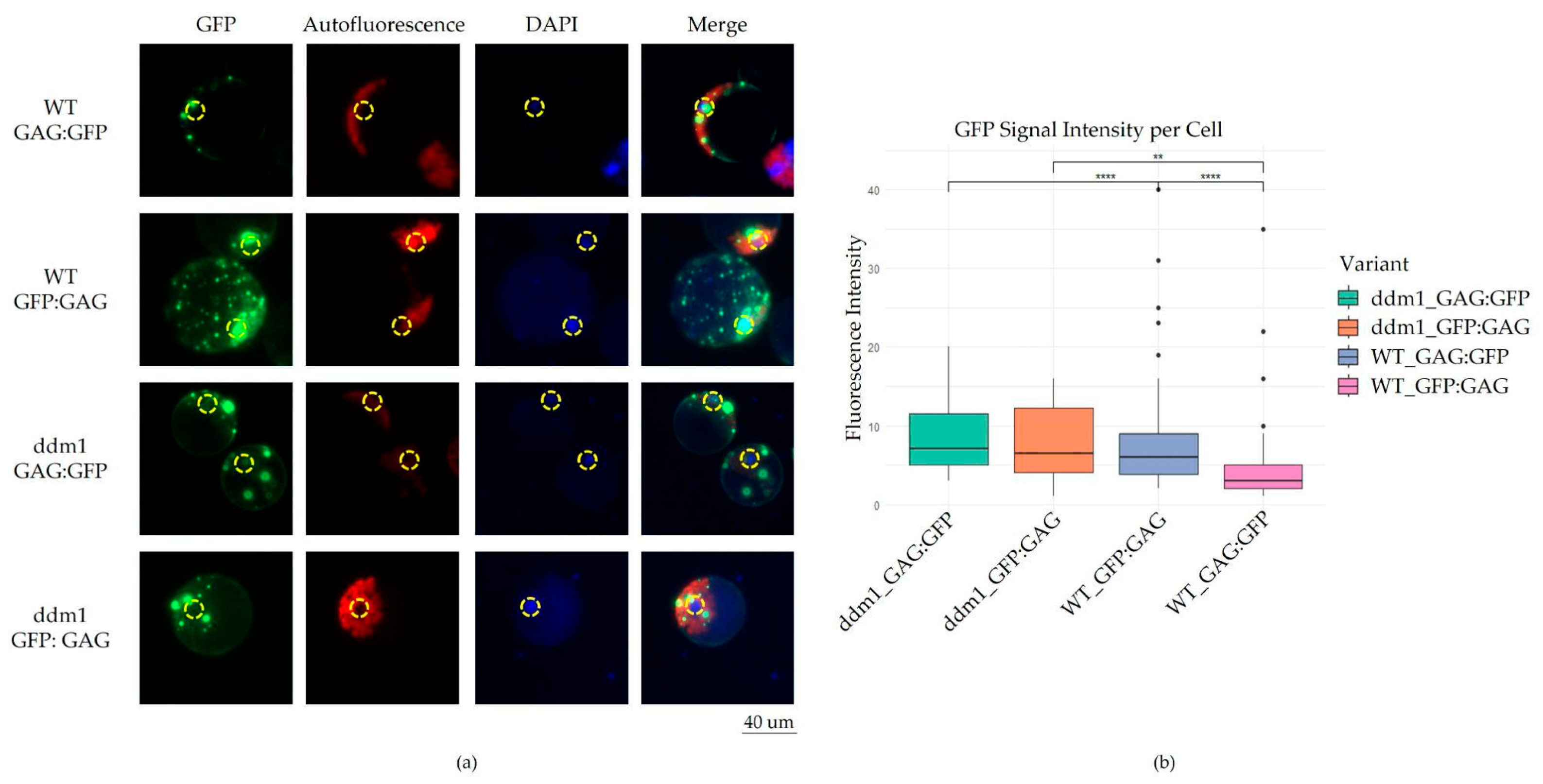
2.1. Structure and Intracellular Localization of A. Thaliana GAG Protein
2.2. Intracellular Localization of evdGAG Protein in Distant Plant Species
2.3. GAG Overexpression Induces Transcriptional Activation of Stress-Responsive Genes in Wild-Type A.Thaliana and ddm1 Mutant
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The impact of transposable elements in adaptive evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.A.; Feldman, M. Evolution and origin of bread wheat. Plant Cell 2022, 34, 2549–2567. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Martienssen, R.A. Regulation of retrotransposition in Arabidopsis. Biochem. Soc. Trans. 2021, 49, 2241–2251. [Google Scholar] [CrossRef] [PubMed]
- Marí-Ordóñez, A.; Marchais, A.; Etcheverry, M.; Martin, A.; Colot, V.; Voinnet, O. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013, 45, 1029–1039. [Google Scholar] [CrossRef]
- Oberlin, S.; Rajeswaran, R.; Trasser, M.; Barragán-Borrero, V.; Schon, M.A.; Plotnikova, A.; Loncsek, L.; Nodine, M.D.; Marí-Ordóñez, A.; Voinnet, O. Innate, translation-dependent silencing of an invasive transposon in Arabidopsis. EMBO Rep. 2022, 23, e53400. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef]
- Oberlin, S.; Sarazin, A.; Chevalier, C.; Voinnet, O.; Marí-Ordóñez, A. A genome-wide transcriptome and translatome analysis of Arabidopsis transposons identifies a unique and conserved genome expression strategy for Ty1/Copia retroelements. Genome Res. 2017, 27, 1549–1562. [Google Scholar] [CrossRef]
- Maldonado, J.O.; Martin, J.L.; Mueller, J.D.; Zhang, W.; Mansky, L.M. New insights into retroviral Gag–Gag and Gag–membrane interactions. Front. Microbiol. 2014, 5, 302. [Google Scholar] [CrossRef]
- Lesage, P.; Maxwell, P.H. A prion-like domain in Gag capsid protein drives retrotransposon particle assembly and mobility. Proc. Natl. Acad. Sci. USA 2023, 120, e2311419120. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, J.R.; Reed, J.C.; Tanaka, M.; Chutiraka, K.; Robinson, B.A. How HIV-1 Gag assembles in cells: Putting together pieces of the puzzle. Virus Res. 2014, 193, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Koonin, E.V. Homologous Capsid Proteins Testify to the Common Ancestry of Retroviruses, Caulimoviruses, Pseudoviruses, and Metaviruses. J. Virol. 2017, 91, e00210-17. [Google Scholar] [CrossRef] [PubMed]
- Bilanchone, V.; Clemens, K.; Kaake, R.; Dawson, A.R.; Matheos, D.; Nagashima, K.; Sitlani, P.; Patterson, K.; Chang, I.; Huang, L.; et al. Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes. PLoS Genet. 2015, 11, e1005528. [Google Scholar] [CrossRef]
- Sandmeyer, S.; Patterson, K.; Bilanchone, V. Ty3, a Position-specific Retrotransposon in Budding Yeast. Microbiol. Spectr. 2015, 3, MDNA3-0057-2014. [Google Scholar] [CrossRef]
- Sandmeyer, S.B.; Clemens, K.A. Function of a retrotransposon nucleocapsid protein. RNA Biol. 2010, 7, 642–654. [Google Scholar] [CrossRef]
- Gumna, J.; Purzycka, K.J.; Ahn, H.W.; Garfinkel, D.J.; Pachulska-Wieczorek, K. Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA. RNA Biol. 2019, 16, 1749–1763. [Google Scholar] [CrossRef]
- Doh, J.H.; Lutz, S.; Curcio, M.J. Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites. PLoS Genet. 2014, 10, e1004219. [Google Scholar] [CrossRef]
- Kim, E.Y.; Wang, L.; Lei, Z.; Li, H.; Fan, W.; Cho, J. Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat. Plants 2021, 7, 303–309. [Google Scholar] [CrossRef]
- McCue, A.D.; Nuthikattu, S.; Reeder, S.H.; Slotkin, R.K. Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA. PLoS Genet. 2012, 8, e1002474. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, S.L.; Nomberg, E.J.; Newman, A.C.; Taylor, J.V.; Guerrero-Ferreira, R.C.; Garfinkel, D.J. An interchangeable prion-like domain is required for Ty1 retrotransposition. Proc. Natl. Acad. Sci. USA 2023, 120, e2303358120. [Google Scholar] [CrossRef]
- Borsetti, A.; Öhagen, Å.; Göttlinger, H.G. The C-Terminal Half of the Human Immunodeficiency Virus Type 1 Gag Precursor Is Sufficient for Efficient Particle Assembly. J. Virol. 1998, 72, 9313–9317. [Google Scholar] [CrossRef]
- Malagon, F.; Jensen, T.H. The T Body, a New Cytoplasmic RNA Granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 2008, 28, 6022–6032. [Google Scholar] [CrossRef]
- Checkley, M.A.; Nagashima, K.; Lockett, S.J.; Nyswaner, K.M.; Garfinkel, D.J. P-Body Components Are Required for Ty1 Retrotransposition during Assembly of Retrotransposition-Competent Virus-Like Particles. Mol. Cell. Biol. 2010, 30, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Lutz, S.; Lesage, P. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, MDNA3-0053-2014. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032813. [Google Scholar] [CrossRef]
- Moon, S.; Namkoong, S. Ribonucleoprotein Granules: Between Stress and Transposable Elements. Biomolecules 2023, 13, 1027. [Google Scholar] [CrossRef]
- Hu, S.; Liang, C.; Guo, F. Stress out the LINEs. Mob. Genet. Elem. 2016, 6, e1133267. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Valiente-Echeverria, F.; Rubilar, P.S.; Garcia-de-Gracia, F.; Ricci, E.P.; Limousin, T.; Décimo, D.; Mouland, A.J.; Ohlmann, T. HIV-2 genomic RNA accumulates in stress granules in the absence of active translation. Nucleic Acids Res. 2014, 42, 12861–12875. [Google Scholar] [CrossRef]
- Valiente-Echeverría, F.; Melnychuk, L.; Mouland, A.J. Viral modulation of stress granules. Virus Res. 2012, 169, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clément, J.-F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010, 123, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Mathur, J.; Koncz, C. PEG-mediated protoplast transformation with naked DNA. Methods Mol. Biol. 1998, 82, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Song, P.; Zhang, S.; Li, J. Co-immunoprecipitation Assays to Detect In Vivo Association of Phytochromes with Their Interacting Partners. Methods Mol. Biol. 2021, 2297, 75–82. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polkhovskiy, A.; Komakhin, R.; Kirov, I. GAG Protein of Arabidopsis thaliana LTR Retrotransposon Forms Retrosome-like Cytoplasmic Granules and Activates Stress Response Genes. Plants 2025, 14, 1894. https://doi.org/10.3390/plants14131894
Polkhovskiy A, Komakhin R, Kirov I. GAG Protein of Arabidopsis thaliana LTR Retrotransposon Forms Retrosome-like Cytoplasmic Granules and Activates Stress Response Genes. Plants. 2025; 14(13):1894. https://doi.org/10.3390/plants14131894
Chicago/Turabian StylePolkhovskiy, Alexander, Roman Komakhin, and Ilya Kirov. 2025. "GAG Protein of Arabidopsis thaliana LTR Retrotransposon Forms Retrosome-like Cytoplasmic Granules and Activates Stress Response Genes" Plants 14, no. 13: 1894. https://doi.org/10.3390/plants14131894
APA StylePolkhovskiy, A., Komakhin, R., & Kirov, I. (2025). GAG Protein of Arabidopsis thaliana LTR Retrotransposon Forms Retrosome-like Cytoplasmic Granules and Activates Stress Response Genes. Plants, 14(13), 1894. https://doi.org/10.3390/plants14131894







