ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis of the ZmHPAT2 Gene
2.2. Subcellular Localization of ZmHPAT2
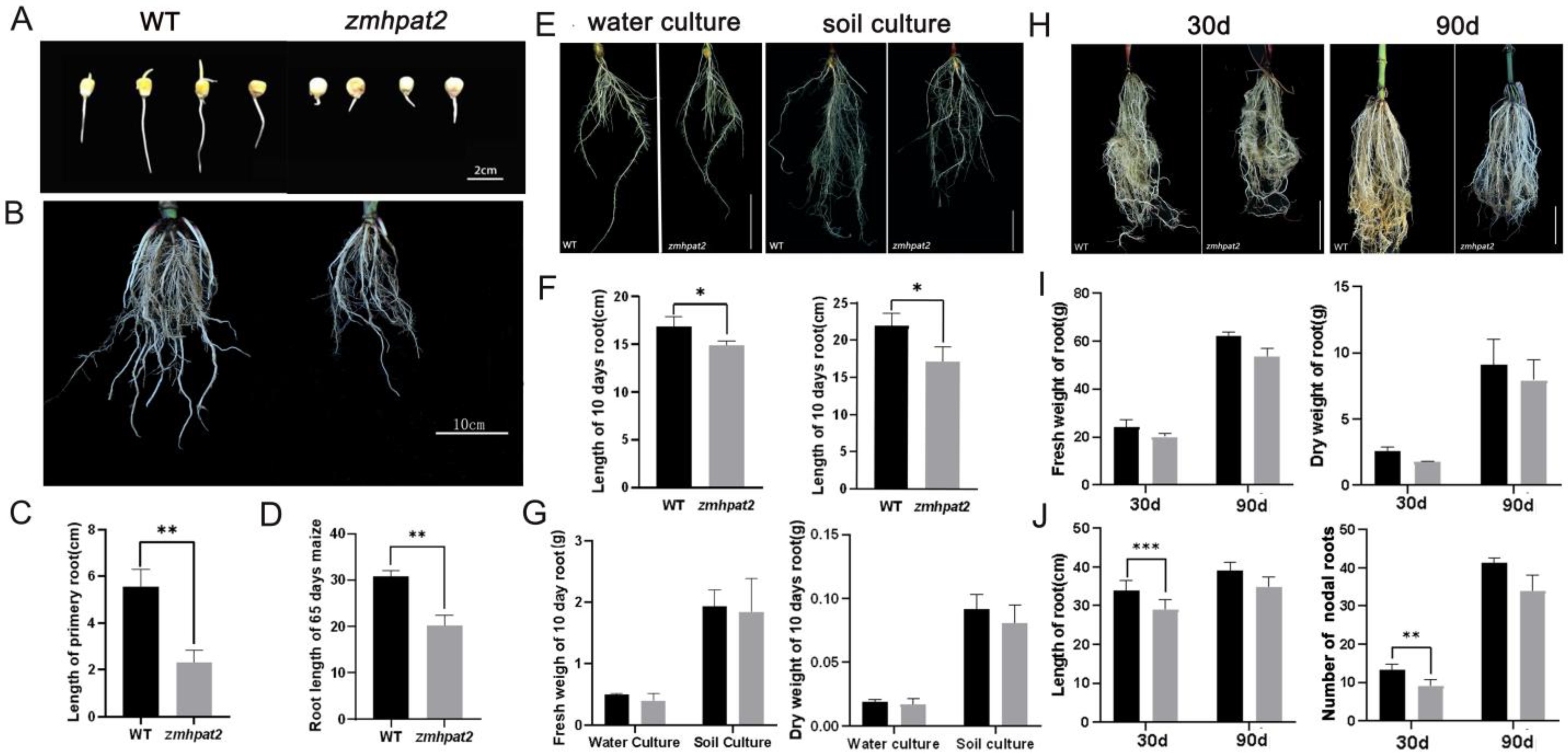
2.3. ZmHPAT2 Positively Regulates Root Development
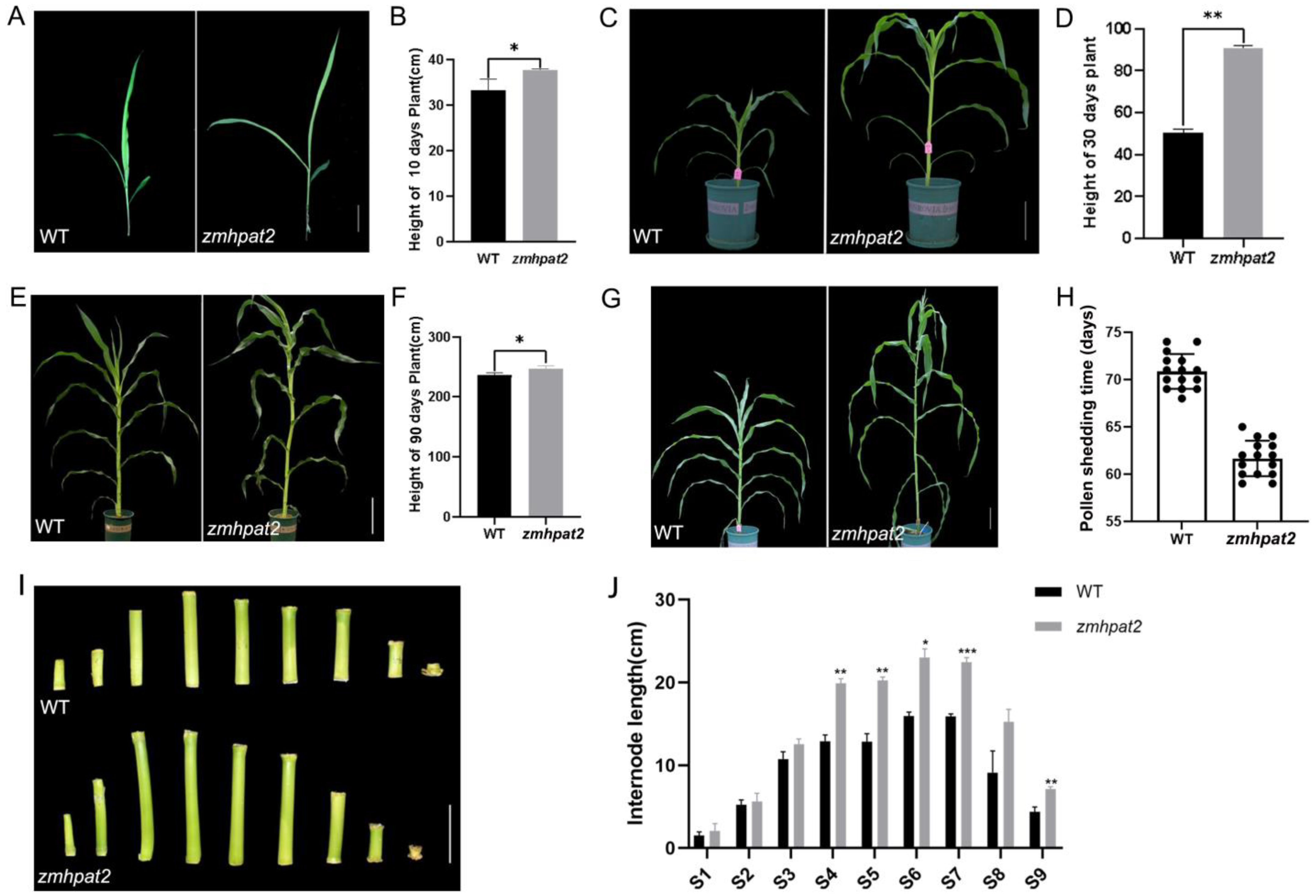
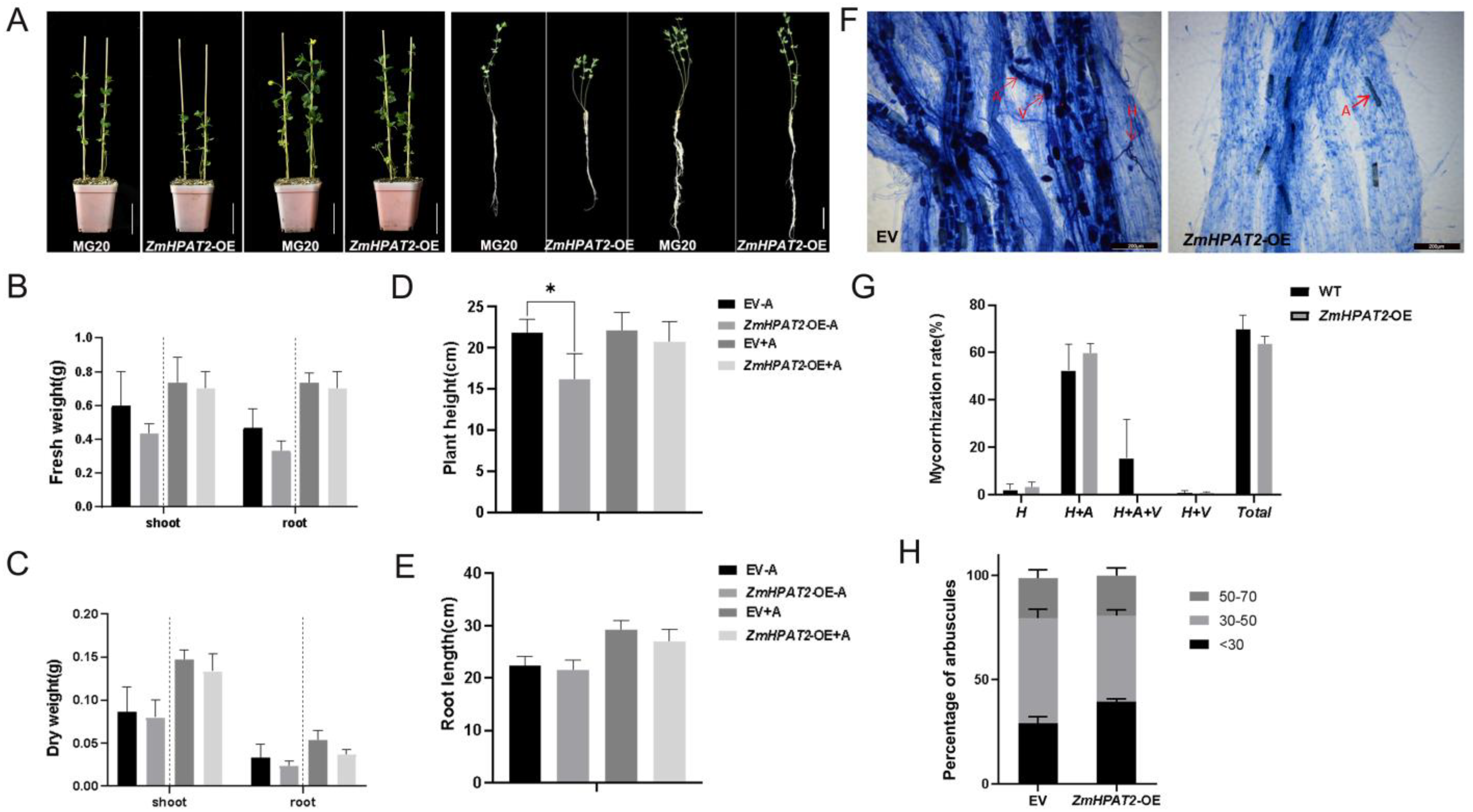
2.4. ZmHPAT2 Negatively Regulated Plant Height and Loose Powder
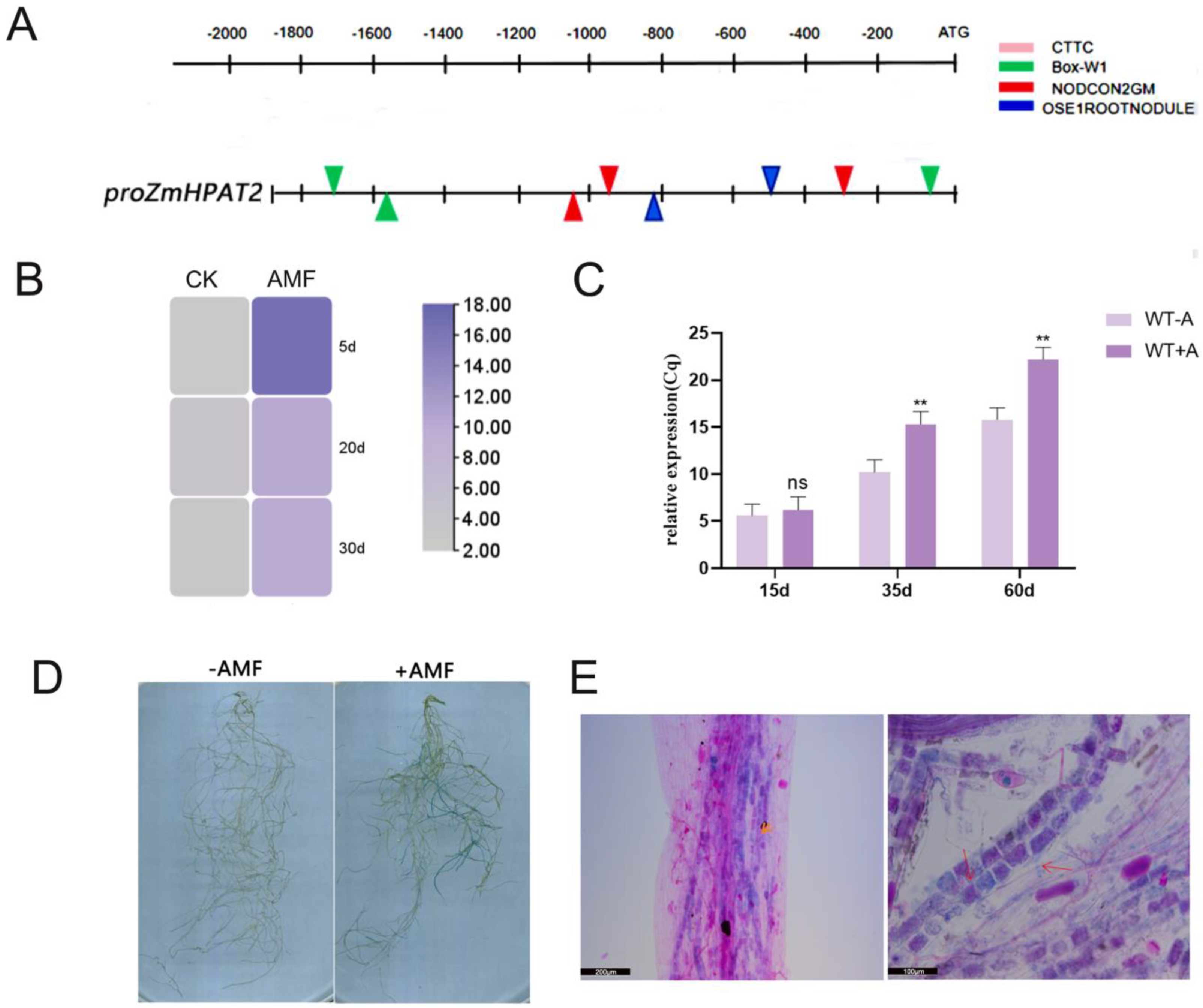
2.5. ZmHPAT2 Is Induced to Express by AM Fungi
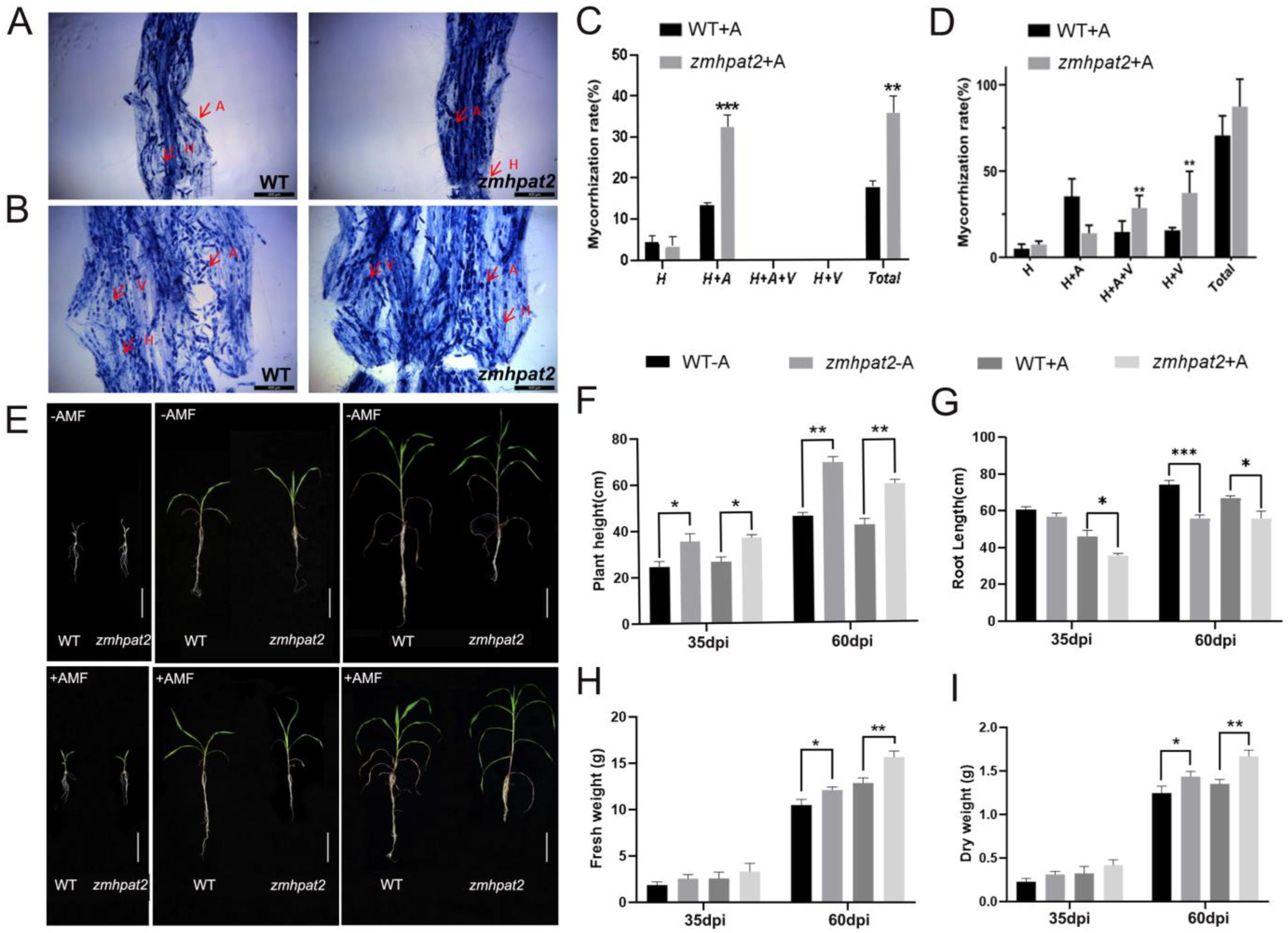
2.6. ZmHPAT2 Negatively Regulates AM Fungi Symbiosis in Maize
2.7. Overexpression of ZmHPAT2 Inhibits Plant Height and Mycorrhizal Symbiosis
2.8. ZmHPAT2 Regulates the Expression of the Auxin-Responsive Gene GH3 and the Branching Degradation-Related Gene ZmMYB1
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Culture Substrates
4.2. Plant Growth and AMF Inoculation
4.3. RNA Isolation and qRT-PCR
4.4. Bioinformatics Analysis
4.5. Subcellular Localization in Maize Protoplasts
4.6. Mycorrhizal Staining and Symbiosis Rate Determination
4.7. Induced Transformation of Hairy Roots of L. japonicus
4.8. Symbiosis Between L. japonicus and AM Fungi, and GUS Staining
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramazan, S.; Nazir, I.; Yousuf, W.; John, R. Environmental stress tolerance in maize (Zea mays): Role of polyamine metabolism. Funct. Plant Biol. FPB 2023, 50, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tan, R.; Zhao, J. Chilling Tolerance in Maize: Insights into Advances-Toward Physio-Biochemical Responses’ and QTL/Genes’ Identification. Plants 2022, 11, 2082. [Google Scholar] [CrossRef]
- Sun, Y.; Zang, H.; Splettstößer, T.; Kumar, A.; Xu, X.; Kuzyakov, Y.; Pausch, J. Plant intraspecific competition and growth stage alter carbon and nitrogen mineralization in the rhizosphere. Plant Cell Environ. 2021, 44, 1231–1242. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Yu, P.; Marcon, C. Genetic Control of Root System Development in Maize. Trends Plant Sci. 2018, 23, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ingram, P.A.; Benfey, P.N.; Elich, T. From lab to field, new approaches to phenotyping root system architecture. Curr. Opin. Plant Biol. 2011, 14, 310–317. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Bruce, W.; Desbons, P.; Crasta, O.; Folkerts, O. Gene expression profiling of two related maize inbred lines with contrasting root-lodging traits. J. Exp. Bot. 2001, 52, 459–468. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sha, Y.; Zhao, S.; Hao, Z.; Liu, Z.; Hu, W.; Feng, G.; Chen, F.; Mi, G. Root Lodging Resistance in Maize (Zea mays L.) Under Conservative Strip-Till Cultivation System. J. Agron. Crop Sci. 2025, 211, e70008. [Google Scholar] [CrossRef]
- Carlson, A.D. Proem. Q. Rev. Biol. 2016, 91, 245–246. [Google Scholar] [CrossRef]
- Rich, M.K.; Nouri, E.; Courty, P.-E.; Reinhardt, D. Diet of Arbuscular Mycorrhizal Fungi: Bread and Butter? Trends Plant Sci. 2017, 22, 652–660. [Google Scholar] [CrossRef]
- Wang, W.; Guo, W.; Le, L.; Yu, J.; Wu, Y.; Li, D.; Wang, Y.; Wang, H.; Lu, X.; Qiao, H.; et al. Integration of high-throughput phenotyping, GWAS, and predictive models reveals the genetic architecture of plant height in maize. Mol. Plant 2023, 16, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yu, Z.; Zhang, M.; Wang, M.; Lu, X.; Liu, X.; Li, Y.; Zhang, X.; Tan, B.C.; Li, C.; et al. ZmTE1 promotes plant height by regulating intercalary meristem formation and internode cell elongation in maize. Plant Biotechnol. J. 2022, 20, 526–537. [Google Scholar] [CrossRef]
- Mu, Q.; Wei, J.; Longest, H.K.; Liu, H.; Char, S.N.; Hinrichsen, J.T.; Tibbs-Cortes, L.E.; Schoenbaum, G.R.; Yang, B.; Li, X.; et al. A MYB transcription factor underlying plant height in sorghum qHT7.1 and maize Brachytic 1 loci. Plant J. Cell Mol. Biol. 2024, 120, 2172–2192. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.-M.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Pingali, P.L. Green Revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef]
- Ogawa-Ohnishi, M.; Matsushita, W.; Matsubayashi, Y. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 2013, 9, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.; Miller, D.H. Hydroxyproline arabinosides in the plant kingdom. Plant Physiol. 1971, 48, 454–456. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Ortiz-Ramirez, C.; Becker, J.D.; Feijo, J.A.; Lippman, Z.B. Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens. Plant J. 2016, 85, 193–208. [Google Scholar] [CrossRef]
- Isidra-Arellano, M.C.; Pozas-Rodríguez, E.A.; Del Rocío Reyero-Saavedra, M.; Arroyo-Canales, J.; Ferrer-Orgaz, S.; Del Socorro Sánchez-Correa, M.; Cardenas, L.; Covarrubias, A.A.; Valdés-López, O. Inhibition of legume nodulation by Pi deficiency is dependent on the autoregulation of nodulation (AON) pathway. Plant J. Cell Mol. Biol. 2020, 103, 1125–1139. [Google Scholar] [CrossRef]
- Yoro, E.; Nishida, H.; Ogawa-Ohnishi, M.; Yoshida, C.; Suzaki, T.; Matsubayashi, Y.; Kawaguchi, M. PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus. J. Exp. Bot. 2019, 70, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Kassaw, T.; Nowak, S.; Schnabel, E.; Frugoli, J. ROOT DETERMINED NODULATION1 Is Required for M. truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol. 2017, 174, 2445–2456. [Google Scholar] [CrossRef] [PubMed]
- Jaffri, S.R.F.; Scheer, H.; MacAlister, C.A. The hydroxyproline O-arabinosyltransferase FIN4 is required for tomato pollen intine development. Plant Reprod. 2023, 36, 173–191. [Google Scholar] [CrossRef]
- Schnabel, E.L.; Kassaw, T.K.; Smith, L.S.; Marsh, J.F.; Oldroyd, G.E.; Long, S.R.; Frugoli, J.A. The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 2011, 157, 328–340. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, W.; Wu, J.; Xie, K.; Li, X. LjAMT2;2 Promotes Ammonium Nitrogen Transport during Arbuscular Mycorrhizal Fungi Symbiosis in Lotus japonicus. Int. J. Mol. Sci. 2022, 23, 9522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liao, J.; Wu, J.; Huang, H.; Yuan, Z.; Yang, W.; Wu, X.; Li, X. Genome-Wide Identification and Characterization of the Soybean DEAD-Box Gene Family and Expression Response to Rhizobia. Int. J. Mol. Sci. 2022, 23, 1120. [Google Scholar] [CrossRef]
- Chen, W.; Ye, T.; Sun, Q.; Niu, T.; Zhang, J. Arbuscular Mycorrhizal Fungus Alters Root System Architecture in Camellia sinensis L. as Revealed by RNA-Seq Analysis. Front. Plant Sci. 2021, 12, 777357. [Google Scholar] [CrossRef]
- Stevens, K.J.; Wall, C.B.; Janssen, J.A. Effects of arbuscular mycorrhizal fungi on seedling growth and development of two wetland plants, Bidens frondosa L., and Eclipta prostrata (L.) L., grown under three levels of water availability. Mycorrhiza 2011, 21, 279–288. [Google Scholar] [CrossRef]
- Wang, C.; Reid, J.B.; Foo, E. The role of CLV1, CLV2 and HPAT homologues in the nitrogen-regulation of root development. Physiol. Plant 2020, 170, 607–621. [Google Scholar] [CrossRef]
- Liao, D.; Wang, S.; Cui, M.; Liu, J.; Chen, A.; Xu, G. Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis. Int. J. Mol. Sci. 2018, 19, 3146. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Pajic, Z.; Radosavljevic, M.; Filipovic, M.; Todorovic, G.; Srdic, J.; Pavlov, M. Breeding of speciality maize for industrial purposes. Genetika 2010, 42, 57–66. [Google Scholar] [CrossRef]
- Fatima, S.; Zeb, S.Z.; Tariq, M.; Nishat, Y.; Mohamed, H.I.; Siddiqui, M.A. Role of CLE peptide signaling in root-knot nematode parasitism of plants. Planta 2025, 261, 3. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.L.; Ishida, T.; Sawa, S. CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 2016, 67, 4813–4826. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod Factor/Nitrate-Induced CLE Genes that Drive HAR1-Mediated Systemic Regulation of Nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef]
- Miyazawa, H.; Oka-Kira, E.; Sato, N.; Takahashi, H.; Wu, G.-J.; Sato, S.; Hayashi, M.; Betsuyaku, S.; Nakazono, M.; Tabata, S.; et al. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 2010, 137, 4317–4325. [Google Scholar] [CrossRef]
- Oka-Kira, E.; Tateno, K.; Miura, K.; Haga, T.; Hayashi, M.; Harada, K.; Sato, S.; Tabata, S.; Shikazono, N.; Tanaka, A.; et al. klavier (klv), A novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 2005, 44, 505–515. [Google Scholar] [CrossRef]
- Krusell, L.; Sato, N.; Fukuhara, I.; Koch, B.E.V.; Grossmann, C.; Okamoto, S.; Oka-Kira, E.; Otsubo, Y.; Aubert, G.; Nakagawa, T.; et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011, 65, 861–871. [Google Scholar] [CrossRef]
- Schnabel, E.; Journet, E.P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef]
- Reid, D.E.; Li, D.; Ferguson, B.J.; Gresshoff, P.M. Structure-function analysis of the GmRIC1 signal peptide and CLE domain required for nodulation control in soybean. J. Exp. Bot. 2013, 64, 1575–1585. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Gresshoff, P.M. Inoculation- and Nitrate-Induced CLE Peptides of Soybean Control NARK-Dependent Nodule Formation. Mol. Plant-Microbe Interact. 2011, 24, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’Haeseleer, K.; Holsters, M.; Goormachtig, S. CLE Peptides Control Medicago truncatula Nodulation Locally and Systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Lodwig, E.M.; Hosie, A.H.F.; Bordès, A.; Findlay, K.; Allaway, D.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Amino-acid cycling drives nitrogen fixation in the legume: Rhizobium symbiosis. Nature 2003, 422, 722–726. [Google Scholar] [CrossRef]
- Mueller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE-SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Le Marquer, M.; Becard, G.; Frey, N.F.D. Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol. 2019, 222, 1030–1042. [Google Scholar] [CrossRef]
- Racolta, A.; Nodine, M.D.; Davies, K.; Lee, C.; Rowe, S.; Velazco, Y.; Wellington, R.; Tax, F.E. A Common Pathway of Root Growth Control and Response to CLE Peptides Through Two Receptor Kinases in Arabidopsis. Genetics 2018, 208, 687–704. [Google Scholar] [CrossRef]
- Wei, L.; Zhang, X.; Zhang, Z.; Liu, H.; Lin, Z. A new allele of the Brachytic2 gene in maize can efficiently modify plant architecture. Heredity 2018, 121, 75–86. [Google Scholar] [CrossRef]
- Teng, F.; Zhai, L.; Liu, R.; Bai, W.; Wang, L.; Huo, D.; Tao, Y.; Zheng, Y.; Zhang, Z. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 2013, 73, 405–416. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, B.-C. New insight in the Gibberellin biosynthesis and signal transduction. Plant Signal. Behav. 2015, 10, e1000140. [Google Scholar] [CrossRef][Green Version]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Toriba, T.; Fujimoto, M.; Tsutsumi, N.; Kitano, H.; Hirano, H.-Y. Conservation and diversification of meristem maintenance mechanism in Oryza sativa Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006, 47, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Yoshida, A.; Hirano, H.-Y. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 2008, 20, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Ohneda, M.; Toriba, T.; Yoshida, A.; Hirano, H.-Y. FON2 SPARE1 Redundantly Regulates Floral Meristem Maintenance with FLORAL ORGAN NUMBER2 in Rice. PLoS Genet. 2009, 5, e1000693. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Sun, M.; Li, H.; Li, Y.; Xiang, H.; Liu, Y.; He, Y.; Qi, M.; Li, T. Tomato YABBY2b controls plant height through regulating indole-3-acetic acid amidosynthetase GH3.8 expression. Plant Sci. 2020, 297, 110530. [Google Scholar] [CrossRef]
- Liao, D.; Chen, X.; Chen, A.; Wang, H.; Liu, J.; Liu, J.; Gu, M.; Sun, S.; Xu, G. The Characterization of Six Auxin-Induced Tomato GH3 Genes Uncovers a Member, SlGH3.4, Strongly Responsive to Arbuscular Mycorrhizal Symbiosis. Plant Cell Physiol. 2015, 56, 674–687. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, K.; Wang, G.; Ni, Y.; Shi, M.; Sun, L.; Cheng, B.; Li, X. ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis. Plants 2025, 14, 1438. https://doi.org/10.3390/plants14101438
Xie K, Wang G, Ni Y, Shi M, Sun L, Cheng B, Li X. ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis. Plants. 2025; 14(10):1438. https://doi.org/10.3390/plants14101438
Chicago/Turabian StyleXie, Kailing, Guoqing Wang, Ying Ni, Minghui Shi, Lixue Sun, Beijiu Cheng, and Xiaoyu Li. 2025. "ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis" Plants 14, no. 10: 1438. https://doi.org/10.3390/plants14101438
APA StyleXie, K., Wang, G., Ni, Y., Shi, M., Sun, L., Cheng, B., & Li, X. (2025). ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis. Plants, 14(10), 1438. https://doi.org/10.3390/plants14101438




