Dual-Phase Severity Grading of Strawberry Angular Leaf Spot Based on Improved YOLOv11 and OpenCV
Abstract
1. Introduction
2. Results
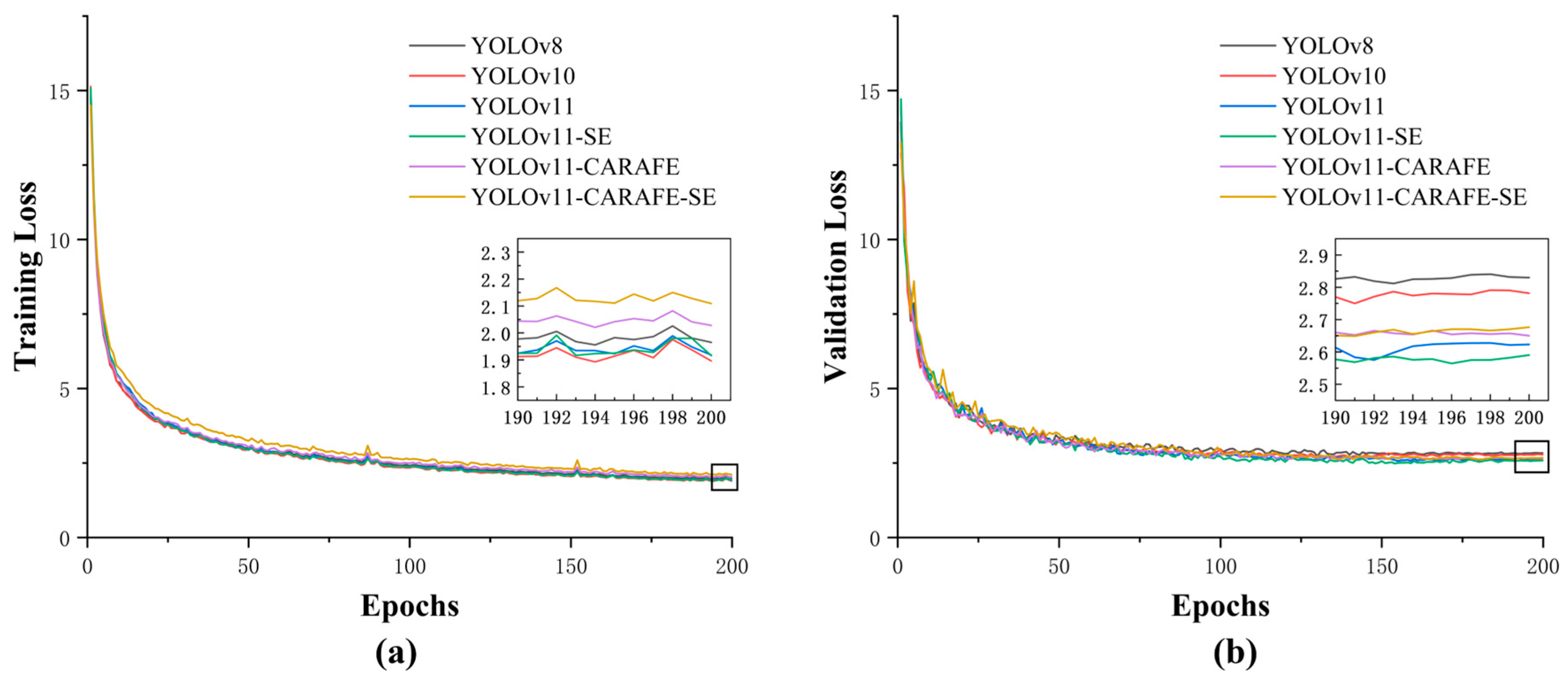
2.1. Model Training
2.2. Ablation Experiments
2.3. Impact of Different Attention Mechanisms on the Model
2.4. Effects of Different Upsampling Methods on the Model
2.5. Performance of the Improved Model in Strawberry Angular Leaf Spot Leaf Segmentation
2.6. Disease Spot Segmentation Based on OpenCV and Disease Severity Classification
3. Discussion
4. Materials and Methods
4.1. Datasets
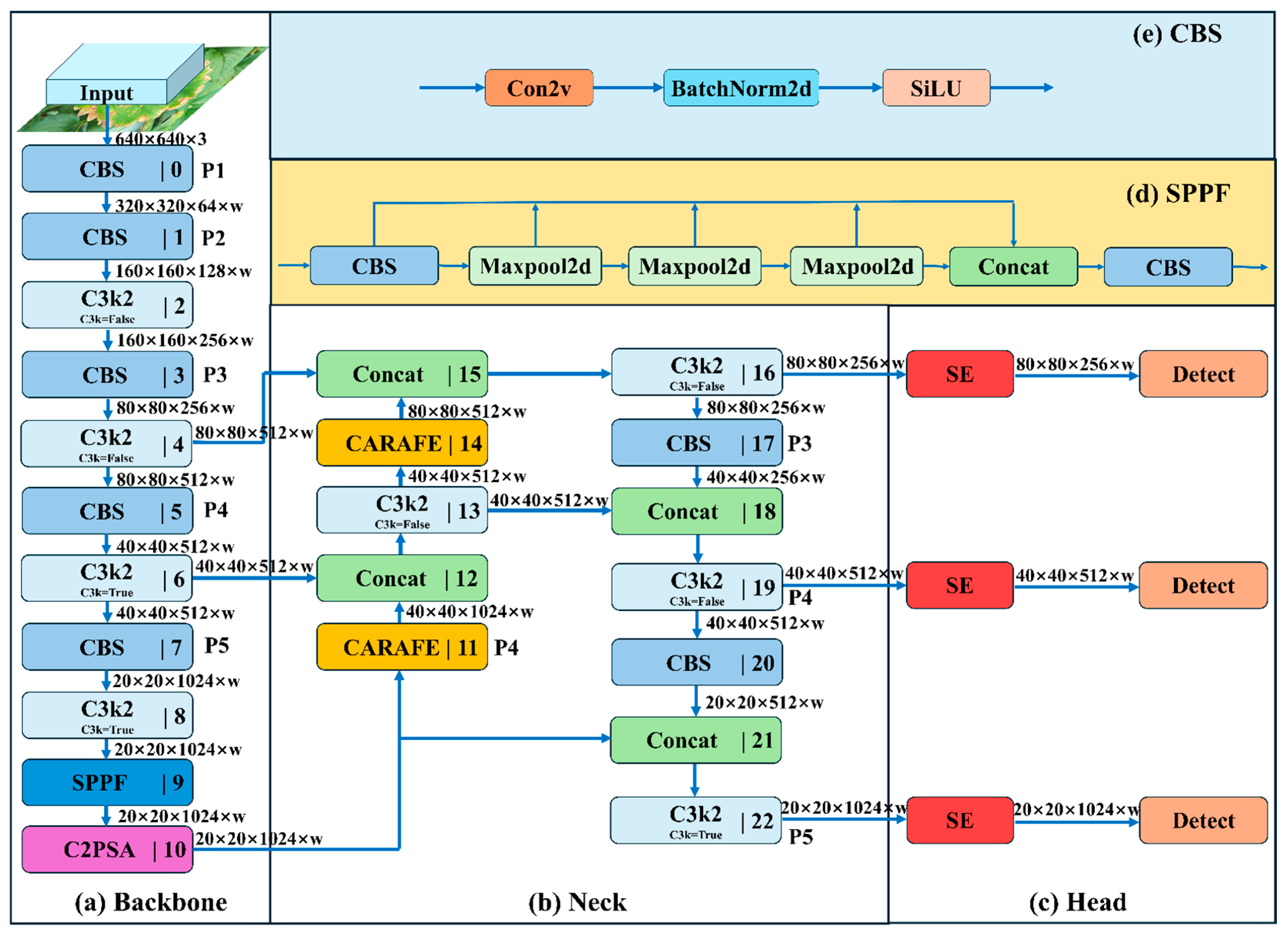
4.2. Strawberry Leaf Segmentation Method Based on Improved YOLOv11
4.2.1. YOLOv11
- (1)
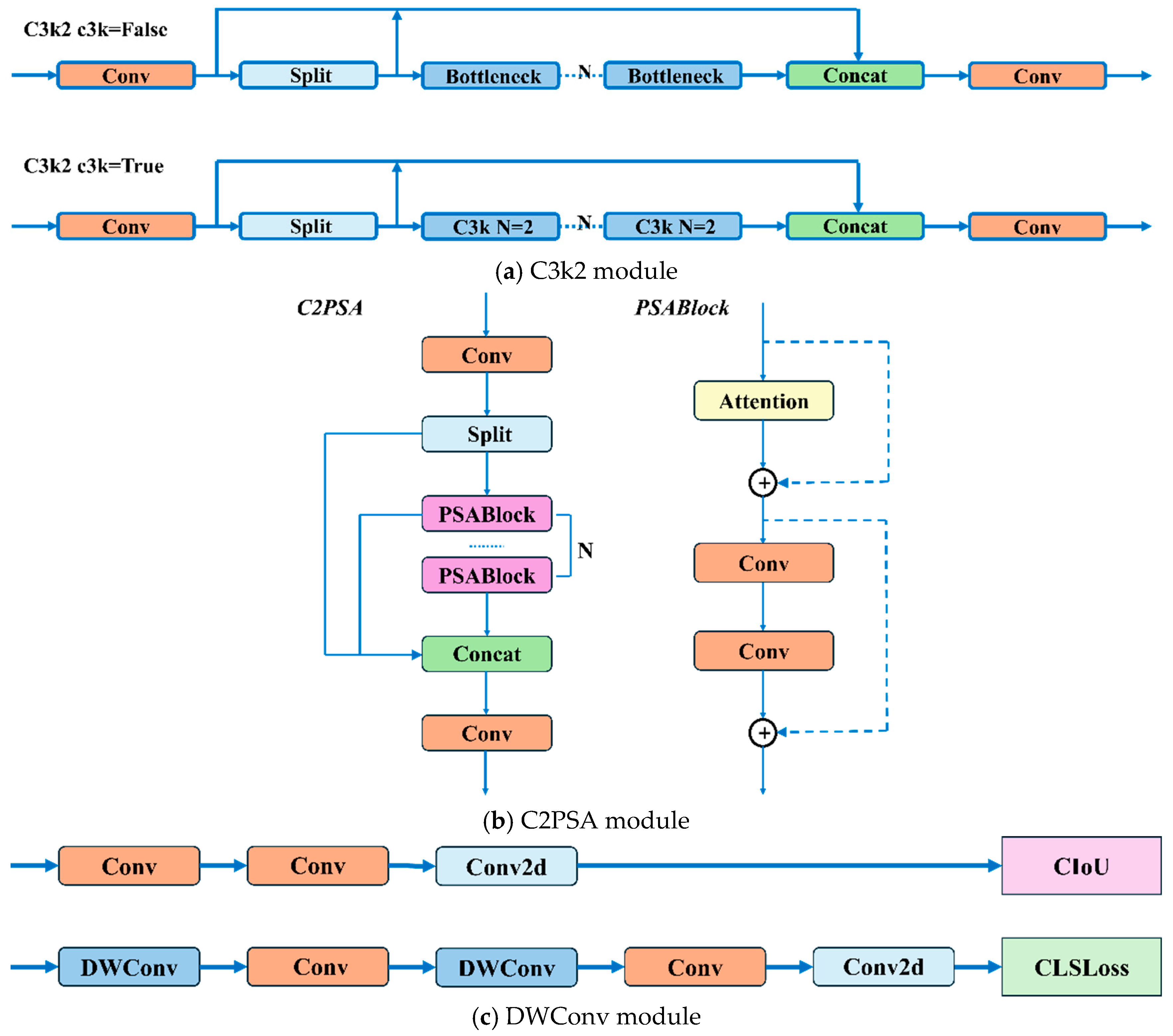
- The C3k2 module is an enhanced design derived from the traditional C3 module. It provides enhanced feature extraction capabilities by integrating variable convolutional kernels and channel separation strategies. In the shallow layers of the network, when the c3k parameter is set to False, the C3k2 module becomes functionally equivalent to the standard C2f module. When the c3k parameter is set to True, the Bottleneck module is replaced with the C3 module, as illustrated in Figure 6a;
- (2)
- The proposal of the C2PSA mechanism integrates a multi-head attention mechanism within the C2 framework. This mechanism is cascaded after the spatial pyramid fast pooling (SPPF) module, as illustrated in Figure 6b;
- (3)
- The classification detection head within the original decoupled head has been enhanced by incorporating two depthwise separable convolutions (DWConvs), resulting in two DWConv layers in total. This modification significantly reduces both parameter count and computational complexity, as shown in Figure 6c;
- (4)
- Significant modifications were made to the model’s depth and width parameters. Furthermore, YOLOv11 offers multiple variants with different scaling factors, allowing the flexibility to meet diverse requirements. In this experiment, the YOLOv11n model was chosen as the base model for further improvements due to its lower parameter count and faster inference speed, making it particularly well suited for deployment in embedded agricultural equipment scenarios. To facilitate the comparison of models, we incorporated YOLOv8 [36], YOLOv9 [37], and YOLOv10 [38].
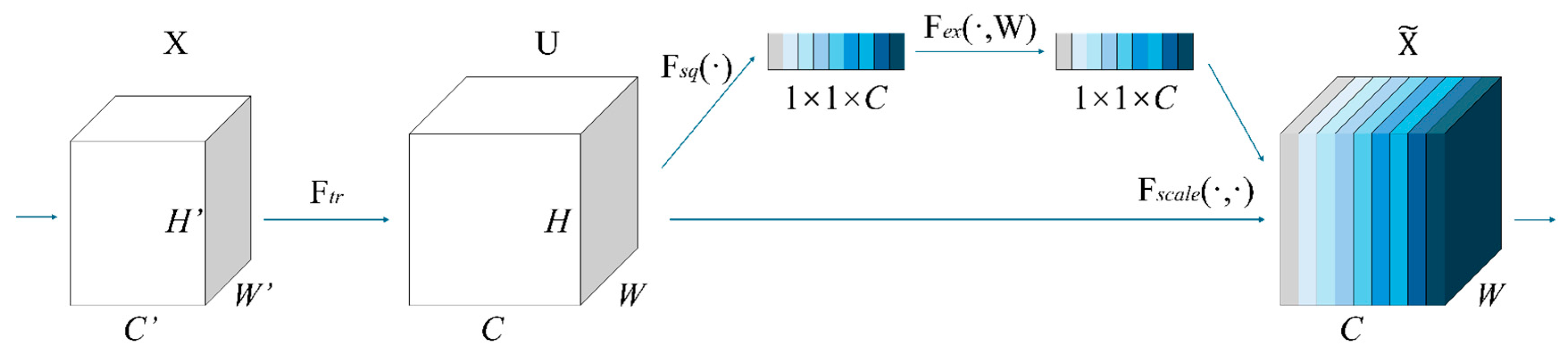
4.2.2. SE Attention
- (1)
- Global statistics extraction: the channel description vector is obtained by applying mean pooling across spatial dimensions, as shown in Equation (1) [39]:where represents the result of global average pooling for the -th channel, and denote the height and width of the feature map, and signifies the feature value at position in the -th channel. The outcome is a vector of length , which encapsulates the global information for each channel.
- (2)
- Dynamic channel calibration: the gating mechanism is used to learn the nonlinear relationship between channels, as shown in Equation (2) [39]:where is the weight matrix of dimension reduction, is the weight matrix of dimension increase, and the scaling factor r is the control parameter quantity.
- (3)
- Feature recalibration: the learned channel weights are applied to the original feature map, as shown in Equation (3) [39]:where denotes the weight for the -th channel, represents the -th channel of the input feature map, and is the adjusted feature map. The output-weighted feature map is given by .
4.2.3. CARAFE Module
4.2.4. Proposed Model
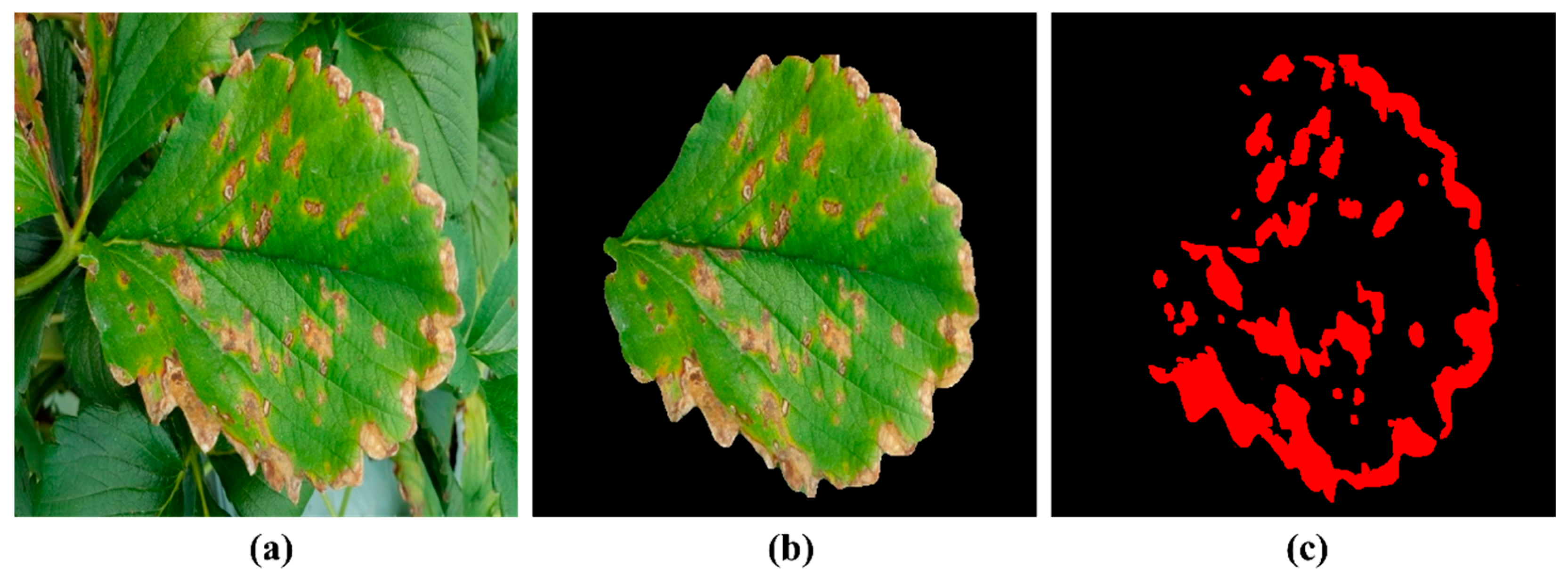
4.3. OpenCV-Based Lesion Segmentation Method and Disease Severity Grading
4.4. A Detection Platform for Strawberry Angular Leaf Spot Severity Based on PyQt5
4.5. Equipment
4.6. Model Evaluation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, F.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Alvarez-Suarez, J.M.; Afrin, S.; Bompadre, S.; Quiles, J.L.; Mezzetti, B.; Battino, M. Strawberry as a health promoter: An evidence based review. Food Funct. 2015, 6, 1386–1398. [Google Scholar] [CrossRef] [PubMed]
- Dholi, P.K.; Khatiwada, P.; Basnet, B.; Bhandari, S. An Extensive Review of Strawberry (Fragaria × ananassa) Diseases and Integrated Management Approaches: Current Understanding and Future Directions. Fundam. Appl. Agric. 2023, 8, 655–667. [Google Scholar] [CrossRef]
- Montarry, J.; Mimee, B.; Danchin, E.G.J.; Koutsovoulos, G.D.; Ste-Croix, D.T.; Grenier, E. Recent Advances in Population Genomics of Plant-Parasitic Nematodes. Phytopathology 2021, 111, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, K.; Wang, Z.; Zhang, H.; Gu, J.; Liu, L.; Yang, J.; Zhang, J. Brassinosteroids function in spikelet differentiation and degeneration in rice. J. Integr. Plant Biol. 2019, 61, 943–963. [Google Scholar] [CrossRef]
- Romero-Oraá, R.; García, M.; Oraá-Pérez, J.; López-Gálvez, M.I.; Hornero, R. Effective Fundus Image Decomposition for the Detection of Red Lesions and Hard Exudates to Aid in the Diagnosis of Diabetic Retinopathy. Sensors 2020, 20, 6549. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Mochida, K.; Koda, S.; Inoue, K.; Hirayama, T.; Tanaka, S.; Nishii, R.; Melgani, F. Computer vision-based phenotyping for improvement of plant productivity: A machine learning perspective. Gigascience 2019, 8, giy153. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Kuska, M.T.; Thomas, S.; Wahabzada, M.; Behmann, J.; Rascher, U.; Kersting, K. Quantitative and qualitative phenotyping of disease resistance of crops by hyperspectral sensors: Seamless interlocking of phytopathology, sensors, and machine learning is needed! Curr. Opin. Plant Biol. 2019, 50, 156–162. [Google Scholar] [CrossRef]
- Prakash, R.M.; Vimala, M.; Ramalakshmi, K.; Prakash, M.B.; Krishnamoorthi, A.; Kumari, R.S.S. Crop Disease Detection and Classification with Transfer learning and hyper-parameters optimized Convolutional neural network. In Proceedings of the 2022 Third International Conference on Intelligent Computing Instrumentation and Control Technologies (ICICICT), Kannur, India, 11–12 August 2022; pp. 1608–1613. [Google Scholar]
- Thyagaraj, R.; Satheesha, T.Y.; Bhairannawar, S. Plant Leaf Disease Classification Using Modified SVM With Post Processing Techniques. In Proceedings of the 2023 International Conference on Applied Intelligence and Sustainable Computing (ICAISC), Dharwad, India, 16–17 June 2023; pp. 1–4. [Google Scholar]
- Selvaraj, A.; Shebiah, N.; Ananthi, S.; Varthini, S. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Arnal Barbedo, J.G. Plant disease identification from individual lesions and spots using deep learning. Biosys. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Wang, P.; Fan, E.; Wang, P. Comparative analysis of image classification algorithms based on traditional machine learning and deep learning. Pattern Recognit. Lett. 2021, 141, 61–67. [Google Scholar] [CrossRef]
- Archana, R.; Jeevaraj, P.S.E. Deep learning models for digital image processing: A review. Artif. Intell. Rev. 2024, 57, 11. [Google Scholar] [CrossRef]
- Attri, I.; Awasthi, L.K.; Sharma, T.P.; Rathee, P. A review of deep learning techniques used in agriculture. Ecol. Inform. 2023, 77, 102217. [Google Scholar] [CrossRef]
- Shafik, W.; Tufail, A.; Liyanage De Silva, C.; Awg Haji Mohd Apong, R.A. A novel hybrid inception-xception convolutional neural network for efficient plant disease classification and detection. Sci. Rep. 2025, 15, 3936. [Google Scholar] [CrossRef]
- Khan, A.I.; Quadri, S.M.K.; Banday, S.; Shah, J.L. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agric. 2022, 198, 107093. [Google Scholar] [CrossRef]
- Roy, A.M.; Bhaduri, J. Real-time growth stage detection model for high degree of occultation using DenseNet-fused YOLOv4. Comput. Electron. Agric. 2022, 193, 106694. [Google Scholar] [CrossRef]
- Cardellicchio, A.; Solimani, F.; Dimauro, G.; Petrozza, A.; Summerer, S.; Cellini, F.; Renò, V. Detection of tomato plant phenotyping traits using YOLOv5-based single stage detectors. Comput. Electron. Agric. 2023, 207, 107757. [Google Scholar] [CrossRef]
- Olisah, C.C.; Trewhella, B.; Li, B.; Smith, M.L.; Winstone, B.; Whitfield, E.C.; Fernández, F.F.; Duncalfe, H. Convolutional neural network ensemble learning for hyperspectral imaging-based blackberry fruit ripeness detection in uncontrolled farm environment. Eng. Appl. Artif. Intell. 2024, 132, 107945. [Google Scholar] [CrossRef]
- Chen, Y.K.; Xu, H.B.; Chang, P.Y.; Huang, Y.Y.; Zhong, F.L.; Jia, Q.; Chen, L.X.; Zhong, H.Q.; Liu, S. CES-YOLOv8: Strawberry Maturity Detection Based on the Improved YOLOv8. Agronomy 2024, 14, 1353. [Google Scholar] [CrossRef]
- Shi, T.; Liu, Y.; Zheng, X.; Hu, K.; Huang, H.; Liu, H.; Huang, H. Recent advances in plant disease severity assessment using convolutional neural networks. Sci. Rep. 2023, 13, 2336. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.K.; Choi, Y.S.; Lee, J.H.; Tran, M.T.; Xin, X. An effective deep learning model for classifying diseases on strawberry leaves and estimating their severity based on the multi-task U-Net. Multimed. Tools Appl. 2024, 1–22. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Tran, T.D.; Nguyen, T.T.; Pham, N.M.; Nguyen Ly, P.H.; Luong, H.H. Strawberry disease identification with vision transformer-based models. Multimed. Tools Appl. 2024, 83, 73101–73126. [Google Scholar] [CrossRef]
- Karki, S.; Basak, J.K.; Tamrakar, N.; Deb, N.C.; Paudel, B.; Kook, J.H.; Kang, M.Y.; Kang, D.Y.; Kim, H.T. Strawberry disease detection using transfer learning of deep convolutional neural networks. Sci. Hortic. 2024, 332, 113241. [Google Scholar] [CrossRef]
- Kumar, R.R.; Chauhan, R.; Dhondiyal, S.A.; Singh, A. Deep Learning-Driven Diagnosis A CNN-SVM Hybrid Approach for Automated Detection of Strawberry Leaf Diseases. In Proceedings of the 2024 IEEE 3rd World Conference on Applied Intelligence and Computing (AIC), Gwalior, India, 27–28 July 2024; pp. 1467–1470. [Google Scholar]
- Vats, S.; Kukreja, V.; Mehta, S. Tea Leaf Disease Detection: Federated Learning CNN Used for Accurate Severity Analysis. In Proceedings of the 2024 IEEE International Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation (IATMSI), Gwalior, India, 14–16 March 2024; pp. 1–6. [Google Scholar]
- Liu, B.-Y.; Fan, K.-J.; Su, W.-H.; Peng, Y. Two-Stage Convolutional Neural Networks for Diagnosing the Severity of Alternaria Leaf Blotch Disease of the Apple Tree. Remote Sens. 2022, 14, 2519. [Google Scholar] [CrossRef]
- Liu, W.; Chen, Y.; Lu, Z.; Lu, X.; Wu, Z.; Zheng, Z.; Suo, Y.; Lan, C.; Yuan, X. StripeRust-Pocket: A Mobile-Based Deep Learning Application for Efficient Disease Severity Assessment of Wheat Stripe Rust. Plant Phenomics 2024, 6, 0201. [Google Scholar] [CrossRef]
- Afzaal, U.; Bhattarai, B.; Pandeya, Y.R.; Lee, J. An Instance Segmentation Model for Strawberry Diseases Based on Mask R-CNN. Sensors 2021, 21, 6565. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Bargoti, S.; Underwood, J. Image Segmentation for Fruit Detection and Yield Estimation in Apple Orchards. J. Field Robot. 2017, 34, 1039–1060. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Wang, J.; Chen, Y.; Dong, Z.; Gao, M. Improved YOLOv5 network for real-time multi-scale traffic sign detection. Neural Comput. Appl. 2021, 35, 7853–7865. [Google Scholar] [CrossRef]
- Ultralytics. YOLOv8: A State-of-the-Art Object Detection Model. Available online: https://github.com/ultralytics/ultralytics (accessed on 20 August 2023).
- Wang, C.-Y.; Yeh, I.-H.; Liao, H.-Y.M. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. In Proceedings of the Computer Vision—ECCV 2024: 18th European Conference, Milan, Italy, 29 September–4 October 2024; Proceedings, Part XXXI. Springer Nature: Cham, Switzerland, 2024; pp. 1–21. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J.; Ding, G. YOLOv10: Real-Time End-to-End Object Detection. arXiv 2024, arXiv:2405.14458. [Google Scholar]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Gao, P.; Lu, J.; Li, H.; Mottaghi, R.; Kembhavi, A. Container: Context Aggregation Network. arXiv 2021, arXiv:2106.01401. [Google Scholar]
- Wang, J.; Chen, K.; Xu, R.; Liu, Z.; Loy, C.C.; Lin, D. CARAFE: Content-Aware ReAssembly of FEatures. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 3007–3016. [Google Scholar]
- Liu, W.; Lu, H.; Fu, H.; Cao, Z. Learning to Upsample by Learning to Sample. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–6 October 2023; pp. 6027–6037. [Google Scholar]
- Liu, Y. An Improved Faster R-CNN for Object Detection. In Proceedings of the 2018 11th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 8–9 December 2018; Volume 2, pp. 119–123. [Google Scholar]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Padilla, R.; Netto, S.L.; da Silva, E.A.B. A Survey on Performance Metrics for Object-Detection Algorithms. In Proceedings of the 2020 International Conference on Systems, Signals and Image Processing (IWSSIP), Niterói, Brazil, 1–3 July 2020; pp. 237–242. [Google Scholar]
- Sze, V.; Chen, Y.-H.; Yang, T.-J.; Emer, J.S. Efficient Processing of Deep Neural Networks: A Tutorial and Survey. Proc. IEEE 2017, 105, 2295–2329. [Google Scholar] [CrossRef]
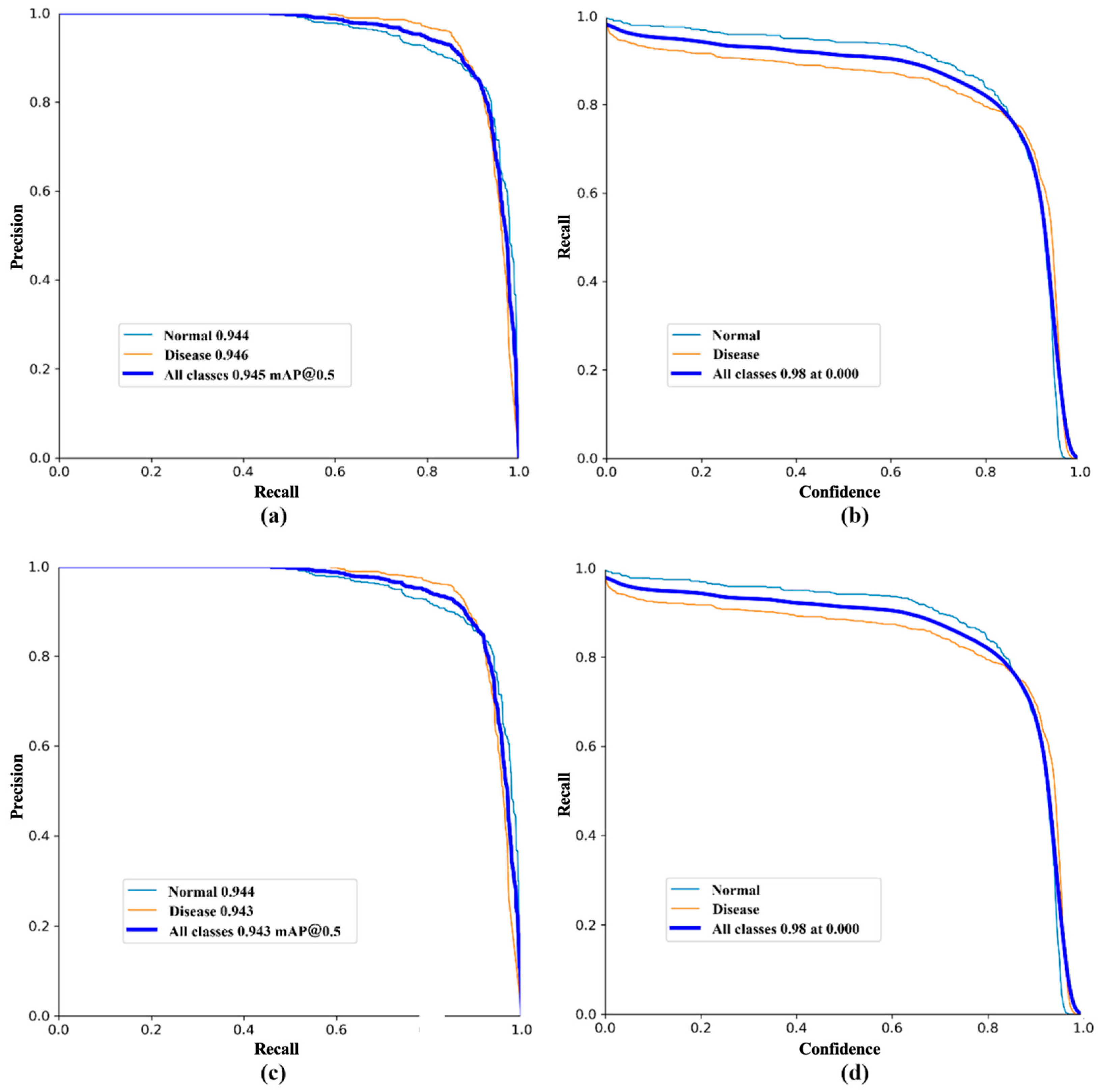





| Methods | Box | Mask | Inference Time/ms | GFLOPS | ||||
|---|---|---|---|---|---|---|---|---|
| Precision /% | Recall /% | mAP@0.5 /% | Precision /% | Recall /% | mAP@0.5 /% | |||
| YOLOv8 | 86.1 | 84.6 | 91.1 | 86.2 | 84.7 | 91.0 | 1.1 | 12.0 |
| YOLOv9 | 88.1 | 81.2 | 91.0 | 88.5 | 80.5 | 90.3 | 1.9 | 53.2 |
| YOLOv10 | 92.5 | 82.2 | 91.7 | 91.5 | 82.9 | 91.9 | 0.9 | 10.6 |
| YOLOv11 | 88.1 | 86.0 | 91.8 | 88.4 | 86.3 | 92.1 | 1.3 | 10.3 |
| YOLOv11-SE | 86.6 | 87.6 | 92.3 | 86.6 | 87.6 | 92.6 | 1.2 | 10.3 |
| YOLOv11-CARAFE | 89.2 | 86.0 | 93.0 | 89.2 | 86.0 | 93.0 | 0.9 | 10.1 |
| YOLOv11-CARAFE-SE | 88.3 | 87.2 | 93.2 | 88.2 | 87.3 | 93.0 | 0.9 | 10.1 |
| Methods | Box | Mask | GFLOPS | ||||
|---|---|---|---|---|---|---|---|
| Precision/% | Recall/% | mAP@0.5/% | Precision/% | Recall/% | mAP@0.5/% | ||
| YOLOv11 (No Attention) | 88.4 | 86.0 | 91.9 | 88.7 | 86.2 | 92.0 | 10.3 |
| SE | 86.7 | 87.6 | 92.5 | 86.7 | 87.6 | 92.6 | 10.4 |
| CBAM | 91.2 | 82.0 | 92.3 | 91.1 | 81.9 | 91.8 | 10.4 |
| Context Aggregation | 90.1 | 84.0 | 92.0 | 91.4 | 83.8 | 91.8 | 10.4 |
| Methods | Box | Mask | GFLOPS | ||||
|---|---|---|---|---|---|---|---|
| Precision/% | Recall/% | mAP@0.5/% | Precision/% | Recall/% | mAP@0.5/% | ||
| YOLOv11 (nearest) | 88.3 | 85.8 | 91.8 | 88.7 | 86.0 | 92.0 | 10.3 |
| YOLOv11 (bilinear) | 90.9 | 80.5 | 91.6 | 90.8 | 80.5 | 91.7 | 10.3 |
| CARAFE | 89.3 | 86.1 | 93.1 | 89.3 | 86.1 | 93.1 | 10.1 |
| DySample | 89.1 | 83.9 | 91.8 | 89.4 | 84.1 | 92.3 | 10.4 |
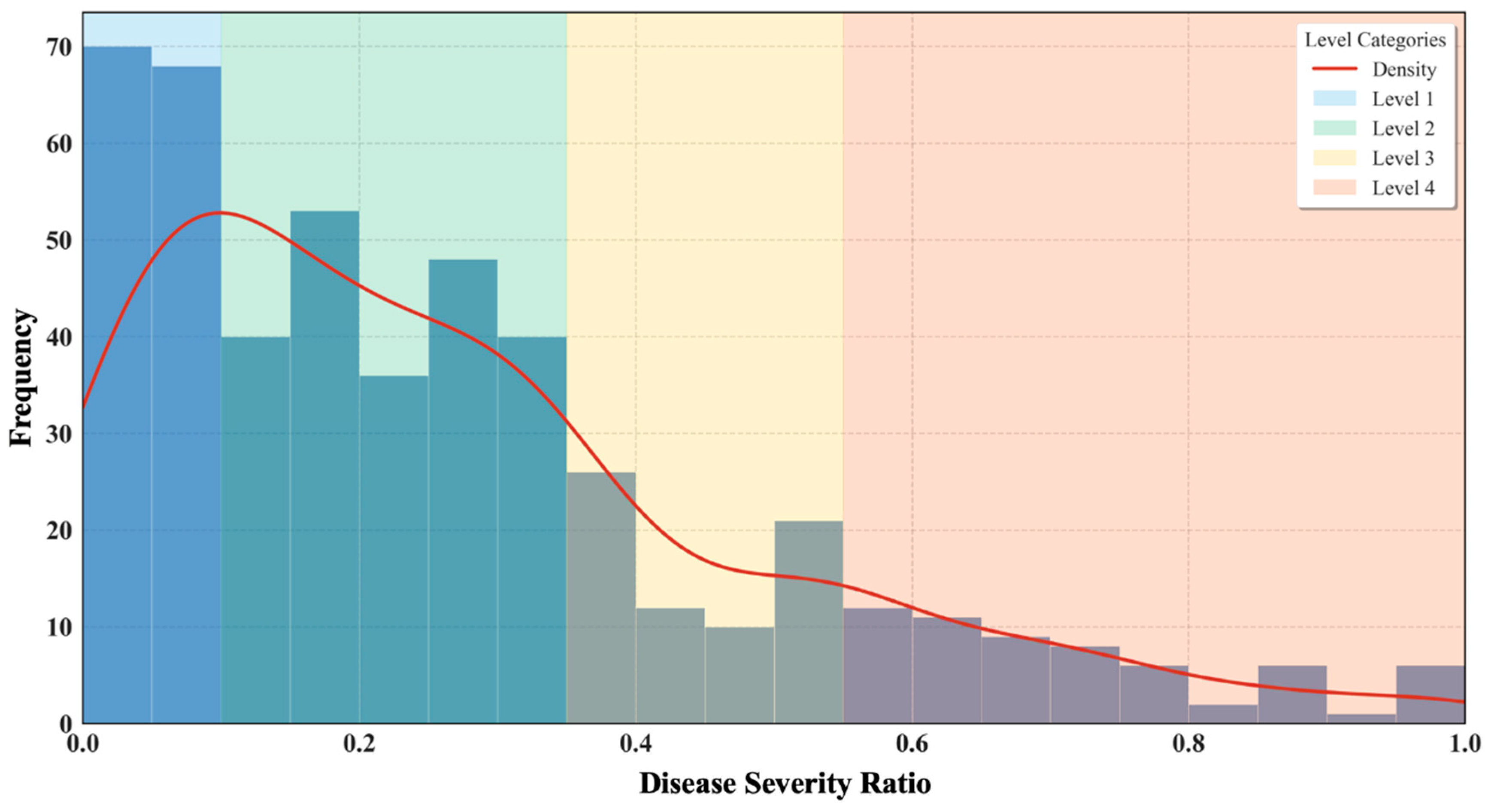
| Severity Level | Symptoms | Disease Area Ratio | Data Quantity |
|---|---|---|---|
| Level 1 | Small water-soaked spots visible on the back of the leaf | (0, 0.10] | 138 |
| Level 2 | Spot area expands, leaf edges appear dried and dehydrated | (0.10, 0.35] | 217 |
| Level 3 | Large disease spots appear but do not completely merge to cover the entire leaf | (0.35, 0.55] | 69 |
| Level 4 | Most of the leaf area is covered with red-brown lesions, which merge into a large patch | (0.55, 1] | 61 |
| Severity Level | Correct Grading | Sample | Accuracy (%) |
|---|---|---|---|
| Level 1 | 134 | 139 | 96.4 |
| Level 2 | 192 | 208 | 92.3 |
| Level 3 | 70 | 75 | 93.3 |
| Level 4 | 61 | 63 | 96.8 |
| Total | 457 | 485 | 94.2 |
| References | Plants | Model | Disease Types/Levels | Accuracy |
|---|---|---|---|---|
| Nguyen et al. [24] | Strawberry | MT-UNet (VGG16 backbone) | Gray Mold, Powdery Mildew, Tip Burn, Healthy | 98.9% |
| Nguyen et al. [25] | Strawberry | Vision Transformer | Anthracnose Fruit Rot, Flower Blight, Gray Mold, Leaf Spot Disease, Powdery Mildew On Leaves, Powdery Mildew On Fruits | 92.7% |
| Karki et al. [26] | Strawberry | Resnet-50 | Angular Leaf Spot, Anthracnose, Gray Mold, and Powdery Mildew on Both Fruit and Leaves | 94.4% |
| Kumar et al. [27] | Strawberry | CNN-SVM | Powdery Mildew, Leaf Scorch, Leaf Blight | 95.0% |
| Vats et al. [28] | Tea | CNN | (1_V Low) 1–20%, (2_Low) 21–40%, (3_Med) 41–60%, (4 High) 61–80%, (5_V High) 81–100% | 97.0% |
| Liu et al. [29] | Apple | DeepLabV3+, PSPNet, UNet | 0 (Healthy), 1 (Mild), 2 (Moderate), 3 (Severe) | 92.8% |
| Liu et al. [30] | Wheat | MobileNetV2-DeepLabV3+ + ResNet50-DeepLabV3+ | IoU score based on health category (IoU-H) | 86.08% |
| Proposed method | Strawberry | YOLOv11-based | 0 (Healthy), 1 (0, 10%], 2 (10%, 35%], 3 (35%, 55%], 4 (55%, 100%] | 94.2% |
| Angular Leafspot | Training Images | Val Images | Test Images | Total Images |
|---|---|---|---|---|
| Diseased | 5) | 5) | 79 | 2744 |
| Healthy | 5) | 5) | 13 | 533 |
| H_max | H_min | S_max | S_min | V_max | V_min | |
|---|---|---|---|---|---|---|
| Diseased | 37 | 1 | 210 | 30 | 244 | 100 |
| Healthy | 64 | 38 | 255 | 100 | 200 | 57 |
| Name | Information |
|---|---|
| CPU | Intel® Core™ i9 14900K @6.00 GHz |
| GPU | NVIDIA GeForce RTX 4080 16G |
| Operating System | Windows 11 |
| Deep Learning Framework | Pytorch 2.5.0 |
| Programming Language | Python 3.12.7 |
| Integrated Development Environment | VScode 1.92 |
| Package Management Tools | Anaconda 2.5.2 |
| Hyperparameter | Value |
|---|---|
| Input image size | 640 × 640 |
| Batch size | 16 |
| Epoch | 200 |
| Maximum learning rate | 0.001 |
| Optimizer | AdamW |
| Weight decay | 0.0005 |
| Thread count | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.-X.; Yu, X.-H.; Yi, Q.; Zhang, Q.-Y.; Su, W.-H. Dual-Phase Severity Grading of Strawberry Angular Leaf Spot Based on Improved YOLOv11 and OpenCV. Plants 2025, 14, 1656. https://doi.org/10.3390/plants14111656
Xu Y-X, Yu X-H, Yi Q, Zhang Q-Y, Su W-H. Dual-Phase Severity Grading of Strawberry Angular Leaf Spot Based on Improved YOLOv11 and OpenCV. Plants. 2025; 14(11):1656. https://doi.org/10.3390/plants14111656
Chicago/Turabian StyleXu, Yi-Xiao, Xin-Hao Yu, Qing Yi, Qi-Yuan Zhang, and Wen-Hao Su. 2025. "Dual-Phase Severity Grading of Strawberry Angular Leaf Spot Based on Improved YOLOv11 and OpenCV" Plants 14, no. 11: 1656. https://doi.org/10.3390/plants14111656
APA StyleXu, Y.-X., Yu, X.-H., Yi, Q., Zhang, Q.-Y., & Su, W.-H. (2025). Dual-Phase Severity Grading of Strawberry Angular Leaf Spot Based on Improved YOLOv11 and OpenCV. Plants, 14(11), 1656. https://doi.org/10.3390/plants14111656







