Site-Directed Mutagenesis Mediated by Molecular Modeling and Docking and Its Effect on the Protein–Protein Interactions of the bHLH Transcription Factors SPATULA, HECATE1, and INDEHISCENT
Abstract
1. Introduction
2. Results
2.1. Protein Structure Modeling and Molecular Docking
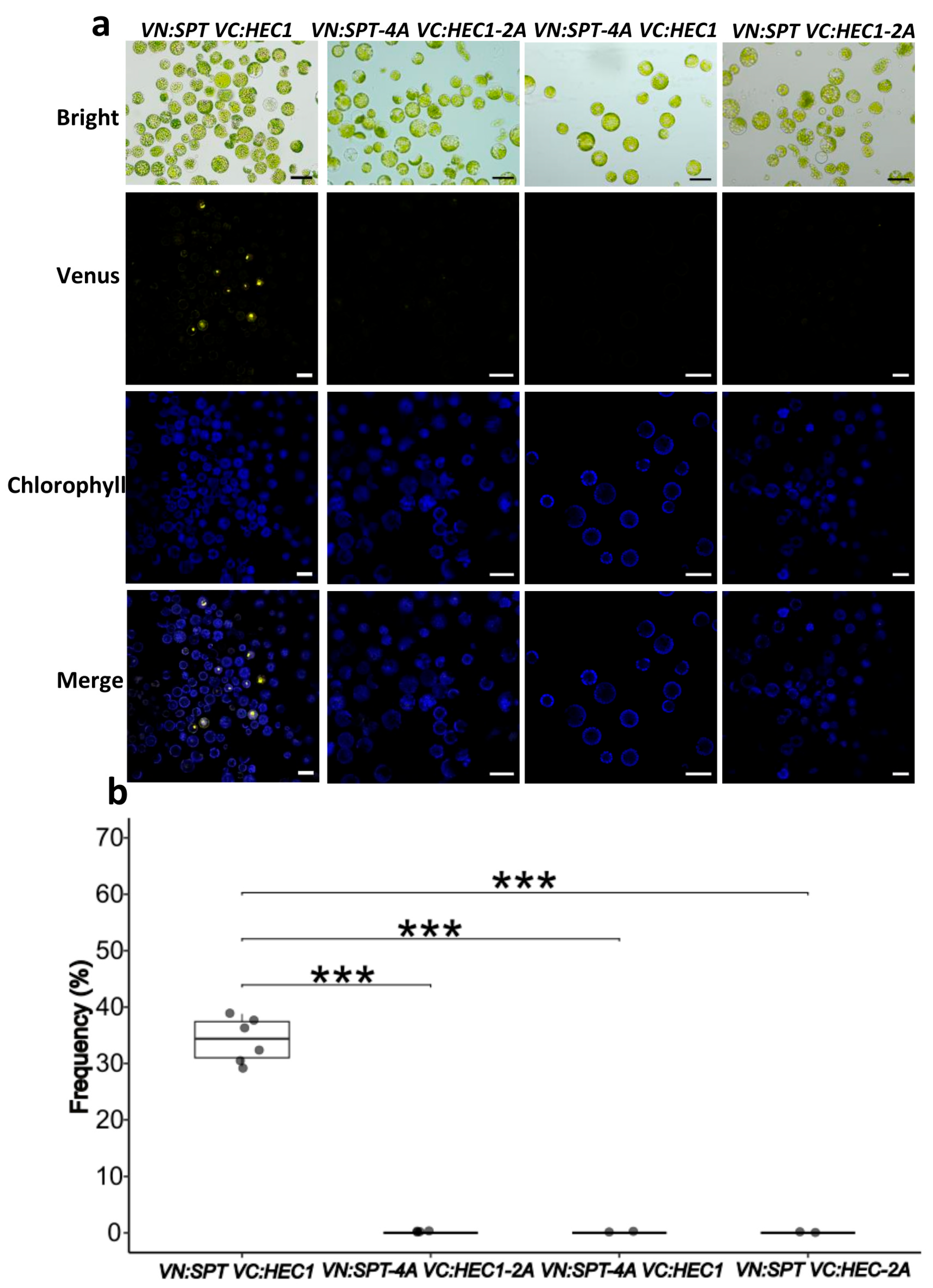
2.2. Protein–Protein Interaction Analyses Using the BiFC Assay
2.3. Generation of Overexpressing Lines of Site-Directed Mutagenized Genes
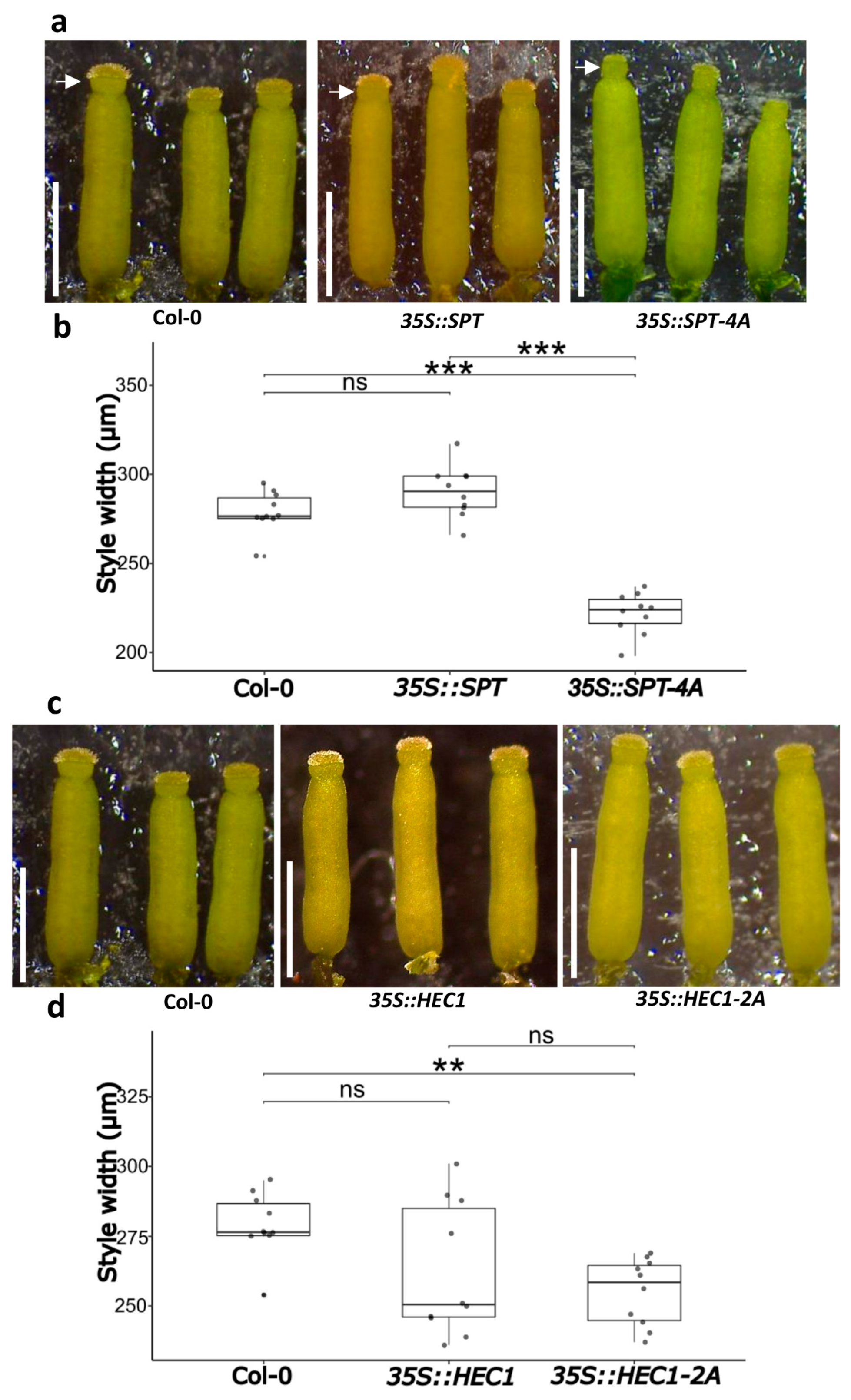
2.4. Functional Characterization of the 35S::SPT-4A Line
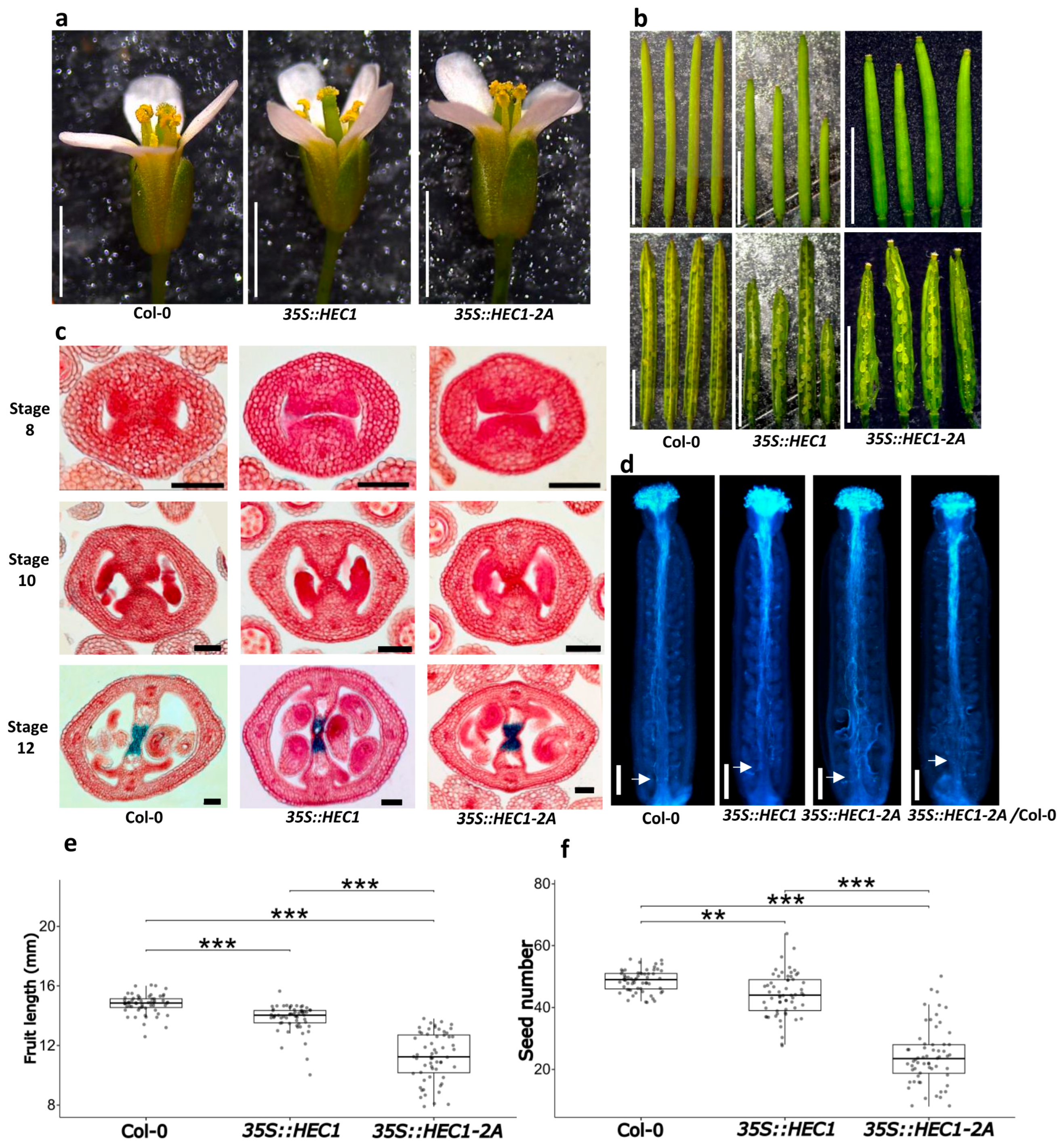
2.5. Functional Characterization of the 35S::HEC1-2A Line
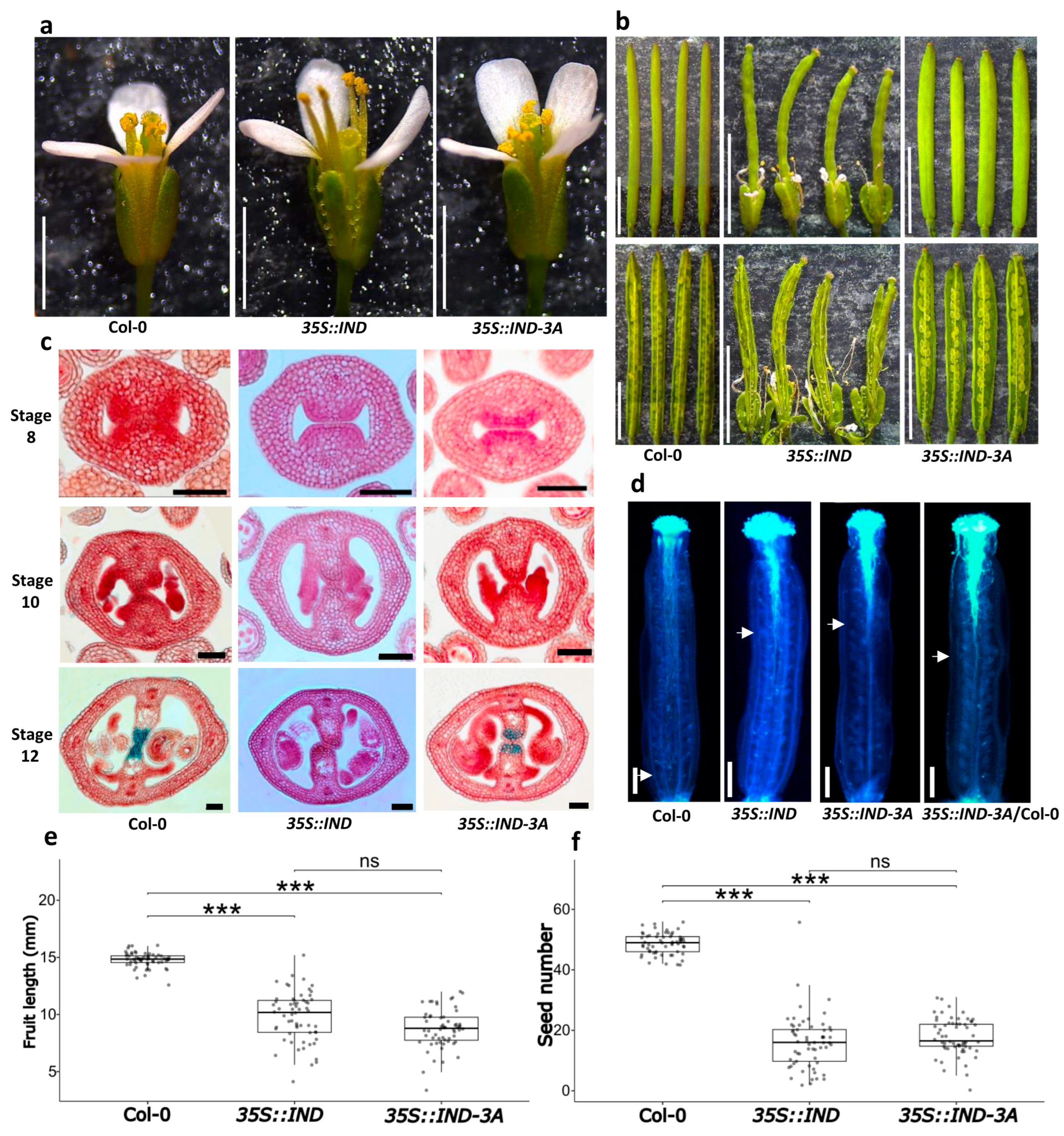
2.6. Functional Characterization of the 35S::IND-3A Line
2.7. Morphological Analysis of the Style Width in the Overexpression Lines
3. Discussion
3.1. Molecular Modeling and Docking for the Prediction of Protein–Protein Interaction Sites
3.2. The Mutated Sites Are Important for the Formation of SPT-HEC1 and SPT-IND Dimers
3.3. Morphological Effects Due to Overexpression of Mutated Gene Versions
4. Materials and Methods
4.1. Structure Modeling and Molecular Docking
4.2. Site-Directed Mutagenesis
4.3. BiFC Assay
4.4. Generation of Arabidopsis Overexpression Lines
4.5. RNA Extraction and RT-PCR Analysis
4.6. Statistical Analysis
4.7. Histology and Microscopy Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reyes-Olalde, J.I.; Zuñiga-Mayo, V.M.; Chávez Montes, R.A.; Marsch-Martínez, N.; de Folter, S. Inside the Gynoecium: At the Carpel Margin. Trends Plant Sci. 2013, 18, 644–655. [Google Scholar] [CrossRef]
- Moubayidin, L.; Østergaard, L. Gynoecium Formation: An Intimate and Complicated Relationship. Curr. Opin. Genet. Dev. 2017, 45, 15–21. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Gómez-Felipe, A.; Herrera-Ubaldo, H.; de Folter, S. Gynoecium Development: Networks in Arabidopsis and Beyond. J. Exp. Bot. 2019, 70, 1447–1460. [Google Scholar] [CrossRef]
- Robles, P.; Pelaz, S. Flower and Fruit Development in Arabidopsis thaliana. Int. J. Dev. Biol. 2005, 49, 633–643. [Google Scholar] [CrossRef] [PubMed]
- de Folter, S.; Immink, R.G.H.; Kieffer, M.; Pařenicová, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive Interaction Map of the Arabidopsis MADS Box Transcription Factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- de Folter, S.; Immink, R.G.H. Yeast Protein–Protein Interaction Assays and Screens. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2011; Volume 754, pp. 145–165. ISBN 9781617791536. [Google Scholar]
- Herrera-Ubaldo, H.; de Folter, S. Gynoecium and Fruit Development in Arabidopsis. Development 2022, 149k, dev200120. [Google Scholar] [CrossRef] [PubMed]
- De Las Rivas, J.; Fontanillo, C. Protein-Protein Interactions Essentials: Key Concepts to Building and Analyzing Interactome Networks. PLoS Comput. Biol. 2010, 6, 1–8. [Google Scholar] [CrossRef]
- Braun, P.; Gingras, A.C. History of Protein-Protein Interactions: From Egg-White to Complex Networks. Proteomics 2012, 12, 1478–1498. [Google Scholar] [CrossRef]
- Berggård, T.; Linse, S.; James, P. Methods for the Detection and Analysis of Protein-Protein Interactions. Proteomics 2007, 7, 2833–2842. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein-Protein Interaction Detection: Methods and Analysis. Int. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef]
- Dill, K.A.; MacCallum, J.L. The Protein-Folding Problem, 50 Years On. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and Its Applications in the Fields of Biology and Medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, B.; Bradley, P. Advances in Protein Structure Prediction and Design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular Docking as a Tool for the Discovery of Molecular Targets of Nutraceuticals in Diseases Management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Huang, S.Y.; Zou, X. MDockPP: A Hierarchical Approach for Protein-Protein Docking and Its Application to CAPRI Rounds 15-19. Proteins Struct. Funct. Bioinform. 2010, 78, 3096–3103. [Google Scholar] [CrossRef]
- Venkatraman, V.; Yang, Y.D.; Sael, L.; Kihara, D. Protein-Protein Docking Using Region-Based 3D Zernike Descriptors. BMC Bioinform. 2009, 10, 407. [Google Scholar] [CrossRef]
- Esquivel-Rodríguez, J.; Yang, Y.D.; Kihara, D. Multi-LZerD: Multiple Protein Docking for Asymmetric Complexes. Proteins Struct. Funct. Bioinform. 2012, 80, 1818–1833. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro Web Server for Protein-Protein Docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Weng, G.; Wang, E.; Wang, Z.; Liu, H.; Zhu, F.; Li, D.; Hou, T. HawkDock: A Web Server to Predict and Analyze the Protein-Protein Complex Based on Computational Docking and MM/GBSA. Nucleic Acids Res. 2019, 47, W322–W330. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. Carpel Development Genes in Arabidopsis. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Heisler, M.G.B.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a Gene That Controls Development of Carpel Margin Tissues in Arabidopsis, Encodes a BHLH Protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA Genes Regulate Growth and Pattern Formation during Gynoecium Development in Arabidopsis thaliana. Int. J. Plant Sci. 2002, 163, 17–41. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The HECATE Genes Regulate Female Reproductive Tract Development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef]
- Liljegren, S.J.; Roeder, A.H.K.; Kempin, S.A.; Gremski, K.; Østergaard, L.; Guimil, S.; Reyes, D.K.; Yanofsky, M.F. Control of Fruit Patterning in Arabidopsis by INDEHISCENT. Cell 2004, 116, 843–853. [Google Scholar] [CrossRef]
- Girin, T.; Paicu, T.; Stephenson, P.; Fuentes, S.; Körner, E.; O’Brien, M.; Sorefan, K.; Wood, T.A.; Balanzá, V.; Ferrándiz, C.; et al. INDEHISCENT and SPATULA Interact to Specify Carpel and Valve Margin Tissue and Thus Promote Seed Dispersal in Arabidopsis. Plant Cell 2011, 23, 3641–3653. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE Genes Function in Phytohormone Control during Gynoecium Development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.; Martínez-Godoy, M.A.; Ezquerro, M.; Navarrete-Gómez, M.; Trigueros, M.; Rodríguez-Concepción, M.; Ferrándiz, C. A Transcriptional Complex of NGATHA and BHLH Transcription Factors Directs Stigma Development in Arabidopsis. Plant Cell 2021, 33, 3645–3657. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Ubaldo, H.; Campos, S.E.; López-Gómez, P.; Luna-García, V.; Zúñiga-Mayo, V.M.; Armas-Caballero, G.E.; González-Aguilera, K.L.; DeLuna, A.; Marsch-Martínez, N.; Espinosa-Soto, C.; et al. The Protein–Protein Interaction Landscape of Transcription Factors during Gynoecium Development in Arabidopsis. Mol. Plant 2023, 16, 260–278. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Cunningham, B.C.; Wells, J.A. High-Resolution Epitope Mapping of HGH-Receptor Interactions by Alanine-Scanning Mutagenesis. Science 1989, 244, 1081–1085. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis Mesophyll Protoplasts: A Versatile Cell System for Transient Gene Expression Analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Wu, F.-H.; Shen, S.-C.; Lee, L.-Y.; Lee, S.-H.; Chan, M.-T.; Lin, C.-S. Tape-Arabidopsis Sandwich—A Simpler Arabidopsis Protoplast Isolation Method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef]
- López-Gómez, P.; de Folter, S. A Detailed Protocol for the Isolation of Arabidopsis thaliana Mesophyll Protoplasts and Their Applications. In Plant MicroRNAs. Methods in Molecular Biology; de Folter, S., Ed.; Humana: New York, NY, USA, 2025; Volume 2900, pp. 341–350. [Google Scholar]
- Penfield, S.; Josse, E.M.; Kannangara, R.; Gilday, A.D.; Halliday, K.J.; Graham, I.A. Cold and Light Control Seed Germination through the BHLH Transcription Factor SPATULA. Curr. Biol. 2005, 15, 1998–2006. [Google Scholar] [CrossRef]
- Groszmann, M.; Paicu, T.; Smyth, D.R. Functional Domains of SPATULA, a BHLH Transcription Factor Involved in Carpel and Fruit Development in Arabidopsis. Plant J. 2008, 55, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Sorefan, K.; Girin, T.; Liljegren, S.J.; Ljung, K.; Robles, P.; Galván-Ampudia, C.S.; Offringa, R.; Friml, J.; Yanofsky, M.F.; Østergaard, L. A Regulated Auxin Minimum Is Required for Seed Dispersal in Arabidopsis. Nature 2009, 459, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Dosztanyi, Z.; Tosatto, S.C.E. MobiDB-Lite: Fast and Highly Specific Consensus Prediction of Intrinsic Disorder in Proteins. Bioinformatics 2017, 33, 1402–1404. [Google Scholar] [CrossRef]
- Lee, D.; Grant, A.; Marsden, R.L.; Orengo, C. Identification and Distribution of Protein Families in 120 Completed Genomes Using Gene3D. Proteins Struct. Funct. Genet. 2005, 59, 603–615. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Teichmann, S.A.; Babu, M.M. Gene Regulatory Network Growth by Duplication. Nat. Genet. 2004, 36, 492–496. [Google Scholar] [CrossRef]
- Van Dijk, A.D.J.; Ter Braak, C.J.F.; Immink, R.G.; Angenent, G.C.; Van Ham, R.C.H.J. Predicting and Understanding Transcription Factor Interactions Based on Sequence Level Determinants of Combinatorial Control. Bioinformatics 2008, 24, 26–33. [Google Scholar] [CrossRef][Green Version]
- Van Dijk, A.D.J.; Morabito, G.; Fiers, M.; Van Ham, R.C.H.J.; Angenent, G.C.; Immink, R.G.H. Sequence Motifs in MADS Transcription Factors Responsible for Specificity and Diversification of Protein-Protein Interaction. PLoS Comput. Biol. 2010, 6, e1001017. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-Domain Transcription Factor Complexes in Arabidopsis Flower Development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein-Nucleic Acid Interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Blanc-Mathieu, R.; Janeau, A.; Paul, M.; Lucas, J.; Xu, X.; Ye, H.; Lai, X.; Le Hir, S.; Guillotin, A.; et al. SEPALLATA-Driven MADS Transcription Factor Tetramerization Is Required for Inner Whorl Floral Organ Development. Plant Cell 2024, 36, 3435–3450. [Google Scholar] [CrossRef]
- Tan, X.M.; Li, Y.R.; Song, M.R.; Yuan, L.N.; Zhao, Z.X.; Liu, Y.; Meng, Q.; Huang, X.; Ma, Y.Y.; Xu, Z.Q. The Molecular Mechanism of Interaction Between SEPALLATA3 and APETALA1 in Arabidopsis thaliana. Plant Direct 2025, 9, e70052. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.; Levy Karin, E.; Kim, H.; Moriwaki, Y.; Ovchinnikov, S.; Steinegger, M.; Mirdita, M. Easy and Accurate Protein Structure Prediction Using ColabFold. Nat. Protoc. 2025, 20, 620–642. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 3.0. 2015. Available online: https://www.pymol.org (accessed on 29 September 2024).
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two Distinct Forms of M-Locus Protein Kinase Localize to the Plasma Membrane and Interact Directly with S-Locus Receptor Kinase to Transduce Self-Incompatibility Signaling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef]
- de Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G.H. A Bsister MADS-Box Gene Involved in Ovule and Seed Development in Petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef]
- Clough, S.; Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Radovich, C.; Welty, N.; Hsu, J.; Li, D.; Meulia, T.; van der Knaap, E. Integration of Tomato Reproductive Developmental Landmarks and Expression Profiles, and the Effect of SUN on Fruit Shape. BMC Plant Biol. 2009, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Ubaldo, H.; Lozano-Sotomayor, P.; Ezquer, I.; Di Marzo, M.; Montes, R.A.C.; Gómez-Felipe, A.; Pablo-Villa, J.; Diaz-Ramirez, D.; Ballester, P.; Ferrańdiz, C.; et al. New Roles of NO TRANSMITTING TRACT and SEEDSTICK during Medial Domain Development in Arabidopsis Fruits. Development 2019, 146, dev172395. [Google Scholar] [CrossRef]
- Zúñiga-Mayo, V.M.; Marsch-Martínez, N.; de Folter, S. JAIBA, a Class-II HD-ZIP Transcription Factor Involved in the Regulation of Meristematic Activity, and Important for Correct Gynoecium and Fruit Development in Arabidopsis. Plant J. 2012, 71, 314–326. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, S.L.; Xie, L.F.; Puah, C.S.; Zhang, X.Q.; Yang, W.C.; Sundaresan, V.; Ye, D. VANGUARD1 Encodes a Pectin Methylesterase That Enhances Pollen Tube Growth in the Arabidopsis Style and Transmitting Tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef]
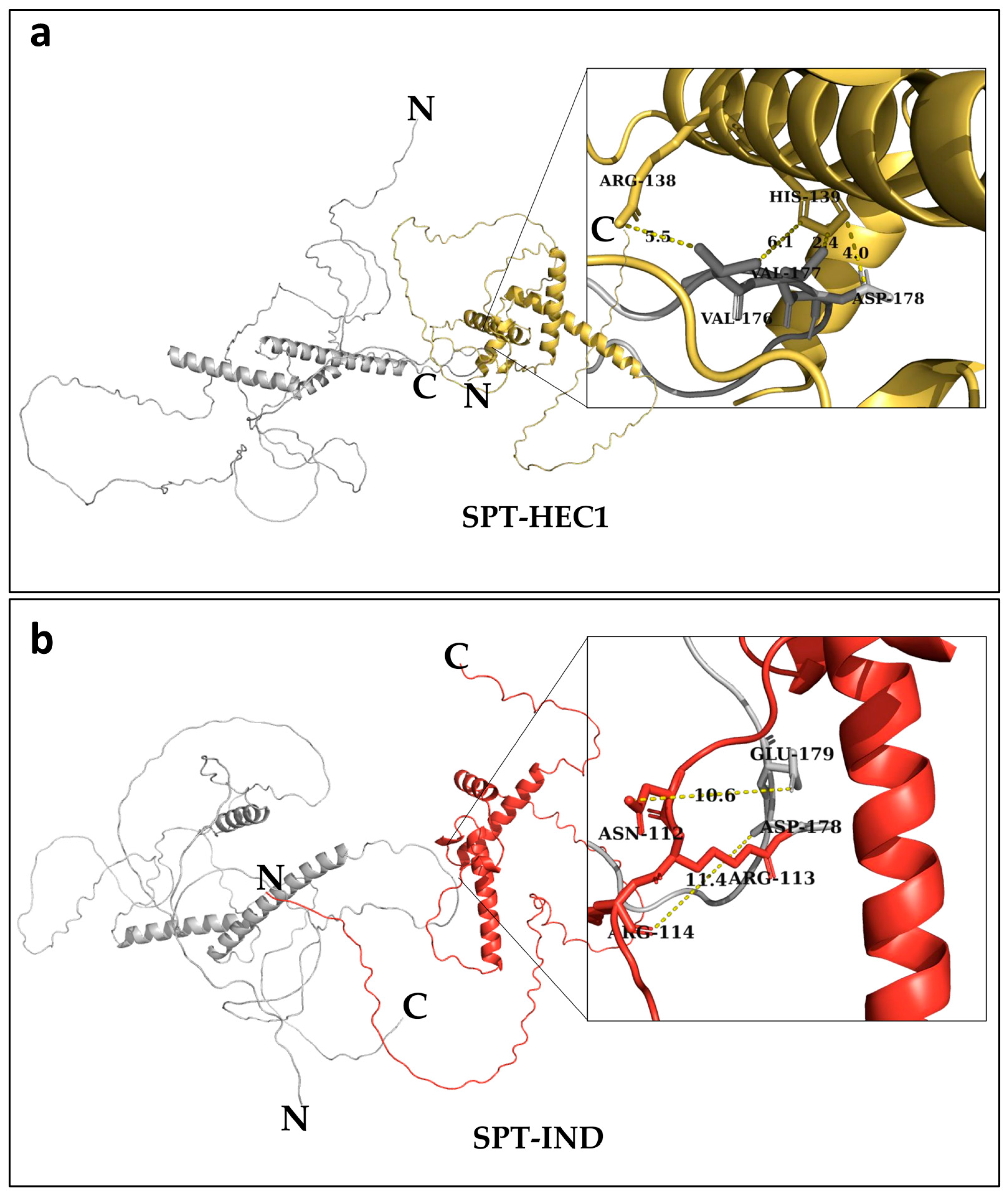


| SPT | Binding Free Energy (Kcal mol−1) | HEC1 | Binding Free Energy (Kcal mol−1) |
|---|---|---|---|
| Val177 | −5.56 | Arg138 | −5.12 |
| Val176 | −4.29 | Arg143 | −4.75 |
| Asp178 | −3.44 | His139 | −3.04 |
| Ile372 | −2.06 | Arg123 | −3.0 |
| Glu171 | −1.95 | Lys130 | −2.91 |
| SPT | Binding Free Energy (Kcal mol−1) | IND | Binding Free Energy (Kcal mol−1) |
|---|---|---|---|
| Glu179 | −3.79 | Arg113 | −9.52 |
| Asp178 | −3.27 | Arg139 | −8.25 |
| Phe130 | −2.71 | Ser135 | −5.14 |
| Ser133 | −1.42 | Arg131 | −2.58 |
| Ala180 | −1.11 | Lys142 | −2.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Gómez, P.; De La Mora-Franco, D.; Herrera-Ubaldo, H.; Díaz-Quezada, C.; Brieba, L.G.; de Folter, S. Site-Directed Mutagenesis Mediated by Molecular Modeling and Docking and Its Effect on the Protein–Protein Interactions of the bHLH Transcription Factors SPATULA, HECATE1, and INDEHISCENT. Plants 2025, 14, 1756. https://doi.org/10.3390/plants14121756
López-Gómez P, De La Mora-Franco D, Herrera-Ubaldo H, Díaz-Quezada C, Brieba LG, de Folter S. Site-Directed Mutagenesis Mediated by Molecular Modeling and Docking and Its Effect on the Protein–Protein Interactions of the bHLH Transcription Factors SPATULA, HECATE1, and INDEHISCENT. Plants. 2025; 14(12):1756. https://doi.org/10.3390/plants14121756
Chicago/Turabian StyleLópez-Gómez, Pablo, Daniela De La Mora-Franco, Humberto Herrera-Ubaldo, Corina Díaz-Quezada, Luis G. Brieba, and Stefan de Folter. 2025. "Site-Directed Mutagenesis Mediated by Molecular Modeling and Docking and Its Effect on the Protein–Protein Interactions of the bHLH Transcription Factors SPATULA, HECATE1, and INDEHISCENT" Plants 14, no. 12: 1756. https://doi.org/10.3390/plants14121756
APA StyleLópez-Gómez, P., De La Mora-Franco, D., Herrera-Ubaldo, H., Díaz-Quezada, C., Brieba, L. G., & de Folter, S. (2025). Site-Directed Mutagenesis Mediated by Molecular Modeling and Docking and Its Effect on the Protein–Protein Interactions of the bHLH Transcription Factors SPATULA, HECATE1, and INDEHISCENT. Plants, 14(12), 1756. https://doi.org/10.3390/plants14121756






