Mapping Quantitative Trait Loci in Arabidopsis MAGIC Lines Uncovers Hormone-Responsive Genes Controlling Adventitious Root Development
Abstract
1. Introduction
2. Results
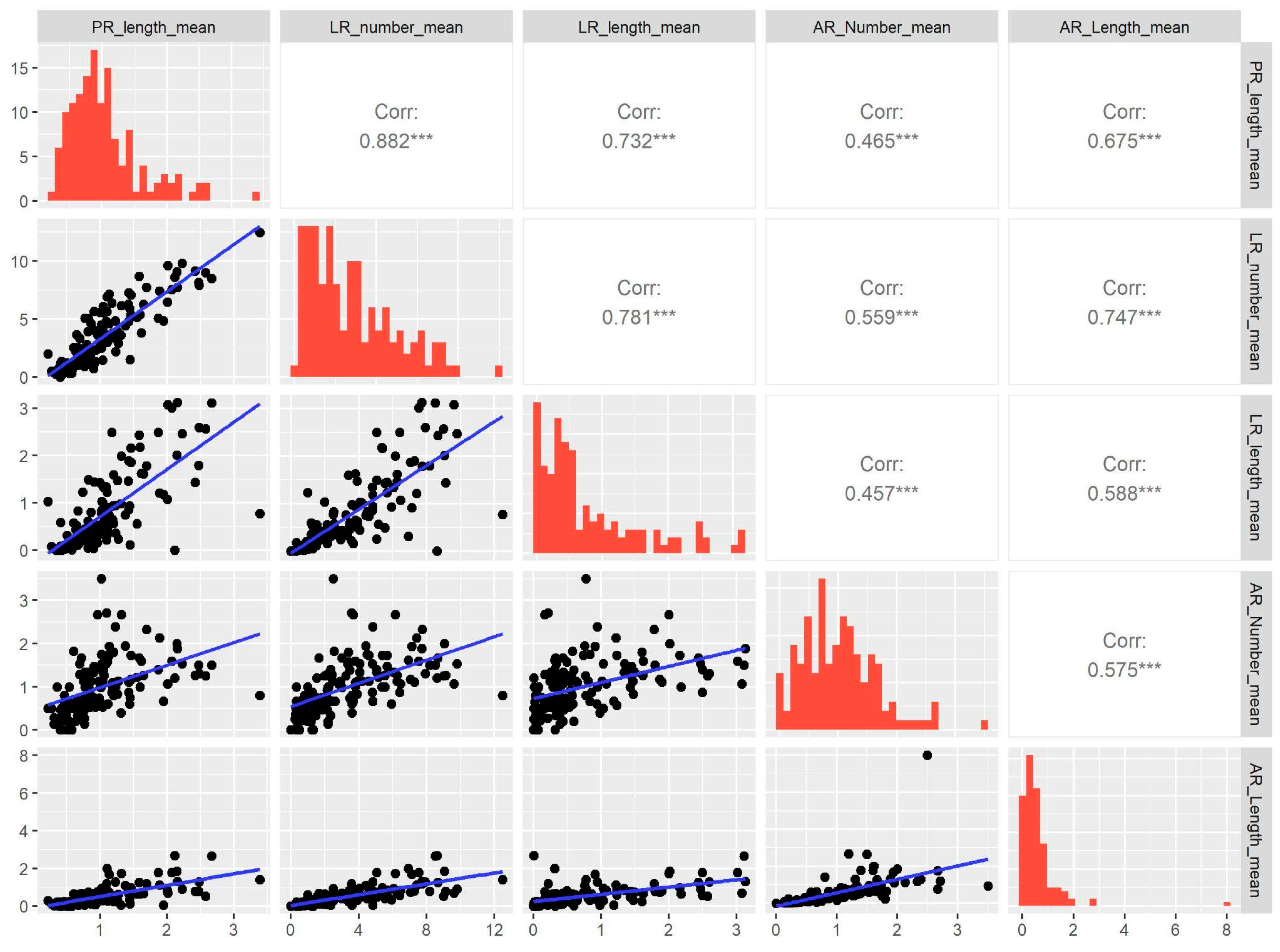
2.1. Root Variation in Arabidopsis MAGIC Lines
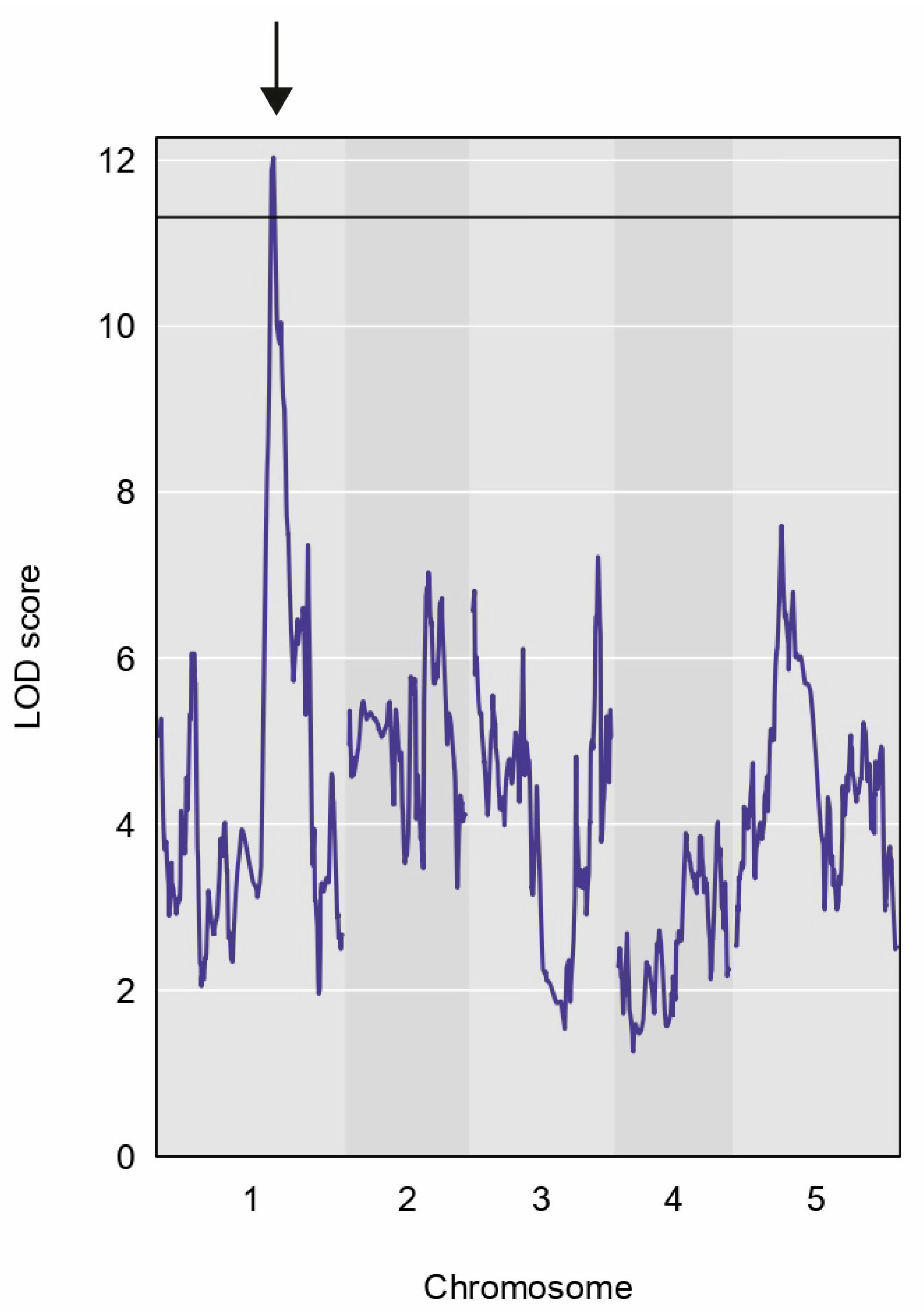
2.2. QTLs Accounted for Adventitious Root Number and Candidate Genes
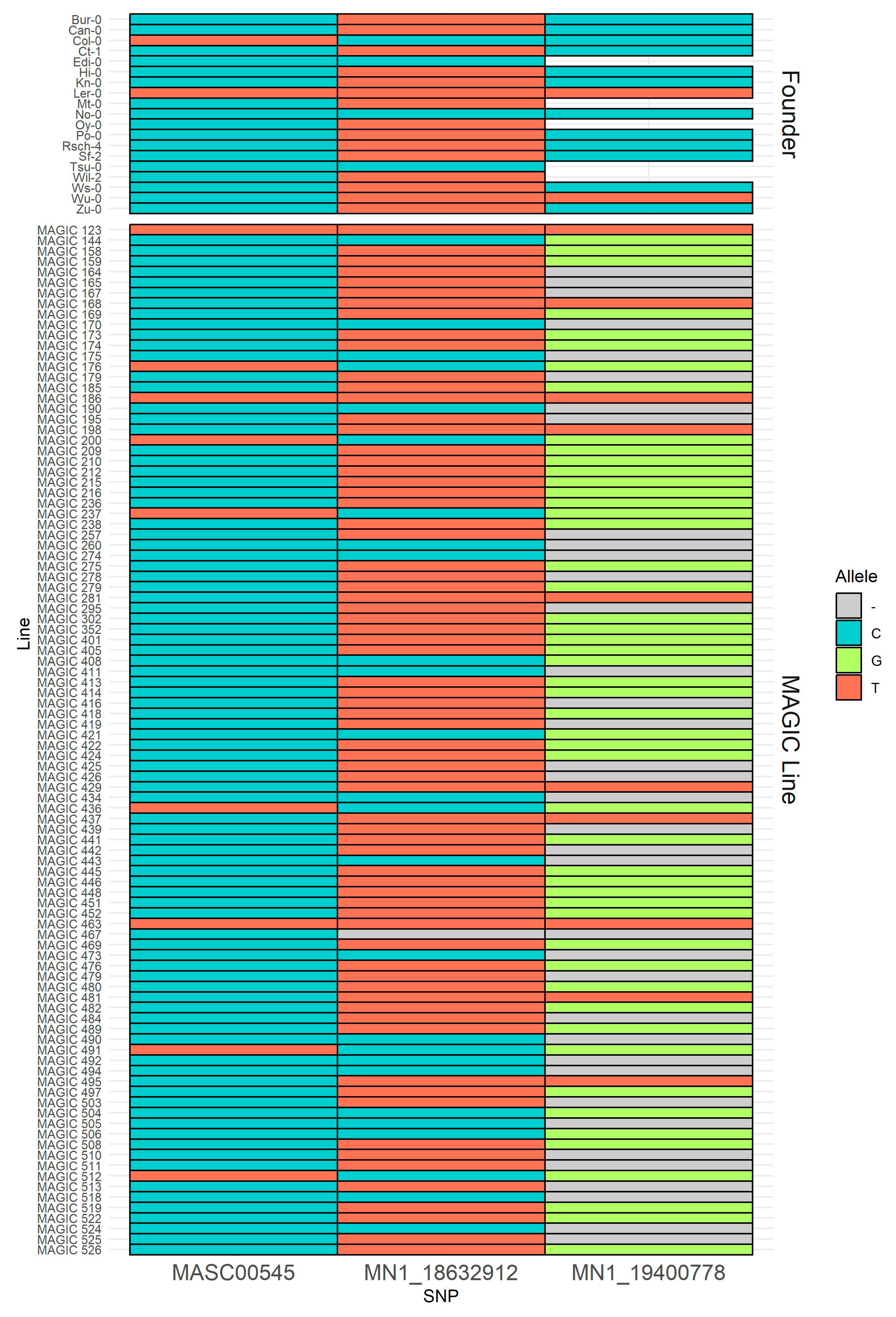
2.3. Allelic Variants in Five Candidate Genes of the Founder Accessions
3. Discussion
4. Materials and Methods
4.1. Growth Conditions and Phenotyping
4.2. Data Visualization and Statistical Analysis
4.3. QTL Mapping
4.4. Search for Candidate Genes in the QTL Peak in Chromosome 1
4.5. Polymorphism Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis Root Development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Benfey, P.N.; Schiefelbein, J.W. Getting to the Root of Plant Development: The Genetics of Arabidopsis Root Formation. Trends Genet. 1994, 10, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, S.; Sarkel, E.; Kleine-Vehn, J. Same Same, but Different: Growth Responses of Primary and Lateral Roots. J. Exp. Bot. 2020, 71, 2397–2411. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Fang, X.; Liu, W.; Sheng, L.; Xu, L. Adventitious Lateral Rooting: The Plasticity of Root System Architecture. Physiol. Plant. 2019, 165, 39–43. [Google Scholar] [CrossRef]
- De Smet, I.; Lau, S.; Mayer, U.; Jürgens, G. Embryogenesis—The Humble Beginnings of Plant Life. Plant J. 2010, 61, 959–970. [Google Scholar] [CrossRef]
- Dubrovsky, J.G.; Doerner, P.W.; Colón-Carmona, A.; Rost, T.L. Pericycle Cell Proliferation and Lateral Root Initiation in Arabidopsis. Plant Physiol. 2000, 124, 1648–1657. [Google Scholar] [CrossRef]
- Xu, L. De Novo Root Regeneration from Leaf Explants: Wounding, Auxin, and Cell Fate Transition. Curr. Opin. Plant Biol. 2018, 41, 39–45. [Google Scholar] [CrossRef]
- Zluhan-Martínez, E.; López-Ruíz, B.A.; García-Gómez, M.L.; García-Ponce, B.; de la Paz Sánchez, M.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Integrative Roles of Phytohormones on Cell Proliferation, Elongation and Differentiation in the Arabidopsis Thaliana Primary Root. Front. Plant Sci. 2021, 12, 659155. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Zhao, D.; Tang, Z.; Zhang, T.; Zhang, K.; Dong, J.; Zhang, H. Genetic Regulation of Lateral Root Development. Plant Signal. Behav. 2023, 18, 2081397. [Google Scholar] [CrossRef]
- Mhimdi, M.; Pérez-Pérez, J.M. Understanding of Adventitious Root Formation: What Can We Learn From Comparative Genetics? Front. Plant Sci. 2020, 11, 582020. [Google Scholar] [CrossRef]
- López-Ruiz, B.A.; Quezada-Rodríguez, E.H.; Piñeyro-Nelson, A.; Tovar, H.; García-Ponce, B.; Sánchez, M.D.L.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Combined Approach of GWAS and Phylogenetic Analyses to Identify New Candidate Genes That Participate in Arabidopsis Thaliana Primary Root Development Using Cellular Measurements and Primary Root Length. Plants 2022, 11, 3162. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef] [PubMed]
- Kover, P.X.; Mott, R. Mapping the Genetic Basis of Ecologically and Evolutionarily Relevant Traits in Arabidopsis Thaliana. Curr. Opin. Plant Biol. 2012, 15, 212–217. [Google Scholar] [CrossRef]
- Scott, M.F.; Ladejobi, O.; Amer, S.; Bentley, A.R.; Biernaskie, J.; Boden, S.A.; Clark, M.; Dell’Acqua, M.; Dixon, L.E.; Filippi, C.V.; et al. Multi-Parent Populations in Crops: A Toolbox Integrating Genomics and Genetic Mapping with Breeding. Heredity 2020, 125, 396–416. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.L.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.-W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-Parent Advanced Generation Inter-Cross (MAGIC) Populations in Rice: Progress and Potential for Genetics Research and Breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef]
- Kover, P.X.; Valdar, W.; Trakalo, J.; Scarcelli, N.; Ehrenreich, I.M.; Purugganan, M.D.; Durrant, C.; Mott, R. A Multiparent Advanced Generation Inter-Cross to Fine-Map Quantitative Traits in Arabidopsis Thaliana. PLoS Genet. 2009, 5, e1000551. [Google Scholar] [CrossRef]
- Gnan, S.; Priest, A.; Kover, P.X. The Genetic Basis of Natural Variation in Seed Size and Seed Number and Their Trade-Off Using Arabidopsis Thaliana MAGIC Lines. Genetics 2014, 198, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Yang, S.; Wu, C.; Shao, Q.; Feng, X. The Genetic Control of Leaf and Petal Allometric Variations in Arabidopsis Thaliana. BMC Plant Biol. 2020, 20, 547. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Fang, X.; Tran, S.; Zhai, N.; Yang, Z.; Guo, F.; Chen, L.; Yu, J.; Ison, M.S.; et al. Transcriptional Landscapes of de Novo Root Regeneration from Detached Arabidopsis Leaves Revealed by Time-Lapse and Single-Cell RNA Sequencing Analyses. Plant Commun. 2022, 3, 100306. [Google Scholar] [CrossRef]
- Deng, K.; Dong, P.; Wang, W.; Feng, L.; Xiong, F.; Wang, K.; Zhang, S.; Feng, S.; Wang, B.; Zhang, J.; et al. The TOR Pathway Is Involved in Adventitious Root Formation in Arabidopsis and Potato. Front. Plant Sci. 2017, 8, 784. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Essemine, J.; Pang, X.; Chen, H.; Cai, W. Exogenous Application of Abscisic Acid to Shoots Promotes Primary Root Cell Division and Elongation. Plant Sci. 2020, 292, 110385. [Google Scholar] [CrossRef] [PubMed]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Sousa Barros, J.A.; Gude, I.; Perianez-Rodriguez, J.; Micol, J.L.; Del Pozo, J.C.; Moreno-Risueno, M.A.; Pérez-Pérez, J.M. Regulation of Hormonal Control, Cell Reprogramming, and Patterning during De Novo Root Organogenesis. Plant Physiol. 2018, 176, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
- Gonin, M.; Bergougnoux, V.; Nguyen, T.D.; Gantet, P.; Champion, A. What Makes Adventitious Roots? Plants 2019, 8, 240. [Google Scholar] [CrossRef]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA Proteins as Dose-Dependent Master Regulators of Arabidopsis Root Development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins Inhibit Adventitious Rooting in Hybrid Aspen and Arabidopsis by Affecting Auxin Transport. Plant J. 2014, 78, 372–384. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.D.; Amasino, R.M. Overexpression of a Novel Class of Gibberellin 2-Oxidases Decreases Gibberellin Levels and Creates Dwarf Plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef]
- King, J.J.; Stimart, D.P. Genetic Analysis of Variation for Auxin-Induced Adventitious Root Formation among Eighteen Ecotypes of Arabidopsis Thaliana L. Heynh. J. Hered. 1998, 89, 481–487. [Google Scholar] [CrossRef]
- Deja-Muylle, A.; Parizot, B.; Motte, H.; Beeckman, T. Exploiting Natural Variation in Root System Architecture via Genome-Wide Association Studies. J. Exp. Bot. 2020, 71, 2379–2389. [Google Scholar] [CrossRef]
- Wintermans, P.C.A.; Bakker, P.A.H.M.; Pieterse, C.M.J. Natural Genetic Variation in Arabidopsis for Responsiveness to Plant Growth-Promoting Rhizobacteria. Plant Mol. Biol. 2016, 90, 623–634. [Google Scholar] [CrossRef]
- Esau, K. Anatomy of Seed Plants, 2nd ed.; Wiley: New York, NY, USA, 1977; ISBN 978-0-471-24520-9. [Google Scholar]
- Falasca, G.; Altamura, M.M. Histological Analysis of Adventitious Rooting in Arabidopsis Thaliana (L.) Heynh Seedlings. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2003, 137, 265–273. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Welander, M.; Geier, T.; Smolka, A.; Ahlman, A.; Fan, J.; Zhu, L. Origin, Timing, and Gene Expression Profile of Adventitious Rooting in Arabidopsis Hypocotyls and Stems. Am. J. Bot. 2014, 101, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Della Rovere, F.; Fattorini, L.; Ronzan, M.; Falasca, G.; Altamura, M.M. The Quiescent Center and the Stem Cell Niche in the Adventitious Roots of Arabidopsis Thaliana. Plant Signal. Behav. 2016, 11, e1176660. [Google Scholar] [CrossRef]
- Ibáñez, S.; Ruiz-Cano, H.; Fernández, M.Á.; Sánchez-García, A.B.; Villanova, J.; Micol, J.L.; Pérez-Pérez, J.M. A Network-Guided Genetic Approach to Identify Novel Regulators of Adventitious Root Formation in Arabidopsis Thaliana. Front. Plant Sci. 2019, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sheen, J. The Role of Target of Rapamycin Signaling Networks in Plant Growth and Metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Santuari, L.; Sanchez-Perez, G.F.; Luijten, M.; Rutjens, B.; Terpstra, I.; Berke, L.; Gorte, M.; Prasad, K.; Bao, D.; Timmermans-Hereijgers, J.L.P.M.; et al. The PLETHORA Gene Regulatory Network Guides Growth and Cell Differentiation in Arabidopsis Roots. Plant Cell 2016, 28, 2937–2951. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W.; et al. 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis Thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]


| Gene | Founder Accessions | Chr: bp | Alleles | Class | Conseq. Type | AA | |
|---|---|---|---|---|---|---|---|
| At1G50030 TOR | Rsch-4 | 1:18523147-18523149 | TCT/- | Deletion | Inframe deletion | - | ED/D |
| Kn-0 | 1:18539595 | C/T | SNP | Missense variant | Tolerated | V/M | |
| No-0 | 1:18539612 | G/T | SNP | Missense variant | Tolerated | T/N | |
| At1g50960 GA2ox7 | Can-0, Hi-0, Kn-0, No-0, Rsch4, Sf-2 | 1:18889623 | G/T | SNP | Missense variant | Tolerated | E/D |
| Can-0, Sf-2 | 1:18889660 | A/T | SNP | Missense variant | Tolerated | I/L | |
| Hi-0 | 1:18889744 | G/A | SNP | Missense variant | Deleterious | A/T | |
| Oy-0 | 1:18889756 | T/A | SNP | Missense variant | Deleterious | W/R | |
| Oy-0 | 1:18889890 | T/A | SNP | Missense variant | Tolerated | N/K | |
| Mt-0 | 1:18889922 | C/G | SNP | Missense variant | Tolerated | S/C | |
| At1G51190 PLT2 | Bur-0, Can-0, Ct-1, Edi-0, Hi-0, Kn-0, Mt-0, Oy-0, Po-0, Rsch-4, Sf-2, Tsu-0, Wil-2, Ws-0, Wu-0, and Zu-0 | 1:18978006 | A/C/T | SNP | Missense variant | Tolerated | E/P |
| Kn-0 | 1:18978071 | A/T | SNP | Missense variant | Tolerated | Q/H | |
| Can-0, Sf-0 | 1:18978257-18978259 | CAC/- | Deletion | Inframe deletion | - | ST/S | |
| Kn-0 | 1:18979010 | G/C | SNP | Missense variant | Tolerated | E/D | |
| Ct-1 | 1:18980072 | C/T/A | SNP | Missense variant | Deleterious | L/I | |
| No-0 | 1:18980238 | G/T | SNP | Missense variant | Deleterious | G/V | |
| At1G51950 IAA18 | Kn-0 | 1:19305779 | C/A | SNP | Missense variant | Tolerated | T/K |
| Mt-0 | 1:19305855 | G/A | SNP | Missense variant | Tolerated | M/I | |
| Mt-0 | 1:19305901 | T/C | SNP | Missense variant | Tolerated | Y/H | |
| Zu-0 | 1:19306234 | G/C | SNP | Missense variant | Deleterious | G/V | |
| Can-0, Hi-0, Kn-0, Ler-0, Mt-0, No-0, Wil-2, Ws-0Zu-0 | 1:19306355 | C/A | SNP | Missense variant | Tolerated | A/D | |
| AT1G49330 Hydroxyproline- rich glycoprotein family protein | Can-0, Po-0, Tsu-0, Zu-0 | 1:18250209 | C/A | SNP | Missense variant | Deleterious | S/Y |
| Po-0 | 1:18250260 | A/C | SNP | Missense variant | Deleterious | E/A | |
| No-0 | 1:18250338 | C/G/T | SNP | Missense variant | Tolerated | P/R | |
| Bur-0, Ler-0, Edi-0 | 1:18250388 | C/T | SNP | Missense variant | Tolerated | P/S | |
| No-0 | 1:18250417 | C/A | SNP | Missense variant | Tolerated | F/L | |
| Edi-0, Oy-0 | 1:18250605 | A/C | SNP | Missense variant | Tolerated | E/A | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ruiz, B.A.; Banta, J.; Salazar-Hernández, P.; Espinoza-Gutiérrez, D.; Alfaro-Mendoza, A.; Rosas, U. Mapping Quantitative Trait Loci in Arabidopsis MAGIC Lines Uncovers Hormone-Responsive Genes Controlling Adventitious Root Development. Plants 2025, 14, 1574. https://doi.org/10.3390/plants14111574
López-Ruiz BA, Banta J, Salazar-Hernández P, Espinoza-Gutiérrez D, Alfaro-Mendoza A, Rosas U. Mapping Quantitative Trait Loci in Arabidopsis MAGIC Lines Uncovers Hormone-Responsive Genes Controlling Adventitious Root Development. Plants. 2025; 14(11):1574. https://doi.org/10.3390/plants14111574
Chicago/Turabian StyleLópez-Ruiz, Brenda Anabel, Joshua Banta, Perla Salazar-Hernández, Daniela Espinoza-Gutiérrez, Andrea Alfaro-Mendoza, and Ulises Rosas. 2025. "Mapping Quantitative Trait Loci in Arabidopsis MAGIC Lines Uncovers Hormone-Responsive Genes Controlling Adventitious Root Development" Plants 14, no. 11: 1574. https://doi.org/10.3390/plants14111574
APA StyleLópez-Ruiz, B. A., Banta, J., Salazar-Hernández, P., Espinoza-Gutiérrez, D., Alfaro-Mendoza, A., & Rosas, U. (2025). Mapping Quantitative Trait Loci in Arabidopsis MAGIC Lines Uncovers Hormone-Responsive Genes Controlling Adventitious Root Development. Plants, 14(11), 1574. https://doi.org/10.3390/plants14111574







