Genome-Wide Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Peanut and Functional Characterization of AhOPR6 in Salt Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of Peanut OPR Genes
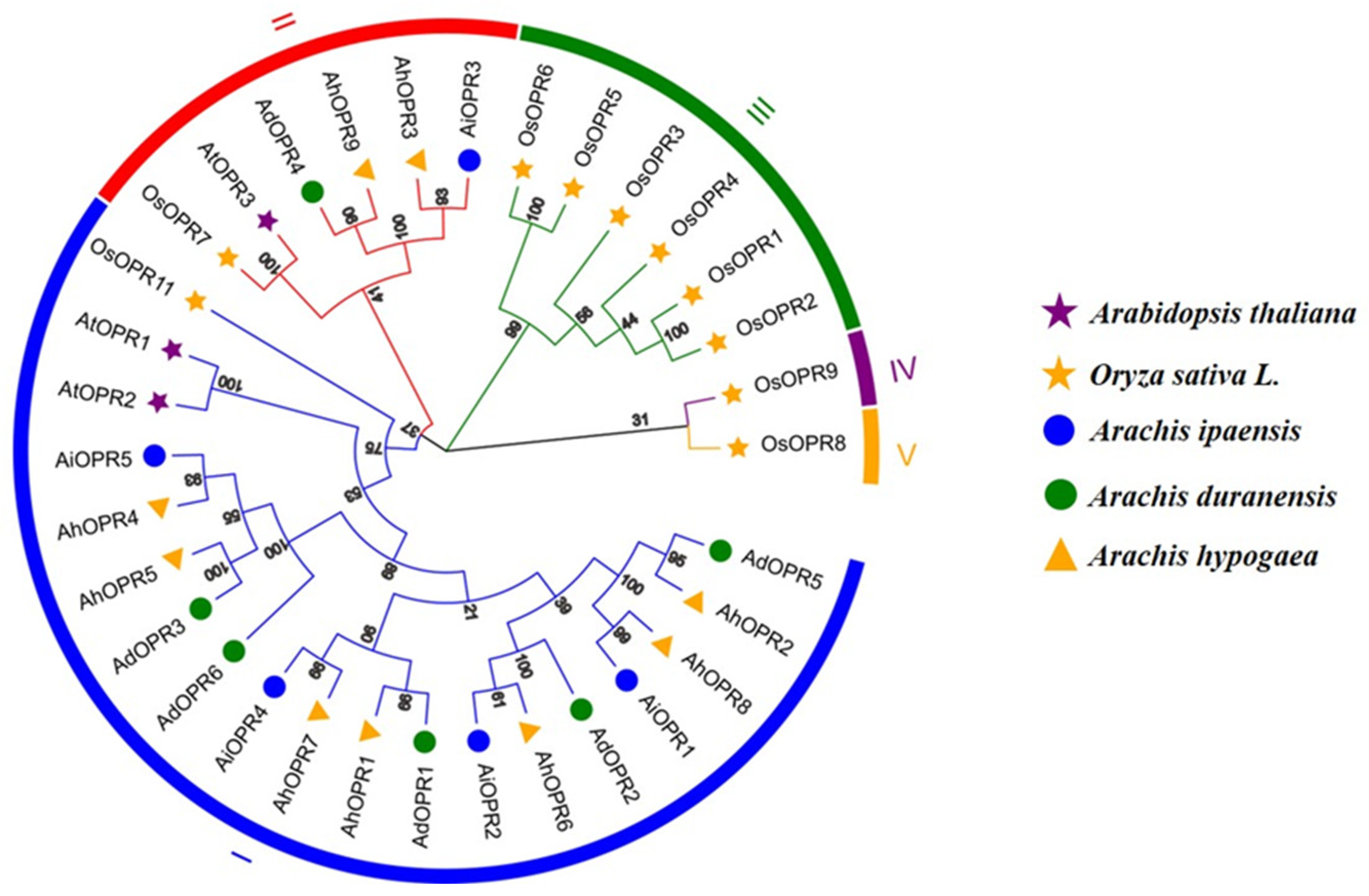
2.2. Phylogenetic Analysis and Classification of OPR Gene Family
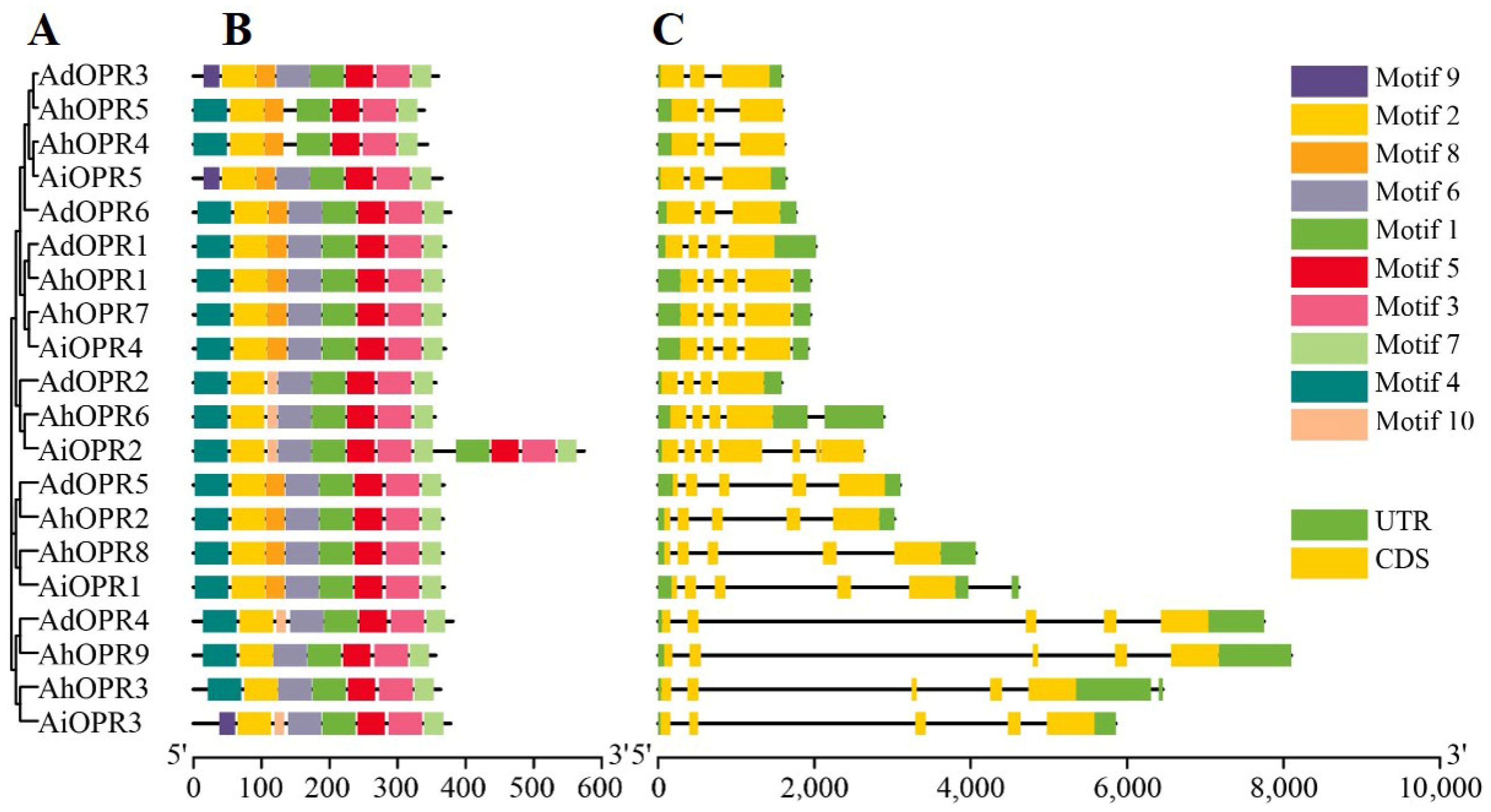
2.3. Conserved Motifs and Gene Structure Analysis of Peanut OPR Genes
2.4. Chromosomal Locations and Synteny Analysis of Peanut OPRs
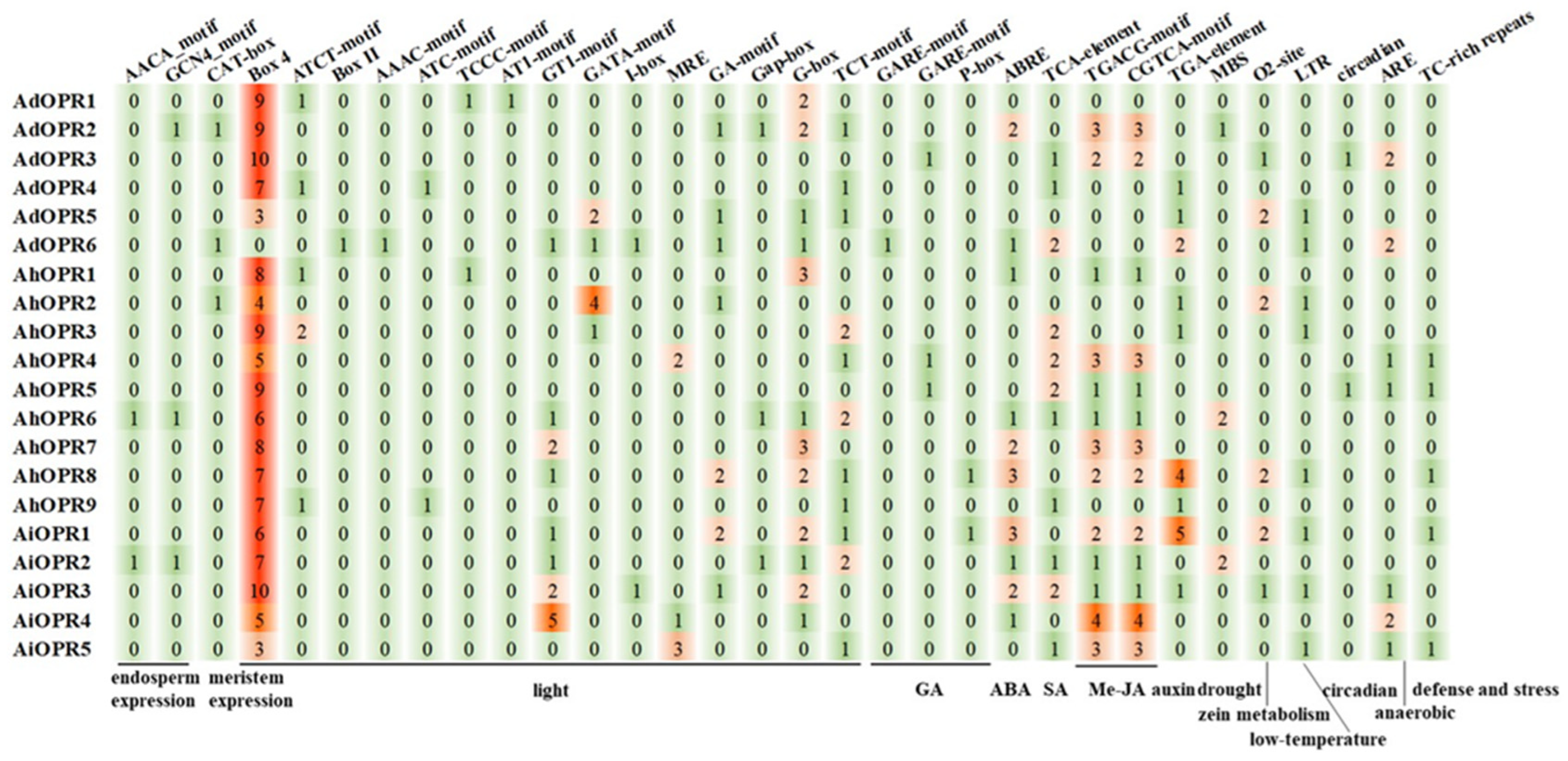
2.5. Prediction and Analysis of Cis-Acting Elements in Promoter Sequences of OPR Genes
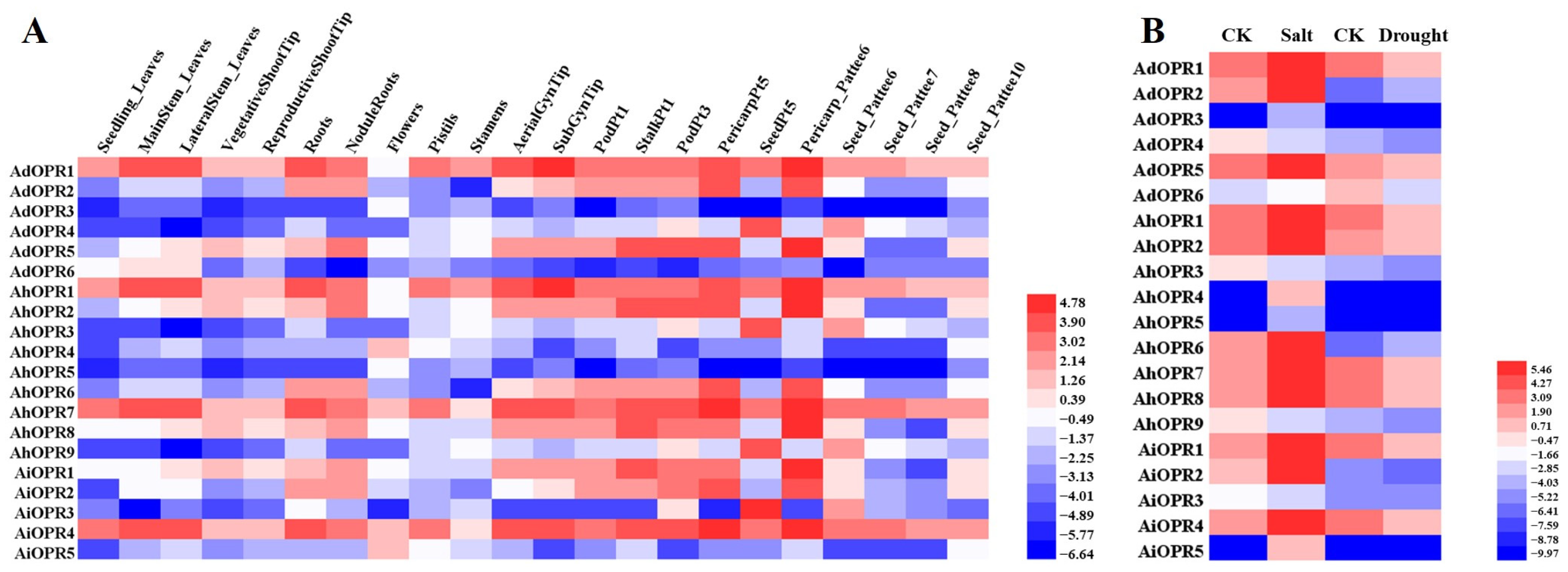
2.6. Expression Patterns of OPR Genes in Different Tissues and in Response to Various Stresses
2.7. Subcellular Location of Peanut OPR Proteins
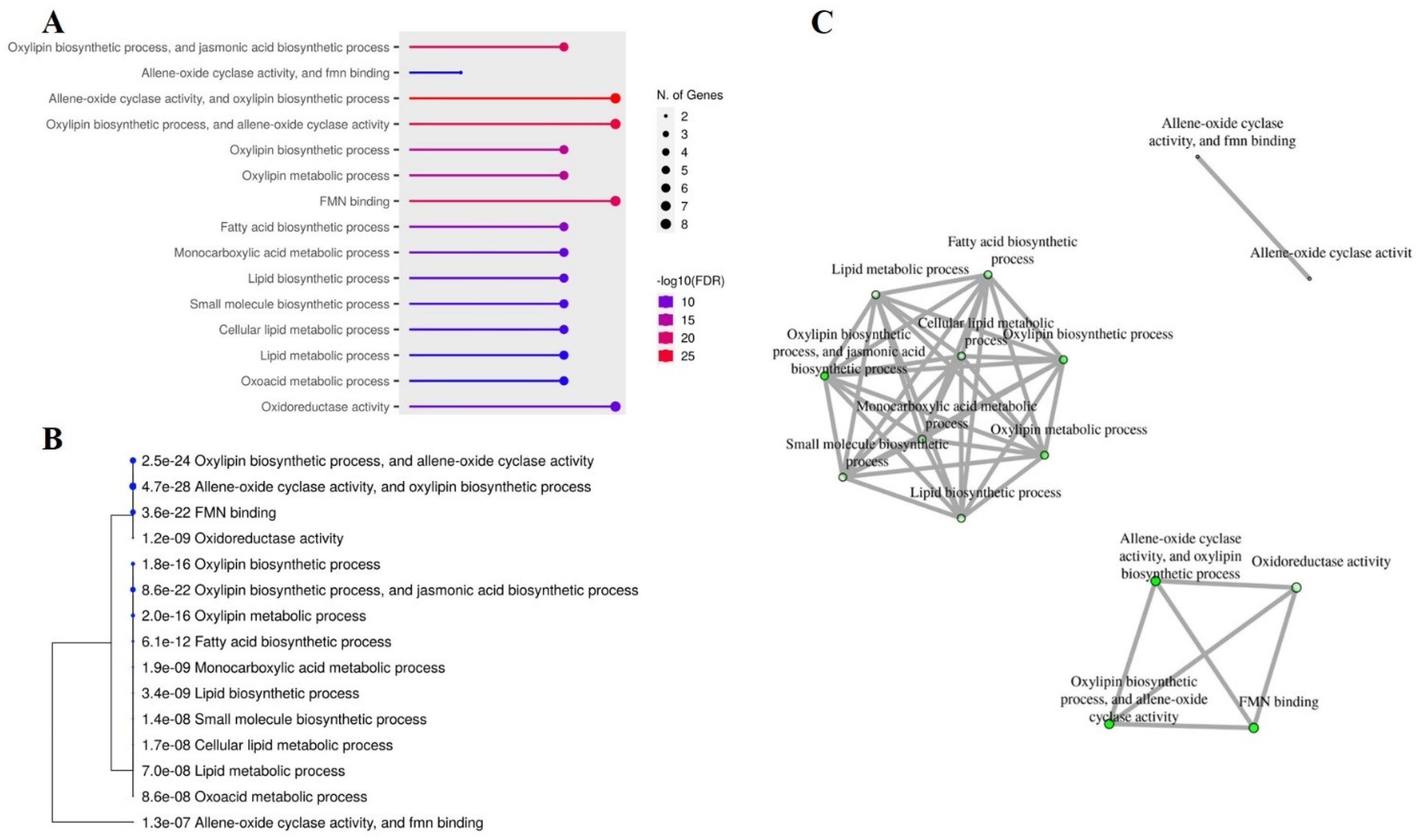
2.8. Gene Ontology (GO) and Interactive Networks of Peanut OPR Genes
2.9. Overexpression of AhOPR6 Enhanced Salt Tolerance in Seedlings
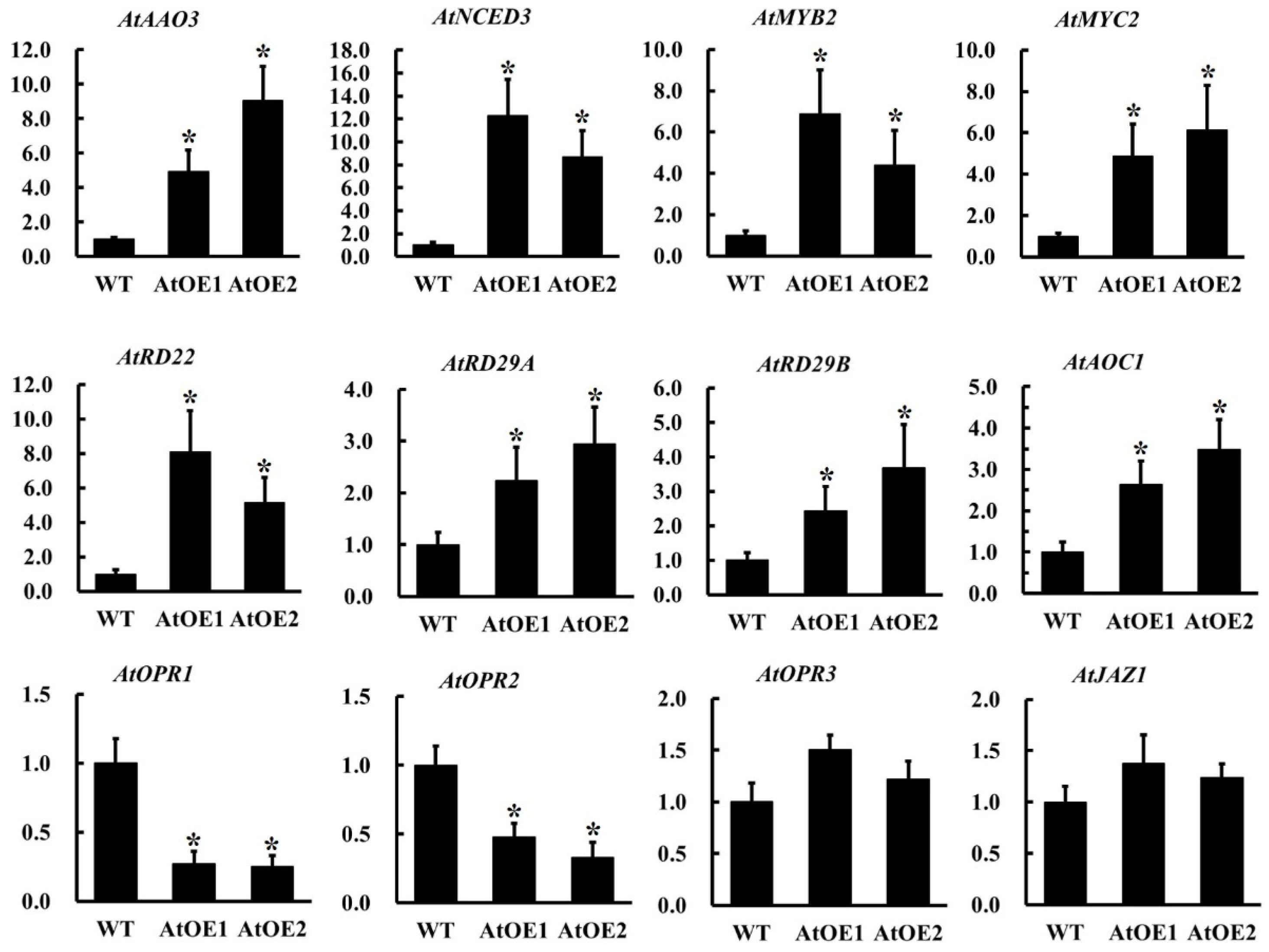
2.10. The Overexpression of AhOPR6 in Arabidopsis Affects the Expression of JA/ABA Pathway-Related Genes
3. Discussion
4. Materials and Methods
4.1. Identification of Peanut OPR Gene Family Members
4.2. Phylogenetic Analysis
4.3. Analysis of Gene Structure and Protein Conserved Domains of OPRs
4.4. Cis-Acting Regulatory Elements and Gene Ontology (GO) Annotation
4.5. Chromosome Localization and Synteny Analysis
4.6. RNA-Seq Data Analysis of OPRs
4.7. Subcellular Localization of AhOPR6 Protein
4.8. Construction of AhOPR6 Vectors and Generation of Transgenic Arabidopsis Plants
4.9. The Treatment and Determination of Salt-Related Physiological Parameters in Transgenic Arabidopsis
4.10. RNA Isolation and Real-Time Quantitative PCR (qRT-PCR) Validation of OPR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Tang, Y.; Yue, Y.; Chen, Y. Advances in the evolution research and genetic breeding of peanut. Gene 2024, 916, 148425. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Crouch, J.H.; Nigam, S.N.; Ferguson, M.E.; Paterson, A.H. Molecular breeding of groundnut for enhanced productivity and food security in the semi- arid tropics: Opportunities and challenges. Adv. Agron. 2003, 80, 153–221. [Google Scholar]
- Sarkar, T.; Thankappan, R.; Kumar, A.; Mishra, G.P.; Dobaria, J.R.; Zhang, J.S. Heterologous Expression of the AtDREB1A Gene in Transgenic Peanut-Conferred Tolerance to Drought and Salinity Stresses. PLoS ONE 2014, 9, e110507. [Google Scholar] [CrossRef] [PubMed]
- Abrol, I.P.; Yadav, J.S.P.; Massoud, F.I. Salt-Affected Soils and Their Management; Food and Agriculture Organization of the United Nations: Rome, Italy, 1988. [Google Scholar]
- Yang, S.; Hao, X.; Xu, Y.; Yang, J.; Su, D. Meta-Analysis of the Effect of Saline-Alkali Land Improvement and Utilization on Soil Organic Carbon. Life 2022, 12, 1870. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, J.; Sun, S.; Cui, F.; Han, Y.; Peng, Z.; Zhang, X.; Wan, S.; Li, G. Defining the function of SUMO system in pod development and abiotic stresses in Peanut. BMC Plant Biol. 2019, 19, 593. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153–S164. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef]
- Li, W.; Liu, B.; Yu, L.; Feng, D.; Wang, H.; Wang, J. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. Bmc Evol. Biol. 2009, 9, 90. [Google Scholar] [CrossRef]
- Breithaupt, C.; Kurzbauer, R.; Schaller, F.; Stintzi, A.; Schaller, A.; Huber, R.; Macheroux, P.; Clausen, T. Structural Basis of Substrate Specificity of Plant 12-Oxophytodienoate Reductases. J. Mol. Biol. 2009, 392, 1266–1277. [Google Scholar] [CrossRef]
- Li, W.; Zhou, F.; Liu, B.; Feng, D.; He, Y.; Qi, K.; Wang, H.; Wang, J. Comparative characterization, expression pattern and function analysis of the 12-oxo-phytodienoic acid reductase gene family in rice. Plant Cell Rep. 2011, 30, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Liu, Y.; Tian, S.; Guo, Q.; Wen, S. Genome-Wide Identification and Characterization of the OPR Gene Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2019, 20, 1914. [Google Scholar] [CrossRef] [PubMed]
- Schaller, F.; Weiler, E.W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana—Structural and functional relationship to yeast old yellow enzyme. J. Biol. Chem. 1997, 272, 28066–28072. [Google Scholar] [CrossRef]
- Schaller, F.; Biesgen, C.; Mussig, C.; Altmann, T.; Weiler, E.W. 12-oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 2000, 210, 979–984. [Google Scholar] [CrossRef]
- Biesgen, C.; Weiler, E.W. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 1999, 208, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Simmons, C.; Yalpani, N.; Crane, V.; Wilkinson, H.; Kolomiets, M. Genomic Analysis of the 12-oxo-phytodienoic Acid Reductase Gene Family of Zea mays. Plant Mol. Biol. 2005, 59, 323–343. [Google Scholar] [CrossRef]
- Scalschi, L.; Sanmartin, M.; Camanes, G.; Troncho, P.; Sanchez-Serrano, J.J.; Garcia-Agustin, P.; Vicedo, B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015, 81, 304–315. [Google Scholar] [CrossRef]
- Strassner, J.; Fürholz, A.; Macheroux, P.; Amrhein, N.; Schaller, A. A homolog of old yellow enzyme in tomato. Spectral properties and substrate specificity of the recombinant protein. J. Biol. Chem. 1999, 274, 35067–35073. [Google Scholar] [CrossRef]
- Dong, W.; Wang, M.; Xu, F.; Quan, T.; Peng, K.; Xiao, L.; Xia, G. Wheat Oxophytodienoate Reductase Gene TaOPR1 Confers Salinity Tolerance via Enhancement of Abscisic Acid Signaling and Reactive Oxygen Species Scavenging. Plant Physiol. 2013, 161, 1217–1228. [Google Scholar] [CrossRef]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-M.; Li, H.-C.; Zhou, S.-R.; Xue, H.-W.; Miao, X.-X. Cis-12-Oxo-Phytodienoic Acid Stimulates Rice Defense Response to a Piercing-Sucking Insect. Mol. Plant 2014, 7, 1683–1692. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Wright, L.P.; Gershenzon, J.; Wasternack, C.; Hause, B.; Schaller, A.; Stintzi, A. Jasmonic Acid and Its Precursor 12-Oxophytodienoic Acid Control Different Aspects of Constitutive and Induced Herbivore Defenses in Tomato. Plant Physiol. 2014, 166, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, OPR3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef]
- Li, S.; Ma, J.; Liu, P. OPR3 is expressed in phloem cells and is vital for lateral root development in Arabidopsis. Can. J. Plant Sci. 2013, 93, 165–170. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, G.; Yuan, S.; Duan, W.; Wang, P.; Bai, J.; Zhang, F.; Gao, S.; Zhang, L.; Zhao, C. TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.). Biochem. Biophys. Res. Commun. 2016, 470, 233–238. [Google Scholar] [CrossRef]
- Ren, W.; Chen, L.; Xie, Z.M.; Peng, X. Combined transcriptome and metabolome analysis revealed pathways involved in improved salt tolerance of Gossypium hirsutum L. seedlings in response to exogenous melatonin application. BMC Plant Biol. 2022, 22, 552. [Google Scholar] [CrossRef]
- Tani, T.; Sobajima, H.; Okada, K.; Chujo, T.; Arimura, S.I.; Tsutsumi, N.; Nishimura, M.; Seto, H.; Nojiri, H.; Yamane, H. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 2008, 227, 517–526. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Seijo, G.; Freitas, F.O.; Valls, J.; Leal-Bertioli, S.; Moretzsohn, M.C. An overview of peanut and its wild relatives. Plant Genet. Resour. 2011, 9, 134–149. [Google Scholar] [CrossRef]
- Lynch, M.; Force, A. The probability of duplicate gene preservation by subfunctionalization. Genetics 2000, 154, 459. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ai, X.; Li, C.; Wang, S.; Zhang, N.; Ren, J.; Wang, J.; Zhong, C.; Zhao, X.; Zhang, H.; et al. A Genome-Wide Analysis of the Jasmonic Acid Biosynthesis Gene Families in Peanut Reveals Their Crucial Roles in Growth and Abiotic Stresses. Int. J. Mol. Sci. 2024, 25, 7054. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, J.; Chen, H.; Luo, H. Genome-wide analysis of ATP-binding cassette transporter provides insight to genes related to bioactive metabolite transportation in Salvia miltiorrhiza. BMC Genom. 2021, 22, 315. [Google Scholar] [CrossRef] [PubMed]
- Nyland, J.; Silbergeld, E. A nanobiological approach to nanotoxicology. Hum. Exp. Toxicol. 2009, 28, 393. [Google Scholar] [CrossRef]
- Carlberg, C.; Molnár, F. Mechanisms of Gene Regulation: How Science Works; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Motallebi, P.; Niknam, V.; Ebrahimzadeh, H.; Enferadi, S.T.; Hashemi, M. The effect of methyl jasmonate on enzyme activities in wheat genotypes infected by the crown and root rot pathogen Fusarium culmorum. Acta Physiol. Plant. 2015, 37, 237. [Google Scholar] [CrossRef]
- Mantri, N.; Patade, V.; Penna, S.; Ford, R.; Pang, E. Abiotic stress responses in plants: Present and future. In Abiotic Stress Responses in Plants; Springer: New York, NY, USA, 2012. [Google Scholar]
- Ahmad, P. Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.). Afr. J. Biotechnol. 2012, 11, 2694–2703. [Google Scholar]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Hossain, A.; Khrupnick, T.; Timsina, J.; Mahboob, M.G.; Hasanuzzaman, M. Agricultural Land Degradation: Processes and Problems Undermining Future Food Security. In Environment, Climate, Plant and Vegetation Growth; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Gu, D.; Liu, X.; Wang, M.; Zheng, J.; Hou, W.; Wang, G.; Wang, J. Overexpression of ZmOPR1 in Arabidopsis enhanced the tolerance to osmotic and salt stress during seed germination. Plant Sci. 2008, 174, 124–130. [Google Scholar] [CrossRef]
- Kim, S.; Kang, J.y.; Cho, D.I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef]
- Yu, D.; Li, X.; Zhao, X.; Du, C.; Chen, J.; Li, C.; Sun, M.; Wang, L.; Lin, J.; Tang, D. RPN1a negatively regulates ABA signaling in Arabidopsis. Plant Physiol. Biochem. PPB 2016, 108, 279–285. [Google Scholar] [CrossRef]
- Gao, F.; Yao, H.; Zhao, H.; Zhou, J.; Luo, X.; Huang, Y.; Li, C.; Chen, H.; Wu, Q. Tartary buckwheat FtMYB10 encodes an R2R3-MYB transcription factor that acts as a novel negative regulator of salt and drought response in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 109, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Abe, H. Role of Arabidopsis MYC and MYB Homologs in Drought and Abscisic Acid-Regulated Gene Expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Zhu, S.; Shi, W.; Jie, Y.; Zhou, Q.; Song, C. A MYB transcription factor, BnMYB2, cloned from ramie (Boehmeria nivea) is involved in cadmium tolerance and accumulation. PLoS ONE 2020, 15, e0233375. [Google Scholar] [CrossRef] [PubMed]
- Urao, T. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar]
- Zhao, P.; Hou, S.; Guo, X.; Jia, J.; Yang, W.; Liu, Z.; Chen, S.; Li, X.; Qi, D.; Liu, G. A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 2019, 19, 564. [Google Scholar] [CrossRef]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2010, 58, 275–286. [Google Scholar] [CrossRef]
- Hiroshi, A.; Takeshi, U.; Takuya, I.; Motoaki, S.; Kazuo, S. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar]
- Liang, X.; Li, Y.; Yao, A.; Liu, W.; Yang, T.; Zhao, M.; Zhang, B.; Han, D. Overexpression of MxbHLH18 Increased Iron and High Salinity Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 8007. [Google Scholar] [CrossRef]
- Flowers, T.J.; Rana, M.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Hasanuzzaman, M.; Li, Y.; Akhtar, K.; Zhang, C.; Zhao, T. Seed Germination Behavior, Growth, Physiology and Antioxidant Metabolism of Four Contrasting Cultivars under Combined Drought and Salinity in Soybean. Antioxidants 2022, 11, 498. [Google Scholar] [CrossRef]
- Liya, M.; Huan, Z.; Lirong, S.; Yiheng, J.; Guozeng, Z.; Chen, M.; Fushun, H. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na/Khomeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012, 63, 305–317. [Google Scholar]
- Xie, Q.; Niu, J.; Xu, X.; Xu, L.; Zhang, Y.; Fan, B.; Liang, X. De novo assembly of the Japanese lawngrass (Zoysia japonica Steud.) root transcriptome and identification of candidate unigenes related to early responses under salt stress. Front. Plant Sci. 2015, 6, 610. [Google Scholar]
- Chang, B.; Ma, K.; Lu, Z.; Lu, J.; Cui, J.; Wang, L.; Jin, B. Physiological, Transcriptomic, and Metabolic Responses of Ginkgo biloba L. to Drought, Salt, and Heat Stresses. Biomolecules 2020, 10, 1635. [Google Scholar] [CrossRef]
- Chen, F.; Fang, P.; Peng, Y.; Zeng, W.; Zhao, X.; Ding, Y.; Zhuang, Z.; Gao, Q.; Ren, B. Comparative Proteomics of Salt-Tolerant and Salt-Sensitive Maize Inbred Lines to Reveal the Molecular Mechanism of Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 4725. [Google Scholar] [CrossRef]
- Xu, L.; Song, J.Q.; Wang, Y.L.; Liu, X.H.; Li, X.L.; Zhang, B.; Li, A.J.; Ye, X.F.; Wang, J.; Wang, P. Thymol improves salinity tolerance of tobacco by increasing the sodium ion efflux and enhancing the content of nitric oxide and glutathione. BMC Plant Biol. 2022, 22, 31. [Google Scholar] [CrossRef]
- Mir, M.A.; John, R.; Alyemeni, M.N.; Alam, P.; Ahmad, P. Jasmonic acid ameliorates alkaline stress by improving growth performance, ascorbate glutathione cycle and glyoxylase system in maize seedlings. Sci. Rep. 2018, 8, 2831. [Google Scholar] [CrossRef]
- El-Beltagi, H.S.; Salama, Z.A.; El Hariri, D.M. Some Biochemical Markers for Evaluation of Flax Cultivars Under Salt Stress Conditions. J. Nat. Fibers 2008, 5, 316–330. [Google Scholar] [CrossRef]
- Koca, H.; Bor, M.; Zdemir, F.; Turkan, I. The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environ. Exp. Bot. 2007, 60, 344–351. [Google Scholar] [CrossRef]
- Claus, W.; Bettina, H. A Bypass in Jasmonate Biosynthesis—The OPR3-independent Formation. Trends Plant Sci. 2018, 23, 276–279. [Google Scholar]
- Takahashi, F.; Yoshida, R.; Ichimura, K.; Mizoguchi, T.; Seo, S.; Yonezawa, M.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 2007, 19, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 Interacts with DELLA Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Zilong, H.; Huangkai, Z.; Shenghan, G.; Lercher, M.J.; Wei-Hua, C.; Songnian, H. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar]
- Timothy, L.B.; Nadya, W.; Chris, M.; Wilfred, W.L. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Oxf. Acad. 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S.; Somers, D.J. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Josh, C.; Ye, C.; Brian, S.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar]
- Zhang, H.; Zhao, X.B.; Sun, Q.X.; Yan, C.X.; Wang, J.; Yuan, C.L.; Li, C.J.; Shan, S.H.; Liu, F.Z. Comparative transcriptome analysis reveals molecular defensive mechanism of Arachis hypogaea in response to salt stress. Int. J. Genom. 2020, 10, 6524093. [Google Scholar] [CrossRef]
- Zhao, X.B.; Li, C.J.; Wan, S.B.; Zhang, T.T.; Yan, C.X.; Shan, S.H. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 2018, 45, 119–131. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D.L. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, Y.; Sun, Q.; Miao, H.; Wang, J.; Wang, Q.; Wang, Q.; Yan, C.; Yuan, C.; Zhao, X.; Li, C.; et al. Genome-Wide Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Peanut and Functional Characterization of AhOPR6 in Salt Stress. Plants 2025, 14, 1408. https://doi.org/10.3390/plants14101408
Mou Y, Sun Q, Miao H, Wang J, Wang Q, Wang Q, Yan C, Yuan C, Zhao X, Li C, et al. Genome-Wide Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Peanut and Functional Characterization of AhOPR6 in Salt Stress. Plants. 2025; 14(10):1408. https://doi.org/10.3390/plants14101408
Chicago/Turabian StyleMou, Yifei, Quanxi Sun, Haocui Miao, Juan Wang, Qi Wang, Qianqian Wang, Caixia Yan, Cuiling Yuan, Xiaobo Zhao, Chunjuan Li, and et al. 2025. "Genome-Wide Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Peanut and Functional Characterization of AhOPR6 in Salt Stress" Plants 14, no. 10: 1408. https://doi.org/10.3390/plants14101408
APA StyleMou, Y., Sun, Q., Miao, H., Wang, J., Wang, Q., Wang, Q., Yan, C., Yuan, C., Zhao, X., Li, C., & Shan, S. (2025). Genome-Wide Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family in Peanut and Functional Characterization of AhOPR6 in Salt Stress. Plants, 14(10), 1408. https://doi.org/10.3390/plants14101408





