Characterization and Expression Analysis of the SABATH Gene Family Under Abiotic Stresses in Cucumber (Cucumis sativus L.)
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CsSABATH Gene Family
2.2. The Conserved Motifs and Gene Structure of CsSABATH Genes
2.3. Phylogenetic Tree of CsSABATH Genes
2.4. Collinearity Analysis of CsSABATH Family Genes
2.5. Analysis of Cis-Acting Elements of CsSABATH Promoter
2.6. Tissue-Specific Expression Profiles of CsSABATH Genes
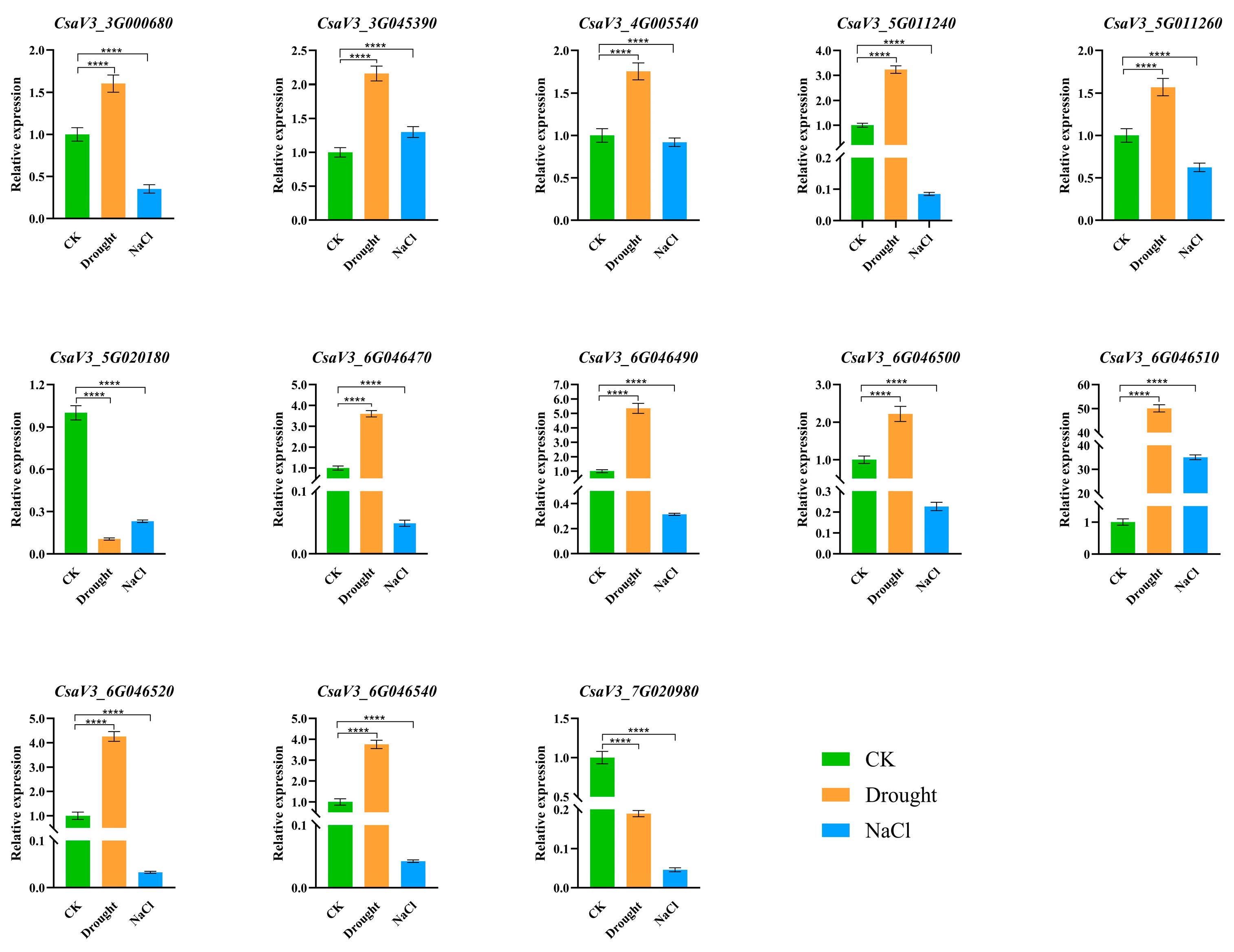
2.7. Relative Expression of the CsSABATH Gene Under Osmotic Stress Treatment
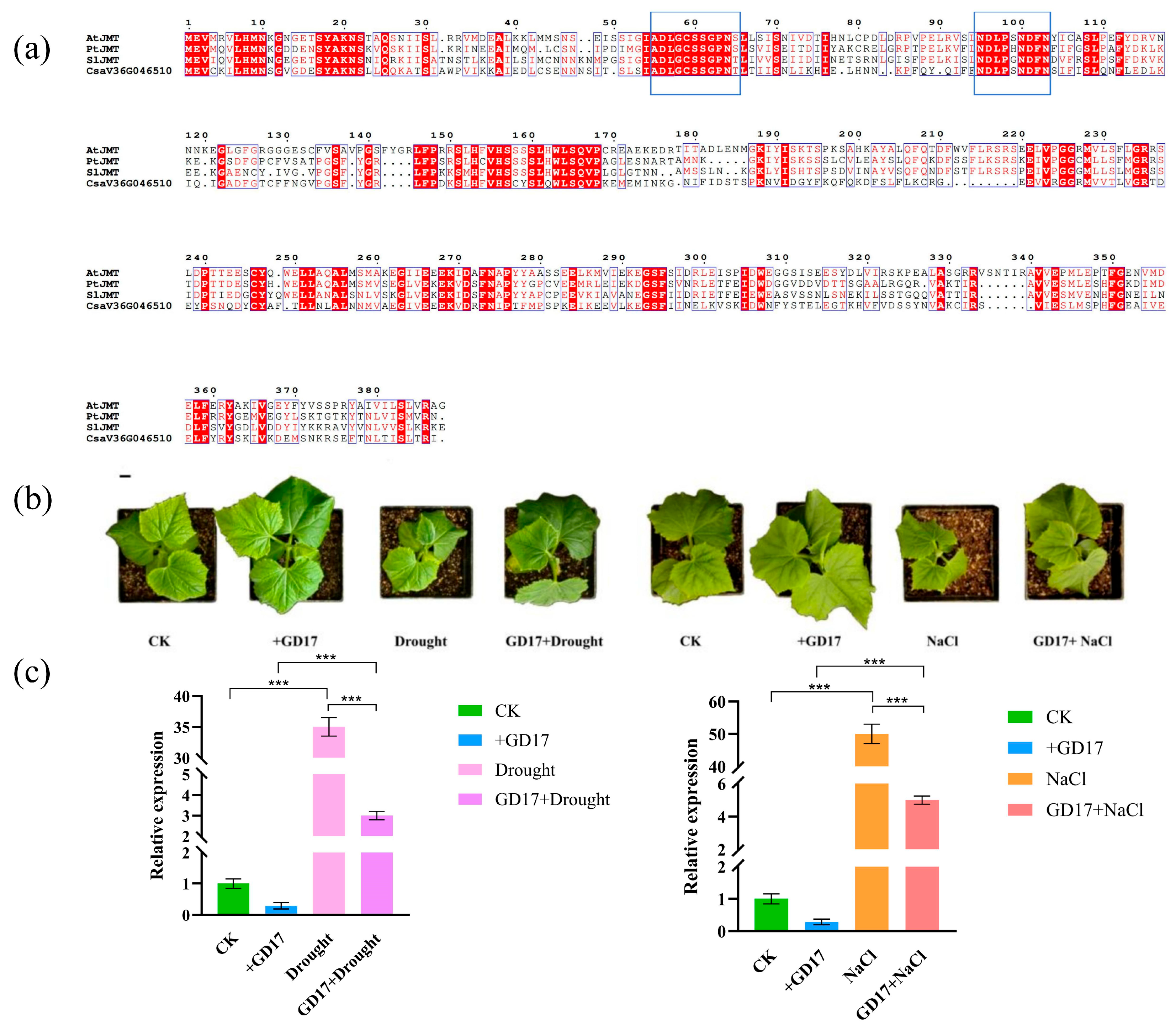
2.8. The Functional Prediction of CsaV3_6G046510 and PGPR-GD17 Can Alleviate the Response of CsaV3_6G046510 to Abiotic Stress
3. Discussion
3.1. Tandem Duplication Might Be One of the Reasons for the Diverse Functions of the CsSABATH Family
3.2. Analysis of Phylogenetic Relationships, Gene Structures, and Cis-Acting Elements
3.3. Functional Speculation of CsaV3_6G046510
4. Materials and Methods
4.1. Plant Culture and Treatment
4.2. Identification of SABATH Genes in Cucumber
4.3. Chromosome Distribution Bioinformatic Analyses of SABATH Gene Family in Cucumber
4.4. Conserved Motif and Gene Structure Analysis
4.5. Construction of Phylogenetic Tree
4.6. Detection of Homologous Gene Pairs and Synteny Analysis
4.7. Analysis of Cis-Acting Elements in SABATH Gene Promoters
4.8. Promoter Cis-Element Analysis
4.9. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
4.10. Identity Comparison
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chaffin, T.A.; Wang, W.; Chen, J.G.; Chen, F. Function and Evolution of the Plant MES Family of Methylesterases. Plants 2024, 13, 3364. [Google Scholar] [CrossRef]
- D’Auria, J.C.; Chen, F.; Pichersky, E. The SABATH family of MTS in Arabidopsis thaliana and other plant species. Recent Adv. Phytochem. 2003, 37, 95–125. [Google Scholar]
- Wang, B.; Li, M.; Yuan, Y.; Liu, S. Genome-Wide Comprehensive Analysis of the SABATH Gene Family in Arabidopsis and Rice. Evol. Bioinform. Online 2019, 15, 1176934319860864. [Google Scholar] [CrossRef]
- Wei, X.; Tao, K.; Zhang, J.; Lu, S.; Chen, S.; Liao, J. Identification of SABATH Family Members in Solanum lycopersicum and Their Expression Patterns Under Abiotic/Biotic Stresses. Plant Mol. Biol. Rep. 2021, 39, 403–418. [Google Scholar] [CrossRef]
- Dubs, N.M.; Davis, B.R.; de Brito, V.; Colebrook, K.C.; Tiefel, I.J.; Nakayama, M.B.; Huang, R.; Ledvina, A.E.; Hack, S.J.; Inkelaar, B.; et al. A Collaborative Classroom Investigation of the Evolution of SABATH Methyltransferase Substrate Preference Shifts over 120 My of Flowering Plant History. Mol. Biol. Evol. 2022, 39, msac007. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef]
- Varbanova, M.; Yamaguchi, S.; Yang, Y.; McKelvey, K.; Hanada, A.; Borochov, R.; Yu, F.; Jikumaru, Y.; Ross, J.; Cortes, D.; et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 2007, 19, 32–45. [Google Scholar] [CrossRef]
- Chung, P.J.; Park, B.S.; Wang, H.; Liu, J.; Jang, I.C.; Chua, N.H. Light-Inducible MiR163 Targets PXMT1 Transcripts to Promote Seed Germination and Primary Root Elongation in Arabidopsis. Plant Physiol. 2016, 170, 1772–1782. [Google Scholar] [CrossRef]
- Wu, R.; Zhang, F.; Liu, L.; Li, W.; Pichersky, E.; Wang, G. MeNA, Controlled by Reversible Methylation of Nicotinate, Is an NAD Precursor that Undergoes Long-Distance Transport in Arabidopsis. Mol. Plant 2018, 11, 1264–1277. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2016, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ferrer, J.L.; Ross, J.; Guan, J.; Yang, Y.; Pichersky, E.; Noel, J.P.; Chen, F. Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol. 2008, 146, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Ament, K.; Kant, M.R.; Sabelis, M.W.; Haring, M.A.; Schuurink, R.C. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 2004, 135, 2025–2037. [Google Scholar] [CrossRef]
- Ament, K.; Krasikov, V.; Allmann, S.; Rep, M.; Takken, F.L.; Schuurink, R.C. Methyl salicylate production in tomato affects biotic interactions. Plant J. 2010, 62, 124–134. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. Ambiguities of PGPR-Induced Plant Signaling and Stress Management. Front. Microbiol. 2022, 13, 899563. [Google Scholar] [CrossRef]
- Giannelli, G.; Potestio, S.; Visioli, G. The Contribution of PGPR in Salt Stress Tolerance in Crops: Unravelling the Molecular Mechanisms of Cross-Talk between Plant and Bacteria. Plants 2023, 11, 2197. [Google Scholar] [CrossRef]
- Sharma, I.; Sharma, S.; Sharma, V.; Singh, A.K.; Sharma, A.; Kumar, A.; Singh, J.; Sharma, A. PGPR-Enabled bioremediation of pesticide and heavy metal-contaminated soil: A review of recent advances and emerging challenges. Chemosphere 2024, 362, 142678. [Google Scholar] [CrossRef]
- Al-Turki, A.; Murali, M.; Omar, A.F.; Rehan, M.; Sayyed, R.Z. Recent advances in PGPR-mediated resilience toward interactive effects of drought and salt stress in plants. Front. Microbiol. 2023, 14, 1214845. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Cao, Y.; Li, G.; Guo, Y.; Ma, L.; Bu, N.; Hao, L. Paraburkholderia sp. GD17 improves rice seedling tolerance to salinity. Plant Soil 2021, 467, 373–389. [Google Scholar] [CrossRef]
- Fu, W.; Zhao, D.; Liu, H.; Wu, L.; Hao, L. Inoculation of Paraburkholderia sp. GD17 improves seedling growth and tolerance to Cadmium in Chinese cabbage. Environ. Exp. Bot. 2024, 227, 105955. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, S.; Liu, Z.; Wan, Z.; Gao, X.; Qiao, Y.; Yu, J.; Zhang, G. Genome-wide identification and expression analysis of the cucumber PYL gene family. PeerJ 2022, 10, e12786. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 2, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yao, J.; Chaiprasongsuk, M.; Li, G.; Guan, J.; Tschaplinski, T.J.; Guo, H.; Chen, F. Molecular and biochemical characterization of the jasmonic acid methyltransferase gene from black cottonwood (Populus trichocarpa). Phytochemistry 2013, 94, 74–81. [Google Scholar] [CrossRef]
- Tieman, D.; Zeigler, M.; Schmelz, E.; Taylor, M.G.; Rushing, S.; Jones, J.B.; Klee, H.J. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 2010, 1, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.; Qualley, A.V.; Xie, Z.; Fridman, E.; Dudareva, N.; Gang, D.R. Evolution of Cinnamate/p-coumarate carboxyl methyltransferases and their role in the biosynthesis of methylcinnamate. Plant Cell 2007, 19, 3212–3229. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, J.S.; Ross, J.; Noel, J.P.; Pichersky, E.; Chen, F. An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 2006, 448, 123–132. [Google Scholar] [CrossRef]
- Wang, W.; Guo, H.; Bowman, J.L.; Chen, F. Plant SABATH Methyltransferases: Diverse Functions, Unusual Reaction Mechanisms and Complex Evolution. Crit. Rev. Plant Sci. 2024, 43, 291–312. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 10, 680–694. [Google Scholar] [CrossRef]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database 2015, 2015, bav017. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Wen, K.; Zhong, W.; Feng, L.; Han, T.; Suo, H.; Ren, H.; Yuan, Q.; Wu, Z.; Chen, Y.; Li, X.; et al. Genome-wide identification of SABATH gene family in soybean relate to salt, aluminum, chromium toxicity. Sci. Rep. 2025, 1, 14030. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiao, D.; Yang, C.; Chen, J.; Li, Y.; Liang, S.; Lin, K.; Chen, Z. Genome-wide identification and expression analysis of SABATH methyltransferases in tea plant (Camellia sinensis): Insights into their roles in plant defense responses. Plant Signal Behav. 2020, 10, 1804684. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.K.; Jie-Ling, H.E. Current reviews of plant SABATH Methyltransferases. Plant Physiol. J. 2011, 47, 840–846. [Google Scholar]
- Gao, R.; Luo, Y.; Pan, X.; Wang, C.; Liao, W. Genome-wide identification of SHMT family genes in cucumber (Cucumis sativus L.) and functional analyses of CsSHMTs in response to hormones and abiotic stresses. 3 Biotech 2022, 11, 305. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, Z.; Luo, S.; Li, X.; Lyu, J.; Liu, Z.; Wan, Z.; Yu, J. Genome-wide identification and expression analysis of the cucumber PP2C gene family. BMC Genom. 2022, 1, 563. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 6, 1349–1359. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 4, 1446. [Google Scholar] [CrossRef] [PubMed]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Anjum, S.A.; Wang, L.; Farooq, M.; Khan, I.; Xue, L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J. Agron. Crop Sci. 2011, 197, 296–301. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 16, 8568. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Pollmann, S.; Müller, M. Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress. Int. J. Mol. Sci. 2023, 6, 5990. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Zhang, R.; Liu, S.; Li, R.; Wang, Y.; Chen, Y.; Hou, H.; Dai, Q. Methyl Jasmonate Alleviates the Deleterious Effects of Salinity Stress by Augmenting Antioxidant Enzyme Activity and Ion Homeostasis in Rice (Oryza sativa L.). Agronomy 2022, 12, 2343. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 3, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, D1, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 11, 1733–1742. [Google Scholar] [CrossRef]
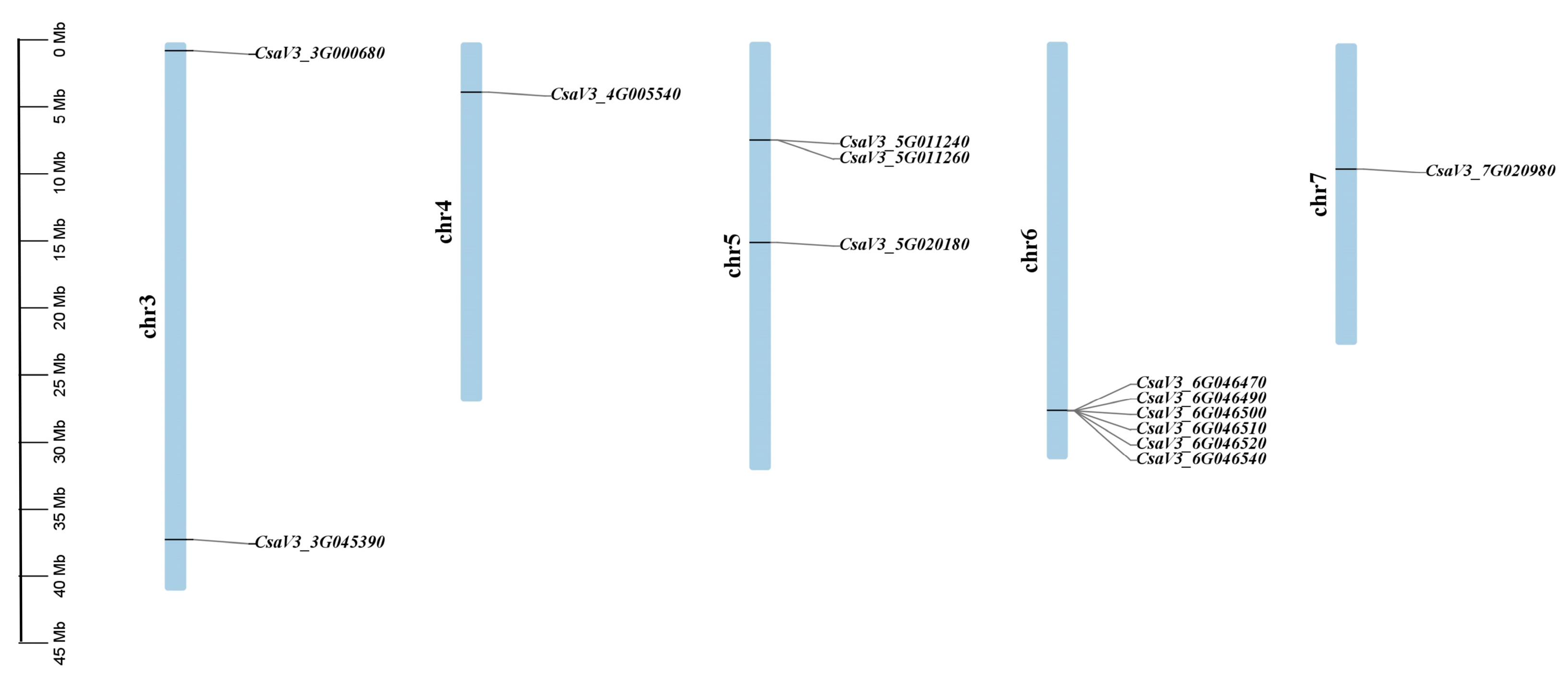





| Gene ID (v2) | Gene ID (v3) | Gene ID (v4) | Location | Protein Length (aa) | MW (Da) | PI | Instability Index | GRAVY Value | SC Localization |
|---|---|---|---|---|---|---|---|---|---|
| Csa3G002690 | CsaV3_3G000680 | CsaV4_3G000075 | 3(572314…574064)- | 373 | 42131.04 | 6.81 | 51.15 | −0.383 | Cytoplasmic |
| Csa3G859730 | CsaV3_3G045390 | CsaV4_3G004520 | 3(37062534…37064406)- | 375 | 42186.99 | 5.92 | 49.29 | −0.260 | Outer Membrane |
| Csa4G046610 | CsaV3_4G005540 | CsaV4_4G000567 | 4(3683812…3688124)- | 368 | 41851.27 | 5.58 | 46.42 | −0.375 | Cytoplasmic/Outer Membrane |
| Csa5G241140 | CsaV3_5G011240 | CsaV4_5G000942 | 5(7294796…7298875)- | 376 | 42357.16 | 5.73 | 44.83 | −0.178 | Cytoplasmic |
| Csa5G241640 | CsaV3_5G011260 | CsaV4_5G000943 | 5(7318739…7325466)- | 361 | 40766.49 | 5.92 | 40.29 | −0.237 | Cytoplasmic |
| Csa5G313340 | CsaV3_5G020180 | CsaV4_5G001567 | 5(14937232…14943388)+ | 459 | 51873.85 | 6.25 | 42.59 | 0.046 | Cytoplasmic /Outer Membrane |
| Csa6G503370 | CsaV3_6G046470 | CsaV4_6G003494 | 6(27448277…27453505)- | 370 | 41317.27 | 6.19 | 41.06 | −0.118 | Outer Membrane /Cytoplasmic/Extracellular |
| Csa6G503870 | CsaV3_6G046490 | CsaV4_6G003496 | 6(27459877…27461717)- | 371 | 42799.93 | 6.67 | 36.34 | −0.326 | Outer Membrane /Extracellular |
| Csa6G503880 | CsaV3_6G046500 | CsaV4_6G003497 | 6(27467723…27469134)- | 295 | 33405.65 | 8.19 | 37.09 | −0.101 | Cytoplasmic/Outer Membrane |
| Csa6G503880 | CsaV3_6G046510 | CsaV4_6G003498 | 6(27470946…27472820)- | 363 | 41359.27 | 5.41 | 43.61 | −0.170 | Outer Membrane /Cytoplasmic |
| Csa6G504380 | CsaV3_6G046520 | CsaV4_6G003499 | 6(27482065…27484009)+ | 364 | 41326.46 | 5.36 | 48.81 | −0.215 | Cytoplasmic |
| Csa6G504400 | CsaV3_6G046540 | CsaV4_6G003501 | 6(27487071…27489858)+ | 312 | 35717.29 | 8.84 | 49.54 | −0.253 | Cytoplasmic/Outer Membrane |
| Csa7G081680 | CsaV3_7G020980 | CsaV4_7G001138 | 7(9311426…9315269)- | 385 | 42441.04 | 5.56 | 42.91 | −0.134 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Li, S.; Zhou, Y.; Chen, M.; Jiang, L.; Fu, W. Characterization and Expression Analysis of the SABATH Gene Family Under Abiotic Stresses in Cucumber (Cucumis sativus L.). Plants 2025, 14, 1748. https://doi.org/10.3390/plants14121748
Zhang X, Li S, Zhou Y, Chen M, Jiang L, Fu W. Characterization and Expression Analysis of the SABATH Gene Family Under Abiotic Stresses in Cucumber (Cucumis sativus L.). Plants. 2025; 14(12):1748. https://doi.org/10.3390/plants14121748
Chicago/Turabian StyleZhang, Xinjie, Shanyu Li, Yang Zhou, Mengxin Chen, Lisi Jiang, and Wei Fu. 2025. "Characterization and Expression Analysis of the SABATH Gene Family Under Abiotic Stresses in Cucumber (Cucumis sativus L.)" Plants 14, no. 12: 1748. https://doi.org/10.3390/plants14121748
APA StyleZhang, X., Li, S., Zhou, Y., Chen, M., Jiang, L., & Fu, W. (2025). Characterization and Expression Analysis of the SABATH Gene Family Under Abiotic Stresses in Cucumber (Cucumis sativus L.). Plants, 14(12), 1748. https://doi.org/10.3390/plants14121748







